Abstract
The NF-κB transcription factor plays essential roles in inflammation and oncogenesis. Its ubiquitous RelA subunit is regulated by several post-translational modifications (PTMs) including phosphorylation, ubiquitination, and acetylation. Ubiquitination promotes the termination of RelA-dependent transcription, but its regulation is incompletely understood. Through mass spectrometry analysis of ubiquitinated RelA, we identified 7 lysines that were attached to degradative and non-degradative forms of polyubiquitin. Interestingly, lysines targeted for acetylation were among the residues identified as ubiquitin acceptor sites. Mutation of these particular sites resulted in decreased polyubiquitination. Acetylation and ubiquitination were found to inhibit each other consistent with their use of overlapping sites. Reconstitution of rela−/− fibroblasts with wild-type and mutant forms of RelA revealed that modifications at these residues can play activating and inhibitory functions depending on the target gene context. Altogether, this study elucidates that ubiquitination and acetylation can modulate each other and regulate nuclear NF-κB function in a gene-specific manner.
Keywords: NF-κB, RelA, p65, Acetylation, Ubiquitination, Transcription
INTRODUCTION
NF-κB is a dimeric transcription factor that plays essential roles in immunity and cell survival (Karin and Greten 2005, Silverman and Maniatis 2001). NF-κB is formed by members of a highly conserved family of proteins that includes 7 different polypeptides in mammals: RelA (also referred to as p65), c-Rel, RelB, p50 and its precursor p105, and p52 and its precursor p100. All NF-κB subunits share a ~ 300 amino acid region termed the Rel Homology Domain (RHD) (Hayden and Ghosh 2008). Canonical NF-κB activity regulates pro-inflammatory and pro-survival gene expression and is primarily mediated by RelA/p50 heterodimers. Non-canonical NF-κB activity, mediated primarily by RelB/p52 heterodimers, regulates the development of various immune organs and differentiation of various lymphoid cell populations.
Given its role in cell survival, abnormal activation of the NF-κB pathway is frequently encountered in cancer (Karin 2006). Mutations in various regulators of NF-κB have been found in several lymphoid malignancies and NF-κB activation is also frequently found in solid tumors. Moreover, the activation of NF-κB in the setting of chronic inflammation is thought to be critically important in the subsequent development of neoplasia (Karin and Greten 2005). Finally, NF-κB activation participates in cancer chemo- and radio-resistance (Basséres and Baldwin 2006). For all these reasons, NF-κB regulation is intensely studied in the context of oncogenesis and cancer therapy.
The regulation of canonical NF-κB activation is mediated by cytosolic and nuclear events. Under basal conditions, NF-κB dimers interact with inhibitory IκB proteins and are localized in the cytosol (Baeuerle and Baltimore 1988). Activation of the IκB kinase (IKK) results in the phosphorylation, ubiquitination and degradation of IκB (Chen et al 1995, Henkel et al 1993), followed by the nuclear translocation of NF-κB dimers, ultimately leading to the induction of target genes (Silverman and Maniatis 2001). While various distinct activators lead to the generation of free DNA-binding subunits, each stimulus results in the induction of a specific subset of genes with peculiar parameters to suit the specific requirements of the activation signal. Therefore, in addition to the control of nuclear translocation, this system requires further mechanisms to regulate NF-κB activity. These include the regulation of RelA activity by PTMs such as phosphorylation (Sakurai et al 1999, Zhong et al 1998), acetylation (Chen et al 2001, Kiernan et al 2003), methylation (Ea and Baltimore 2009, Yang et al 2009, Yang et al 2010), proline isomerization (Ryo et al 2003), and ubiquitination (Maine et al 2007, Saccani et al 2004, Tanaka et al 2007).
Of these various PTMs, ubiquitination has been shown to favor the termination of NF-κB activity by promoting the degradation of a fraction of DNA-bound and active RelA in a gene-specific manner (Natoli and Chiocca 2008, Saccani et al 2004). The ubiquitination and proteasomal degradation of RelA is stimulated by its phosphorylation downstream of the IKK complex (Lawrence et al 2005, Mao et al 2009). Phosphorylation of serine 468 facilitates RelA ubiquitination and degradation (Geng et al 2009) by promoting its binding to GCN5, a factor that in this context serves to tether RelA to a COMMD1-Cul2 containing ubiquitin ligase (Maine et al 2007, Mao et al 2009). Other potential routes for RelA ubiquitination include a SOCS1-Cul2 ligase (Ryo et al 2003), and the nuclear protein PDLIM2 (Tanaka et al 2007), but whether these events are linked to the actions of IKK and COMMD1 has not been fully elucidated. Moreover, whether other PTMs besides phosphorylation can modulate RelA ubiquitination is incompletely understood, but it has been suggested that acetylation and methylation may modulate RelA ubiquitination (Yang et al 2010). Finally, it also unclear whether non-degradative forms of ubiquitination can modulate RelA activity.
In order to better understand the role of RelA ubiquitination in transcriptional control, we mapped the ubiquitin acceptor sites in this protein through mass spectrometry. We found that several of these residues overlap with lysine acetylation sites, and that indeed these two modifications regulate each other. Moreover, non-degradative forms of RelA ubiquitination were also detected, suggesting that this PTM may have broader effects than the release of DNA-bound active RelA. Finally, our studies provide a picture of the complex and gene specific effects of PTMs on these residues, including the repressive effect of RelA acetylation on a number of gene targets, a process that is likely part of the critical chain of events that provide gene specificity to the NF-κB pathway.
RESULTS
RelA ubiquitination is directed to the amino-terminal RHD
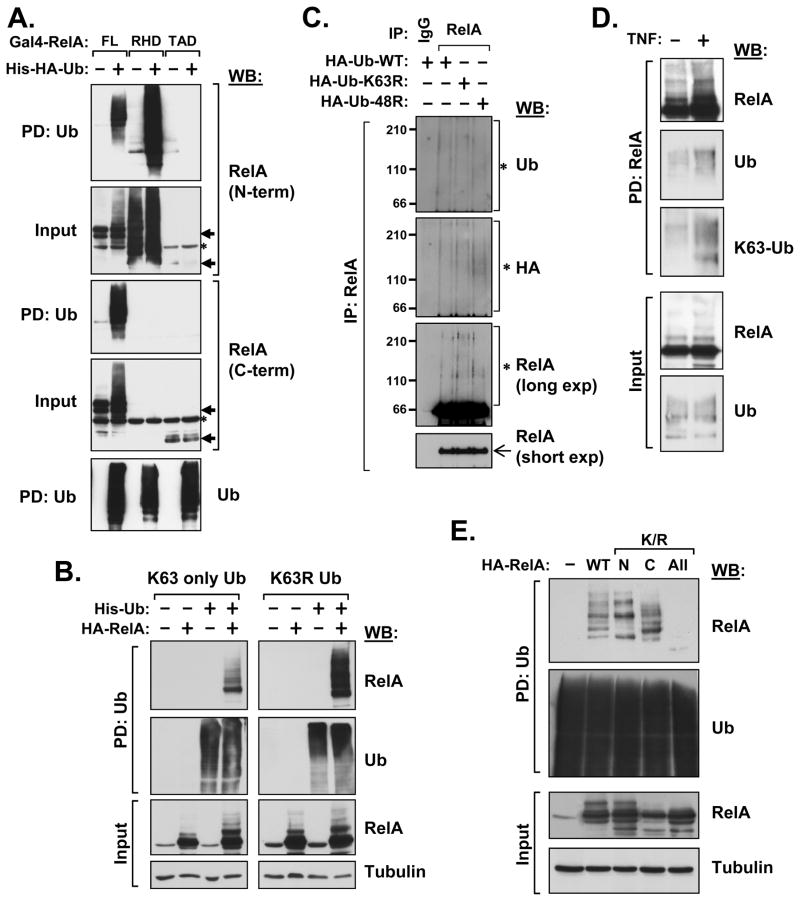
To identify the ubiquitin acceptor residues responsible for RelA ubiquitination, we first investigated the ubiquitination of broad truncation mutants (the amino-terminal RHD or the carboxyl-terminal transactivation domains or TAD). The RHD was strongly ubiquitinated (Fig. 1A, upper panels using an N-terminal specific antibody), while the TAD was not ubiquitinated at all when expressed in isolation of the RHD (Fig. 1A, lower panels). These findings are in keeping with the fact that COMMD1 and SOCS1 both bind to the RHD (Maine et al 2007, Ryo et al 2003) and suggest that the TAD fails to be ubiquitinated either as a result of a lost interaction with the ubiquitin ligase or because it lacks the lysine(s) that serve as acceptor site(s).
Figure 1. RelA ubiquitination is promiscuous and involves non-degradative polyubiquitin chains.
(A) Ubiquitination of truncation mutants of RelA. Gal4-tagged RelA was expressed together with His-tagged ubiquitin in HEK293 cells. After lysis of the cells, ubiquitinated proteins were precipitated with Ni-NTA beads and this material was immunoblotted for RelA using either amino-terminal (F-6) or carboxyl-terminal (C-20) specific antibodies against RelA. Recovery of polyubiquitinated proteins was confirmed by immunoblotting for ubiquitin (P4D1). The non-ubiquitinated RelA proteins are indicated by arrows and non-specific bands are noted by asterisks. PD: pull-down; FL: full-length RelA (amino acids 1–551); RHD: Rel homology domain (amino acids 1–285); TAD: transactivation domains (amino acids 286–551). (B) RelA ubiquitination involves K63 and non-K63 linked polyubiquitin chains. RelA ubiquitination was examined as in (A) after expression of a ubiquitin mutant where all lysines except K63 had been mutated to arginine (K63 only), or after expression of a ubiquitin mutant where only K63 had been mutated to arginine, leaving all other lysines intact (K63R). RelA was detected using an HA antibody. (C) Ubiquitination of RelA by non-K48 and non-K63 linked polyubiquitin chains. U2OS cells were cultured with Doxycycline for 4 days to express Tet-inducible shRNAs against all 4 human ubiquitin genes and Tet-inducible HA-tagged ubiquitin (wild-type, WT, or the lysine mutants K63R or K48R). After 3 h treatment with MG-132 (20μM), denaturing cell lysates were prepared and RelA immunoprecipitation was performed. The presence of ubiquitinated RelA was determined by immunoblotting for ubiquitin or HA. Similarly, ubiquitinated RelA forms were detected after short and long exposures of the RelA immunoblots. (D) RelA is K63-polyubiquitinated after TNF stimulation. HEK293 cells transfected to express His-tagged RelA and HA-tagged ubiquitin were stimulated with TNF (2000 u/mL for 60 min) and then lyzed. RelA was precipitated (Ni-NTA beads) and the material was probed with HA antibody or a K63-specific polyubiquitin antibody. (E) Only complete deficiency of lysines prevents RelA ubiquitination. Lysine residues in RelA were progressively mutated to arginine (K/R) in the amino terminus (N, amino acids 1–218), the carboxyl terminus (C, amino acids 301–551) or the entire RelA polypeptide (All, amino acids 1–551). RelA ubiquitination was assessed as in (B).
RelA can be targeted by non-degradative ubiquitination
While RelA ubiquitination has been shown to be stimulus-dependent and to affect RelA stability, only a small amount of the cellular pool of this protein is degraded in response to pro-inflammatory stimuli. Unlike IκB-α, whose stimulus-induced ubiquitination results in substantial degradation, RelA ubiquitination does not affect overall cellular levels of this protein and is only noticeable when specific cellular compartments are examined or when new protein synthesis is blocked (Geng et al 2009, Maine et al 2007, Yang et al 2009). All this suggested that not all RelA ubiquitination may be for degradative purposes, and thus we set out to examine whether alternative forms of ubiquitination, such as lysine-63 (K63) linked chains, also take place. RelA was expressed together with ubiquitin and the presence of ubiquitinated RelA was detected after enrichment of ubiquitinated proteins on Ni-NTA columns (Fig. 1B). Using a mutant form of ubiquitin that only has the K63 site available for polyubiquitin chain formation, we found evidence that RelA can be ubiquitinated in a K63-dependent manner (Fig. 1B, left panels). Conversely, other lysine linkages were also evident, since a ubiquitin mutant lacking K63 was also able to recover polyubiquitinated RelA (Fig. 1B, right panels).
To confirm these findings and exclude the recovery of mixed polyubiquitin chains containing endogenous and mutant ubiquitin, we utilized a ubiquitin replacement strategy. This was accomplished through recently developed cell lines where endogenous ubiquitin expression is repressed by Tet-inducible shRNA against all 4 human genes encoding ubiquitin, while mutant ubiquitin is concurrently expressed (Xu et al 2009). After Tet-induction, we immunoprecipitated RelA (Fig. 1C) and found that ubiquitinated RelA could be recovered from cells expressing the K63R and K48R ubiquitin mutants, confirming the conjugation of RelA with non-K48 and non-K63 linked chains. Next, we utilized an antibody that is specific for K63-linked polyubiquitin chains and found that cell stimulation with TNF resulted in the accumulation of polyubiquitinated RelA, and this material was immunoreactive with the K63-specific antibody (Fig. 1D).
Redundant lysines can be targeted for ubiquitination
Having identified the RHD as a potential site for ubiquitin conjugation we undertook a mutagenesis approach to try to identify the ubiquitin accepting residue(s). Progressive mutagenesis of all lysines in the RHD (N) failed to abrogate RelA ubiquitination (Fig. 1E). The apparent discrepancy between this result and the findings in Fig. 1A, where the RHD deletion mutant (TAD) was not ubiquitinated, suggested that the prior result was more likely related to the structural alterations inherent to the deletion rather than to the actual loss of absolutely required lysines. Moreover, we found that K/R mutations in the TAD (C) also had persistent ubiquitination. Only a mutant completely devoid of all lysines (All) was ultimately able to abrogate all RelA ubiquitination, indicating the highly redundant nature of lysines as acceptor sites for ubiquitination of this protein.
Mass spectrometry identifies ubiquitin acceptor sites in the amino-terminus of RelA
The ubiquitin acceptor sites proved to be highly redundant precluding the identification of physiological target sites using a mutagenesis approach. Therefore, we decided to utilize mass spectrometry to identify physiologic acceptor sites and polyubiquitin branching in a non-biased manner. To this end, ubiquitinated RelA was purified through a sequential affinity procedure termed bimolecular affinity purification (BAP) and the recovered material was analyzed by mass spectrometry. The procedure was performed 3 times, and in aggregate allowed the identification of 7 ubiquitin conjugated lysines from among the 18 lysines present in RelA (Fig. 2A). Three of the ubiquitin conjugated residues corresponded to lysines previously described as acetylation sites. In addition, these data confirmed the presence of K48-linked polyubiquitin chains, as well as other linkages including K29, K33 and K63 (data not shown).
Figure 2. RelA polyubiquitination occurs at sites of acetylation.
(A) Cartoon depicting the lysine acceptor sites identified by mass spectrometry. Residues indicated by black arrows have not been reported to be acetylated, while red arrows indicate residues previously noted to be acetylated. Two adjacent lysines previously also reported to be acetylated (K122 and K314) are noted by asterisks. (B) Schematic representation of the mutants used in subsequent analysis. K4R-Non Ac: mutation of lysines not known to be acetylation sites (K56,62,79,195R); K4R: mutation of 4 lysines previously noted to be acetylation sites (K122,123,314,315R); K5R: as K4R, but with the additional mutation of K310 (K122,123,310,314,315R); K8R combined mutation of 8 lysine residues (K56,62,79,122, 123,195,314,315R). (C) Acetylation of RelA K4R - NonAc is not significantly affected. HA tagged RelA mild-type (WT) or a mutant targeting sites not previously known as acetylation sites (K4R - NonAc) were co-expressed with p300 in HEK293 cells. RelA was subsequently precipitated from cell lysates (with an HA antibody). This material was immunoblotted with an acetyl-lysine reactive antibody (top panel) or with the HA antibody to detect RelA precipitation (middle panel). The expression of p300 in input lysates was detected by using a FLAG antibody (bottom panel). (D) Acetylation of RelA is directed to the K310 and 4 additional lysines previously reported. RelA mutants fused to HA were expressed in HEK293 cells along with p300 as indicated. After cell lysis, RelA was immunoprecipitated (HA antibody) and the recovered material was immunoblotted with an acetyl-lysine reactive antibody (top panel) or was blotted for RelA (HA antibody, middle panel). Expression of p300 was confirmed in the input (FLAG antibody).
RelA polyubiquitination overlaps with acetylation target lysines
In order to understand the physiologic relevance of RelA ubiquitination at the sites identified, we introduced point mutations in the indicated lysines. We focused further analysis on 4 mutants of RelA which are schematically displayed in Fig. 2B. In the first mutant all 4 lysines not known to be acetylation targets were changed to arginines (K4R-NonAc). The other 3 lysines (K123, K310 and K315) have been reported to be acetylation sites with two of these sites (K123 and K315) being immediately adjacent to a preceding lysine (marked with an asterisk) that can also be acetylated (Buerki et al 2008, Chen et al 2002, Kiernan et al 2003, Rothgiesser et al 2010). Based on this, K122 and K314 were also included in the analysis. Given that K310 has received particular attention as an acetylation site, we generated two mutants affecting the lysines targeted for acetylation excluding or including K310 (K4R and K5R, respectively). Finally, a mutant containing most of the lysines identified was also generated (K8R).
Next, we examined whether these mutations affected RelA acetylation. RelA was co-expressed with p300, the most potent acetyltransferase that targets this protein. After RelA precipitation, the recovered material was immunoblotted with an acetyl-lysine reactive antibody. Wild-type (WT) RelA or the K4R-NonAc mutant targeting sites not known to be acetylation sites were clearly acetylated by p300 (Fig. 2C). On the other hand, the RelA K310R mutant was poorly acetylated, while the K4R mutation also showed strongly impaired RelA acetylation. The combined K5R mutation had the most dramatic reduction with nearly no detectable acetylation (Fig. 2D), indicating that these 5 lysines are indeed the main acetylation sites targeted by p300. Moreover, since the K310R mutation was sufficient to abrogate most acetylation, we concluded that this site is responsible for most of the signal or that the process of acetylation requires cooperativity between the K310 site and the other lysines.
Acetylation acceptor lysines are primarily responsible for RelA polyubiquitination
The ubiquitination status of these RelA mutants was examined next. RelA was co-expressed with His-tagged ubiquitin, followed by enrichment of ubiquitinated proteins on Ni-NTA beads. Western blot analysis revealed intact ubiquitination of the K4R-NonAc mutant, while the K4R mutant was hypo-ubiquitinated compared to the WT protein (Fig. 3A). Additional experiments confirmed that the mutations of K122 and K314 included in the K4R mutant were required for this effect (data not shown), suggesting that they indeed function as redundant sites for both ubiquitination and acetylation.
Figure 3. RelA polyubiquitination is primarily directed to acetylation sites.
(A), (B) and (C) Various RelA mutants were coexpressed with His-tagged ubiquitin and their ubiquitination status was assessed by precipitation and immunoblotting in panels (A) and (C) according to the same approach described for Fig. 1B. In (B), ubiquitinated RelA was detected as a laddered high molecular weight material that has a shifted mobility in His-Ubiquitin transfected cells (noted on direct western blots and marked by an asterisk). (D) Mutation of the acetylation sites impairs TNF-inducible ubiquitination of RelA. Ubiquitination of RelA was detected after PD from cell lysates using the same approach described in Fig. 1D.
Interestingly, the K8R mutant, which includes the acetylation sites and the 4 non-acetylation sites identified by mass spectrometry was ubiquitinated to a similar extent as the WT protein. Moreover, additional lysine mutations (K8R+K310R, and others) often resulted in a paradoxical increase in RelA ubiquitination (data not shown), suggesting that extensive perturbations of RelA after multiple lysine mutations may result in polyubiquitination due to disturbed secondary and tertiary structures.
The K310R and K5R mutants were similarly examined. Expression of RelA along with His-tagged ubiquitin resulted in the upward shift of the high-molecular ladder above RelA, indicating that this laddered material corresponded to ubiquitinated RelA (Fig. 3B, first and second lanes). While K310R showed no differences, the K4R and K5R mutants displayed reduced ubiquitination. Upon precipitation of ubiquitinated proteins, the pulldown was immunoblotted for RelA confirming confirmed that K4R and K5R are hypo-ubiquitinated (Fig. 3C). The K5R mutation not only abrogated the basal ubiquitination of RelA, but also led to dramatic reductions in polyubiquitination after TNF stimulation, which were evident at the level of total RelA ubiquitination and K63-linked polyubiquitination (Fig. 3D).
Polyubiquitinated RelA is not concurrently acetylated
These findings indicated that lysines that are acetylation target sites are also responsible for most polyubiquitin conjugation to RelA, implicating that polyubiquitinated RelA cannot be concurrently acetylated. To test this notion, ubiquitinated RelA was purified using the BAP procedure (Fig. 4A) and the acetylation of ubiquitinated RelA was examined. The acetyl-lysine specific antibody was reactive with the non-ubiquitinated form of RelA, and never produced any signal against ubiquitinated RelA, supporting the hypothesis that acetylated RelA is refractory to ubiquitination.
Figure 4. RelA acetylation and ubiquitination compete with each other.
(A) Ubiquitinated RelA is not concurrently acetylated. RelA fused to a biotinylation tag (TB) was expressed together with His-tagged human ubiquitin. In one case, RelA was purified directly from cell lysates using streptavidin agarose beads (SA). In the other case, ubiquitin-conjugated RelA was purified by first extracting ubiquitinated proteins using nickel-NTA agarose beads and then repurifying RelA from this fraction as before (Ni-SA). The material obtained was immunoblotted for RelA (Streptavidin-HRP) or for acetyl-lysine. Two aliquots from the SA column of increasing quantity were loaded (noted by an asterisk at the bottom). (B) and (C) RelA ubiquitination inhibits its acetylation. GST-tagged RelA was expressed along with FLAG-tagged p300. RelA ubiquitination was stimulated by increasing expression of His-tagged ubiquitin in (B), or by Cul2 and COMMD1 or GCN5 E575Q (acetylation-defective mutant) in (C). Two days later, cells were harvested in PBS and lyzed in a native buffer containing Triton X-100 buffer or in a denaturing buffer containing 8M urea. The native lysate was used for RelA pulldown (with GSH beads) to evaluate RelA acetylation (top panels). The denatured lysate was used for ubiquitin pulldown (with Ni-NTA beads) followed by RelA immunoblotting to assess the ubiquitination status of RelA (bottom panels). (D) RelA acetylation inhibits ubiquitination. His-tagged RelA was expressed together with increasing amounts of CBP as shown. His-tagged RelA was enriched on Ni-NTA beads, followed by immunoblotting with the indicated antibodies (the arrow points to acetylated RelA. The lower part shows the input material.
Ubiquitination inhibits RelA acetylation
Next we extended these findings in vivo, where we set out to promote RelA ubiquitination to examine its effects on the levels of acetylated RelA. p300-dependent acetylation of GST-tagged RelA was monitored by precipitation and immunoblotting with an acetyl-lysine reactive antibody. First, we used increasing expression of His-tagged ubiquitin to drive RelA ubiquitination (Fig. 4B). This led to greater amounts of ubiquitinated RelA (Fig. 4B, bottom panel) and a parallel decrease in the levels of acetylated RelA (Fig 4B, top panel). Interestingly, this effect was observed even at the level of the non-ubiquitinated form of RelA and corresponded to a progressive decrease in RelA-p300 co-precipitation (Fig. 4B, third panel), suggesting that occupation of lysines is not the only mechanism by which these two modifications compete with each other.
To substantiate these results in a complementary setting, RelA ubiquitination was promoted through 2 alternative approaches (Maine et al 2007, Mao et al 2009): the concurrent expression of Cul2 and COMMD1 (components of the ligase complex that targets RelA), and expression of GCN5, a cofactor that binds to this ligase and promotes RelA ubiquitination (to avoid any potential cross-talk with acetylation, the E575Q acetyltransferase deficient mutant of GCN5 was utilized). Both Cul2/COMMD1 and GCN5 promoted strong RelA ubiquitination (Fig. 4C, bottom panel and were associated with a dose-dependent reduction in RelA acetylation (Fig. 4C, top panel), and reduced co-precipitation of p300 (not shown).
Increased acetylation inhibits RelA ubiquitination
These experiments indicated that ubiquitination of RelA inhibits its acetylation, and we set to examine whether the converse could be true (Fig. 4D). RelA acetylation was promoted by expression of CBP, a homolog of p300. Increased RelA acetylation was paralleled by decreased RelA ubiquitination noted by precipitating RelA and immunoblotting for ubiquitin (Fig. 4D, top panel; also evident as high molecular weight material in RelA western blots). Altogether, the data indicated that these two PTMs can target the same sites and antagonize each other in vivo.
Generation of rela−/− reconstituted fibroblasts
In order to examine the functional contribution of RelA modification at these five lysines, mutations of these sites were introduced in the corresponding murine homolog of RelA (designated here as mRelA). It is important to note that all the lysines identified by mass spectrometry are tightly conserved among mammals and for simplicity we will continue to refer to these sites according to their position in human RelA.
We generated K to R mutations, which abrogate all modifications of these residues while preserving the overall charge at these positions, as well as K to Q mutations, which mimic lysine acetylation. These mRelA mutants were stably expressed in rela−/− mouse embryo fibroblasts utilizing lentiviral infection. Comparable expression of the desired mutants as well as relevant NF-κB regulatory proteins was confirmed by immunoblotting (Fig. 5A). Expression of classical IκB proteins and other NF-κB subunits became greater upon reintroduction of mRelA, and this effect was similar across all cell lines developed. Since these proteins are themselves expressed from NF-κB responsive genes (Hayden and Ghosh 2008), these data suggested that the transcriptional activity of the mutants was preserved. Indeed, mRNA expression of Nfkbia, the gene encoding IκB-α, was largely similar across all cell lines (Fig. 5B)
Figure 5. Modifications of RelA acetylation sites have diverse function in endogenous gene expression.
(A) Generation of stably reconstituted rela−/− mouse embryo fibroblasts. The indicated murine RelA mutants were stably expressed after lentiviral infection. Expression of RelA and the other NF-κB regulatory proteins was assessed by immunoblotting as indicated. (B) mRNA expression of the Nfkbia gene encoding IκB-α, was determined by qRT-PCR. (C) Dimerization and IκB binding is not affected in mRelA mutants. Lysates from the stable cell lines described in (A) were used for RelA immunoprecipitation (HA antibody), followed by immunoblotting for p105/p50 as well as IκB-α. (D) DNA binding is not affected in mRelA mutants. Nuclear extracts from unstimulated cells were mixed with a biotinylated double stranded short oligonucleotide probe that was subsequently precipitated using streptavidin beads. Following elution, mRelA binding was determined by immunoblotting. (E) Identification of RelA- and K5-regulated TNF-response genes. Cells stably reconstituted with wild type mRelA (WT) or K5 mutants were stimulated for 1h or 8h with TNF (20ng/ml) or were left untreated in two independent experiments as indicated. Total RNA was extracted and transcriptome-wide microarray experiments were performed. The whole data set was filtered for transcripts that were regulated by TNF in WT-reconstituted cells by more than 2-fold. This data set was further filtered for genes that were differentially regulated by either K5 mutation at least 2-fold on average in both experiments, resulting in 185 genes of interest. Depicted is a hierarchical cluster analysis of this group of genes in which relative changes of expression levels are displayed as color-coded log2-transformed ratio values (see methods section for details). Ratio values from mRelA-deficient cells (Vector, lanes 1–3) were not clustered but are aligned along the gene list for comparison. Gray colors indicate no measurable expression. The average of both experiments are displayed for the vector and WT control lines, while each replicate is shown for the K5R and K5Q cell lines. Red arrow heads point to genes evaluated further by qRT-PCR.
RelA K/R and K/Q mutants are able to dimerize and bind DNA
The RHD is required for dimerization and DNA binding, and this latter function involves indirectly K122 and K123 (Chen et al 1998). Thus, we examined first whether the RelA mutants could dimerize, bind to IκB-α and to their cognate DNA. Immunoprecipitated mRelA from the stable cell lines demonstrated co-precipitation of endogenous p105, p50 and IκB-α (Fig. 5C) indicating that they can form heterodimers and bind to IκB-α. The K4Q and K5Q mutants displayed slight predilection for p50 compared to IκB-α binding, consistent with prior reports that acetylation has an inhibitory effect on RelA-IκB-α interactions (Chen et al 2001). Similarly, nuclear levels of RelA K5Q were slightly higher in unstimulated cells, while post-stimulation levels were comparable to WT and K5R stable cells (Fig. S1). Next, we assessed the ability of these mutants to bind to DNA using a DNA/protein co-precipitation assay using nuclear extract and biotinylated κB-containing oligonucleotides (Fig. 5D). All mutants were coprecipitated to a similar extent by DNA in unstimulated cells (Fig. 5D) and in extracts from TNF stimulated cells (data not shown). Altogether, the K/R or K/Q mutations did not impair dimerization or DNA binding, a pre-requisite to examine the contribution of these residues to gene expression.
Lysine modifications play gene-specific roles in transcription
Next, we examined the effects of modifications at these residues on global TNF-inducible gene expression in K5R and K5Q reconstituted fibroblasts. After filtering the data for genes regulated by at least 2-fold in response to TNF in WT cells, we identified 185 genes that were differentially regulated by a factor of 2-fold in either the K5R or K5Q cells compared to the WT cells (Fig. 5E). Contrary to our initial expectation, the acetyl-mimicking K5Q mutation did not lead to a global activation of gene expression. Rather, many TNF-inducible and RelA-dependent genes in these cells were specifically underexpressed in K5Q cells. Nevertheless, an activating effect of the K5Q mutation was also seen (lower part of the gene cluster in Fig. 5E).
Key findings from this analysis were confirmed by qRT-PCR, such as the dramatic repression of Vcam1, Il6 and Lamb3 expression in K5Q reconstituted fibroblasts (Fig. 6A and S2); the converse increase of Mmp13, Mmp9, Expi and Ccl9 in K5Q expressing cells was also observed (Fig. 6F and S2). Moreover, similar alterations were noted for genes whose expression levels were too low to pass filter criteria of the microarray analysis such as Icam1, Saa3, and Cxcl10, which demonstrated repressed expression in K5Q cells (Fig. 6B and S2) and Mmp3, which demonstrated exaggerated late induction in K5Q cells (Fig. S2).
Figure 6. The function of RelA modifications on transcription and chromatin association is gene-specific.
(A), (B) and (F) Expression of the Vcam1 (A), Icam1 (B) and Mmp13 genes was examined further in the indicated by qRT-PCR. TNF stimulation for 1 or 8 h was used; error bars show standard deviations from three independent experiments. (C) Promoter-associated RelA ubiquitination is stimulated by TNF. Using U2OS cells with inducible replacement of endogenous ubiquitin by HA-tagged ubiquitin we performed a ChIP/re-ChIP experiment. Ubiquitin was immunoprecipitated next (using an HA antibody) and RelA was re-precipitated after elution. The human ICAM1 promoter region surrounding the κB site was detected by qPCR. The signal was normalized by the background precipitation (IgG control IP) and compared to the unstimulated sample (set as 1). (D), (E) and (G). RelA recruitment and histone acetylation at the Vcam1, Icam1 and Mmp13 promoters were determined by ChIP. As before, the respective DNA regions were detected by qPCR; the signal was normalized by the background precipitation (IgG control IP) and compared to the unstimulated sample (set as 1).
Inducible RelA ubiquitination can be detected on chromatin
Vcam1 and Icam1 expression was evaluated in further detail by qRT-PCR, which confirmed the repressive effect of RelA K5Q observed in the microarray analysis (Fig. 6A and 6B). In addition, the K4Q mutation was similarly repressive, while isolated K310 mutations had minimal effects on these and other genes examined (Vcam1, Icam1, Mmp13, Mmp3, Csf2, Saa3, Il6, Ptgs2 and Cxcl10 - Fig. 6 and data not shown).
Cells reconstituted with K5R, a mutant unable to be modified on any of these residues, displayed increased expression of Vcam1 and Icam1. We initially speculated that this was due to loss of degradative ubiquitination, given the inhibitory effects of ubiquitination on Icam1 expression (Burstein et al 2005, Maine et al 2007, Mao et al 2009). However, the unexpected inhibitory effects of the K5Q mutant, suggested that loss of acetylation could also account for the increased expression in the K5R expressing fibroblasts. To clarify this question, the presence of ubiquitinated RelA at this promoter was examined by chromatin IP (ChIP) and re-ChIP using ubiquitin and RelA sequential immunoprecipitations. This confirmed the physiologic induction of RelA ubiquitination on the human ICAM1 promoter (Fig. 6C).
Histone acetylation correlates with the transcriptional effects of K5 mutations
We next evaluated the potential mechanism(s) for the transcriptional effects of K5 mutations. ChIP analysis of the Vcam1 or the Icam1 promoters indicated that RelA recruitment was not impaired even for the repressive RelA K5Q mutant (Fig. 6D and 6E). In fact, the recruitment of RelA K5R and K5Q to the Icam1 promoter persisted at 5 h compared to RelA WT, consistent with their decreased ubiquitination. Similarly, recruitment of CBP to this region could not account for differences in expression (Fig. S3). However, histone acetylation of the Vcam1 and Icam1 promoters had a general correlation with gene expression, being decreased in RelA K5Q reconstituted fibroblasts and somewhat elevated -particularly after TNF- in RelA K5R cells (Fig. 6D and 6E). The converse findings were made for Mmp13, a gene that was selectively induced in RelA K5Q cells (Fig. 6F). Again, RelA or CBP recruitment did not correlate with differences in Mmp13 expression (Fig. 6G and S3). However, histone acetylation in this promoter region was dramatically increased in RelA K5Q expressing fibroblasts, consistent with their increased expression of Mmp13 (Fig. 6G).
DISCUSSION
The present study identifies the ubiquitination sites in the NF-κB subunit RelA. While our mapping studies utilizing a mass spectrometry approach identified 7 acceptor sites, we find that ubiquitination of this protein is promiscuous and only the elimination of all lysines abrogates ubiquitination completely. However, such extensive mutagenesis produces a non-functional protein, unable to bind DNA (data not shown). Consistent with another report (Fan et al 2009), the residue K195 was identified in this study as a ubiquitin acceptor site. However, we found no evidence that this site serves as the dominant or preferential site of ubiquitination. Rather, we found that 4 lysines that are also acetylation target sites (K122, K123, K314 and K315) are primarily responsible for RelA polyubiquitination. Indeed, other recent studies have highlighted the importance of K314 and K315 in RelA ubiquitination, as mutation of these two residues results in increased protein stability (Yang et al 2009). Our studies would suggest that this result may be explained in part by the fact that these residues are ubiquitin acceptor sites.
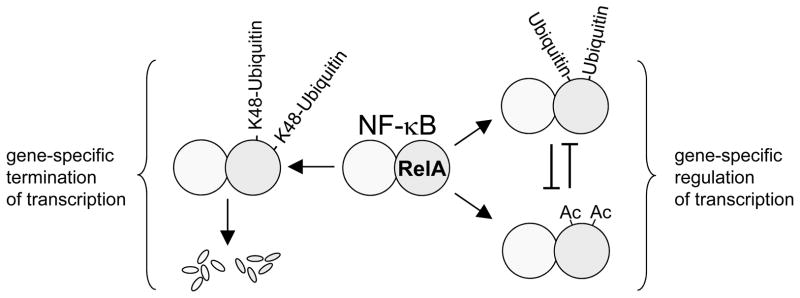
Using mass spectrometry and other approaches, we demonstrate that RelA ubiquitination can involve degradative and non-degradative polyubiquitination. RelA ubiquitination has been shown to promote its release from chromatin through proteasomal degradation (Burstein et al 2005, Geng et al 2009, Maine et al 2007, Mao et al 2009, Saccani et al 2004), and indeed using ChIP/re-ChIP we show that TNF induces ubiquitinated RelA on the human ICAM1 promoter. The specific nature of chromatin-associated RelA ubiquitination could not be directly ascertained by ChIP/re-ChIP due to limitations imposed by currently available reagents. Nevertheless, based on the recruitment of COMMD1 and GCN5 to specific gene promoters, and the effects of these factors on RelA-chromatin interactions, we believe that degradative forms of RelA ubiquitination occur at least in part on chromatin. The role of non-degradative RelA ubiquitination remains to be fully elucidated, but it is possible that this PTM may modulate modifications of the targeted lysines, including acetylation, without necessarily triggering protein degradation (Fig. 7).
Figure 7. Model depicting the relationship between acetylation and ubiquitination of RelA in transcriptional control.
Several complementary experimental approaches provide evidence for an active interplay between ubiquitination and acetylation of RelA (Fig. 7), a phenomenon that has been previously described for other transcription factors such as p53 (Li et al 2002). Ubiquitination can inhibit RelA acetylation as least by two mechanisms: direct competition for lysine acceptor sites, and indirectly through reduced interactions between p300 and RelA. The latter phenomenon remains unexplained at this point but suggests that ubiquitinated RelA can act in trans on non-ubiquitinated RelA. In this regard, it has been previously suggested that p300 interacts simultaneously with two molecules of RelA through dual domains (Gerritsen et al 1997). In that model it is conceivable that ubiquitinated RelA may prevent binding to non-modified RelA. Alternatively, ubiquitination may affect intramolecular interactions between the amino- and carboxyl-termini of RelA, akin to the effect of serine 276 phosphorylation on p300/RelA binding (Zhong et al 2002).
In addition, we find that RelA acetylation can conversely decrease RelA ubiquitination. This is in agreement with the finding that Sirt1 deficiency, encoding a deacetylase that targets RelA, leads to increased RelA acetylation and concurrent decrease in RelA ubiquitination and degradation (Yang et al 2010). Moreover, the ability of acetylation to inhibit RelA ubiquitination may explain in part the original observation that RelA acetylation leads to prolonged nuclear RelA accumulation (Chen et al 2001), a phenomenon seen when RelA ubiquitination and degradation are impaired (Maine et al 2007, Mao et al 2009). Additionally, it is conceivable that other PTMs on these lysines, such as methylation (Yang et al 2009, Yang et al 2010), could affect RelA ubiquitination through similar mechanisms.
Finally, our studies highlight the complex interplay of modifications of these lysine residues and their effects on endogenous gene expression. While the K310 residue clearly plays an important role in RelA acetylation and accounts for most of the p300-induced signal, the effects observed on the expression of a panel of endogenous genes were less pronounced than previous observations made using luciferase reporter constructs (Chen et al 2001, Chen et al 2002). However, modifications of the K4 lysine group appear to have more dramatic effects on gene expression. The effects of K/Q substitutions indicate that acetylation is not only an activating modification, but can inhibit the expression of many target genes, consistent with other reports (Kiernan et al 2003). The ultimate mechanism that links RelA acetylation to transcriptional repression in a gene-specific manner remains to be elucidated, but our data suggest that recruitment of acetylated RelA to these gene promoters can initiate a set of repressive events on the chromatin environment. For other genes that are stimulated by RelA acetylation, such as Mmp13, the converse set of events can be demonstrated. Thus, acetylation acts more as a modification that provides gene specificity to the NF-κB response, rather than simply a PTM that amplifies the activation signal as has been more commonly regarded. Other modifications of these lysine residues, including ubiquitination and methylation, could potentially play similar roles in the specification of the NF-κB transcriptional response. This hypothesis is in line with recent studies documenting that RelA modifications, alone or in combination, function to direct transcription in a highly target gene-specific fashion (Hayden and Ghosh 2008, Hoffmann et al 2006, Moreno et al 2010).
MATERIALS AND METHODS
Cell culture, transfection and generation of stable cell lines
Human embryonic kidney (HEK) 293 cells were obtained from ATCC (Mannassas, Virginia, USA). HEK 293T cells and rela−/− mouse embryo fibroblasts were kindly provided by Maria S. Soengas and by Marty W. Mayo, respectively. U2OS Ubiquitin replacement cells were kindly provided by Zhijian Chen (Xu et al 2009). All cell lines were cultured in DMEM supplemented with 10% FBS and L-glutamine (2mM). A standard calcium phosphate transfection protocol was used to transfect HEK 293 cells (Maine et al 2009). The generation of stable cell lines expressing various mutant forms of RelA was achieved through lentiviral infection and antibiotic selection utilizing an FG9-based vector system as previously described (Lois et al 2002, Mao et al 2009, Wright and Duckett 2009).
Immunoblotting and protein precipitation
Cell lysate preparation, immunoprecipitations, GSH precipitations and immunoblotting were performed as previously described (Burstein et al 2004, Burstein et al 2005).
BAP (bimolecular affinity purification) and mass spectrometry-based mapping of ubiquitin acceptor sites
This procedure has been described in detail elsewhere (Maine et al 2010). Briefly, HEK 293 cells were transfected with His6-tagged human ubiquitin and RelA fused to TB (a biotinylation target peptide). The cells were grown in biotin-supplemented media and after transient proteasomal blockade with MG-132 at 40μM for 5 hours (Boston Biochem; Cambridge, Massachusetts, USA), the culture was lysed in a protein denaturing buffer containing 8M urea. Ubiquitinated RelA was isolated through sequential affinity purification using nickel-NTA and streptavidin columns (Invitrogen; Carlsbad, California, USA). The obtained material was separated by SDS-PAGE, and bands corresponding to ubiquitinated RelA were excised for tryptic digestion, liquid chromatography separation and tandem mass spectrometry. The presence of a mass shift of 114 Da (corresponding to a diglycine signature) was considered evidence of ubiquitin conjugation to a lysine residue.
Quantitative RT-PCR (qRT-PCR)
Cells were stimulated with 1000u/mL of TNF (Roche; Indianapolis, Indiana, USA) and total RNA was extracted using the RNAeasy kit (Qiagen; Valencia, California, USA) at the indicated time points. One μg of RNA was used for cDNA synthesis from Oligo (dT)20 primers using the Superscript first strand synthesis system (Invitrogen). Real-time PCR was performed using specific primers and SYBR Green ROX Mix (ABgene; Schwerte, Germany), utilizing an Applied Biosystems 7300 real time PCR system (Carlsbad, California, USA). Experiments were performed in triplicate, data were normalized to housekeeping genes and the relative abundance of transcripts was calculated by the comparative ΔΔCt method. All primer sequences are available upon request.
κB DNA co-precipitation assay
Sense and antisense oligonucleotides encoding tandem κB sites (sense strand, AGCTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGG) were annealed after heating and slow cooling. The sense strand was biotinylated in the 5′ position, while a non-biotinylated oligonucleotide was used as control. Nuclear extracts were prepared as previously described (Burstein et al 2005) and incubated with annealed oligonucleotides for 60 minutes at 4°C. The oligonucleotides were subsequently precipitated by streptavidin agarose beads and thoroughly rinsed 4 times before proceeding to Western blot analysis.
Supplementary Material
Acknowledgments
We are grateful to Zhijian ‘James’ Chen, Maria S. Soengas and Marty W. Mayo for providing cell lines that were utilized in these studies. We also want to thank Doris Newel and Oliver Dittrich for their technical assistance with the microarray experiments. This work was supported by an NIH R01 DK073639, a CCFA Senior Research Award and a UTSW DOCS Award to E.B. The work of M.L.S. and M.K. is supported by grants from the DFG.
Footnotes
CONFLICT OF INTEREST
The authors do not have any conflict of interest in connection to this work.
Supplementary materials and methods. Detailed information about the plasmids, antibodies and primers utilized in this study is provided in table form as supplementary information. Similarly, detailed description of microarray analysis and chromatin immunoprecipitation is provided there.
References
- Baeuerle PA, Baltimore D. IκB: a specific inhibitor of the NF-κB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Basséres DS, Baldwin AS. Nuclear factor-κB and inhibitor of κB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, et al. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E, Ganesh L, Dick RD, van De Sluis B, Wilkinson JC, Klomp LW, et al. A novel role for XIAP in copper homeostasis through regulation of MURR1. EMBO Journal. 2004;23:244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, et al. COMMD proteins: A novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280:22222–22232. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, et al. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes & Development. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Ea CK, Baltimore D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Mao R, Zhao Y, Yu Y, Sun W, Song P, et al. Tumor necrosis factor-α induces RelA degradation via ubiquitination at lysine 195 to prevent excessive nuclear factor-κB activation. J Biol Chem. 2009;284:29290–29297. doi: 10.1074/jbc.M109.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Wittwer T, Dittrich-Breiholz O, Kracht M, Schmitz ML. Phosphorylation of NF-κB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10:381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci U S A. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Henkel T, Machleidt T, Alkalay I, Krönke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of IκB-α is necessary for activation of transcription factor NF-κB. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-κB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, et al. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-κB subunits through a Cullin-containing ubiquitin ligase. EMBO Journal. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Gluck N, Zaidi IW, Burstein E. Bimolecular Affinity Purification (BAP): Tandem affinity purification using two protein baits. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Li H, Zaidi IW, Basrur V, Elenitoba-Johnson KS, Burstein E. A bimolecular affinity purification method under denaturing conditions for rapid isolation of a ubiquitinated protein for mass spectrometry analysis. Nat Protoc. 2010;5:1447–1459. doi: 10.1038/nprot.2010.109. [DOI] [PubMed] [Google Scholar]
- Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, et al. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev. 2009;23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Sobotzik JM, Schultz C, Schmitz ML. Specification of the NF-κB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKKε. Nucleic Acids Res. 2010;38:6029–6044. doi: 10.1093/nar/gkq439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Chiocca S. Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci Signal. 2008;1:pe1. doi: 10.1126/stke.11pe1. [DOI] [PubMed] [Google Scholar]
- Rothgiesser KM, Fey M, Hottiger MO. Acetylation of p65 at lysine 314 is important for late NF-κB-dependent gene expression. BMC Genomics. 2010;11:22. doi: 10.1186/1471-2164-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- Silverman N, Maniatis T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes & Development. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- Wright CW, Duckett CS. The aryl hydrocarbon nuclear translocator alters CD30-mediated NF-κB-dependent transcription. Science. 2009;323:251–255. doi: 10.1126/science.1162818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XD, Tajkhorshid E, Chen LF. Functional Interplay between Acetylation and Methylation of the RelA Subunit of NF-κB. Mol Cell Biol. 2010 doi: 10.1128/MCB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.