Abstract
The evolutionarily conserved Highwire (Hiw)/Drosophila Fsn E3 ubiquitin ligase complex is required for normal synaptic morphology during development and axonal regeneration after injury. However, little is known about the molecular mechanisms that regulate the Hiw E3 ligase complex. Using tandem affinity purification techniques, we identified Drosophila Rae1 as a previously unknown component of the Hiw/Fsn complex. Loss of Rae1 function in neurons results in morphological defects at the neuromuscular junction that are similar to those seen in hiw mutants. We found that Rae1 physically and genetically interacts with Hiw and restrains synaptic terminal growth by regulating the MAP kinase kinase kinase Wallenda. Moreover, we found that the Rae1 is both necessary and sufficient to promote Hiw protein abundance, and it does so by binding to Hiw and protecting Hiw from autophagy-mediated degradation. These results describe a previously unknown mechanism that selectively controls Hiw protein abundance during synaptic development.
A series of in vivo studies in a variety of organisms have found that the evolutionarily conserved PHR proteins, including Hiw (fly), Phr1 (mouse), RPM-1 (worm) and Esrom (zebrafish), regulate the development and repair of the nervous system1. In Drosophila, hiw mutants exhibit a marked synaptic terminal overgrowth at the larval neuromuscular junctions (NMJs) with an increased number of boutons and a decreased bouton size2,3, and a similar anatomical overgrowth of the giant fiber–tergotrochanteral motoneuron synapse in the CNS4. Aberrant synaptic morphology has also been identified in rpm-1 mutant worms5,6 and Phr1 mutant mice7,8. In addition, disrupting the function of hiw orthologs in worm, zebrafish and mouse results in prominent axon guidance defects5,7,9–11. More recently, hiw, rpm-1 and their downstream target wallenda (fly) or dlk-1 (worm) have been shown to regulate axonal regeneration after nerve injury12–14. Thus, PHR proteins are important in a broad range of neuronal processes, including axon guidance, synaptic development and axonal regeneration. However, it is unclear how these diverse functions of PHR proteins are achieved. An important step toward answering this question is to understand the molecular mechanisms that regulate PHR proteins.
Hiw and its orthologs are enormous proteins that share a number of highly conserved functional domains, including an RCC1 domain15–17, two PHR repeats18, a Myc-binding domain16 and a C-terminal RING-H2 finger E3 ubiquitin ligase domain9,19. Studies in worms, flies and mice have found that the PHR E3 ubiquitin ligases associate with a highly conserved F-box protein—FSN-1 in worm, Fsn in fly or Fbox45 in mouse—and together they function as a SCF-like E3 ubiquitin ligase complex to regulate neural development20–22. An important downstream target of the ubiquitin ligase complex is the MAP kinase kinase kinase (MAPKKK) Wallenda (Wnd), which activates a MAP kinase cascade to control synaptogenesis23,24. Although considerable progress has been made in understanding the PHR-associated ligase complex and its downstream signaling cascade, very little is known about how PHR proteins are regulated. Autophagy can negatively regulate the abundance of Hiw protein25 in fly, but it is unclear how autophagy, a general protein-degradation pathway, can be controlled to modulate Hiw protein levels during synaptic development. Identifying Hiw cofactors that regulate Hiw activity and abundance is necessary to determine the mechanisms by which hiw functions are controlled.
We identified one such Hiw cofactor, Drosophila melanogaster Rae1. Rae1 encodes a 346 amino-acid protein that belongs to an evolutionary conserved WD40 repeat protein family. Three major functions of Rae1 have been reported in different organisms. First, Rae1 associates with microtubules in the cytoplasm and regulates the organization of the cytoskeletal network during mitosis26. Second, Rae1 binds to Nup98 to facilitate the transportation of poly-A RNA from the nucleus to the cytosol27. Third, Rae1 is an anaphase-promoting complex (APC)-associated protein that inhibits the targeting of Securin by the APC ubiquitin ligase complex, thereby regulating entry into anaphase during mitosis28. In cultured Drosophila SL2 cells, knockdown of Rae1 by RNA interference induces arrest in G1 phase, but results in no mRNA transport defects, suggesting that Rae1 is involved in cell cycle regulation, but not in mRNA export, in these cells29. Although Rae1 binds microtubules in cultured mammalian neurons30, the function of Rae1 in the nervous system has not yet been studied. We found that Rae1 is a binding partner and positive regulator of Hiw. Rae1 and Hiw physically and genetically interacted and worked together to restrain synaptic terminal growth. These results indicate that the function of Rae1 in the Hiw E3 ubiquitin ligase complex is to prevent the autophagy-mediated degradation of Hiw protein in post-mitotic neurons. Our findings, and those of others (B. Grill, L. Chen, E.D. Tulgren, S.T. Baker, W. Bienvenut et al., personal communication), suggest an evolutionarily conserved role for Rae1 in the regulation of neural development.
RESULTS
Drosophila Rae1 associates with Hiw
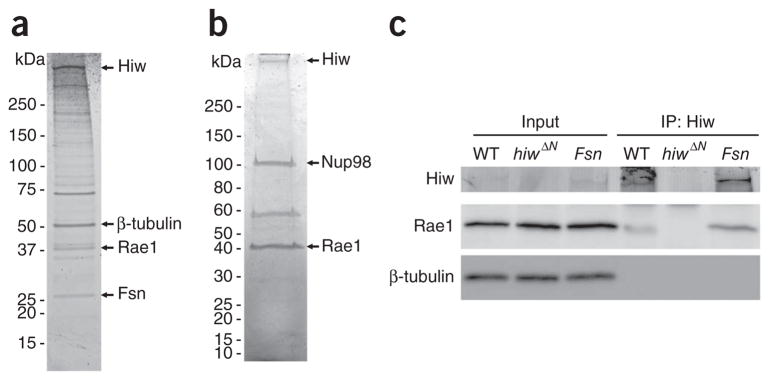
To identify Hiw cofactors, we used a two-step purification method called tandem affinity purification (TAP) to isolate Hiw-interacting proteins from the Drosophila adult head31, and then used liquid chromatography–tandem mass spectrometry (LC-MS/MS) to identify the proteins contained in the Hiw-associated complex (see Online Methods). We previously identified Drosophila Fsn as a Hiw-interacting protein and found that Hiw participates in a SCF-like E3 ubiquitin ligase complex20,21. Here we identified another Hiw-binding partner, Drosophila Rae1. Rae1 was first identified from the ~40-kDa protein band migrating on the SDS-PAGE gel (Fig. 1a) on the basis of the recovery of four unique peptides (MFGATQSTNR, EPNPMMTINLPER, GLIIYSLQNSPTEYK and VAIQYVNPGNPK). In addition, using a shotgun proteomic approach that is capable of identifying all of the trypsin-digested peptides from the Hiw-associated protein complex (see Online Methods), we found numerous peptides from Rae1 (Supplementary Table 1), which further validated the presence of Rae1 in the protein complexes that were pulled down by N-terminal TAP-tagged Hiw. As an independent test of the association between Rae1 and Hiw, we performed a reverse TAP using neuronally expressed transgenic TAP-tagged Rae1 as the bait to isolate Rae1-interacting proteins in the brain. We then resolved Rae1 and its associating proteins by electrophoresis on a SDS-PAGE gel (Fig. 1b) and used shotgun proteomics analysis to identify the associated proteins. As expected, Hiw was identified with very high confidence (Supplementary Table 2), along with Fsn, which further confirmed the association of Rae1 with the Hiw/Fsn E3 ubiquitin ligase complex.
Figure 1.
Rae1 and Hiw interact with each other in neurons. (a) The Hiw-associated complex was purified by TAP (see Online Methods) and analyzed by one-dimensional SDS-PAGE gel followed by Sypro Ruby staining. Mass spectrometry identified the full-length Hiw, β-tubulin, Rae1 and Fsn, indicated by arrows. (b) The Rae1-associated complex purified from fly brains by TAP was analyzed by one-dimensional SDS-PAGE gel followed by Coomassie blue G-250 staining. Mass spectrometry identified Hiw, Nup98 and Rae1, indicated by arrows. (c) Larval brain lysate of wild-type (WT), hiw null mutant (hiwΔN) and Fsn mutant (Fsnf06595/Df(2R)7872) flies was subject to co-immunoprecipitation with antibody to Hiw (Hiw2b). Both the input and the immunoprecipitated complexes were analyzed by western blots with antibodies to Hiw or Rae1. The membrane was also blotted with antibody to β-tubulin to ensure equal amount of total proteins in the input samples. Full-length blots are presented in Supplementary Figure 8.
We then tested the interaction between endogenous Hiw and Rae1 proteins in 3rd instar larval brains. We immunoprecipitated endogenous Hiw using an antibody to Hiw (Hiw2b)19 and found that Rae1 co-immunoprecipitated with Hiw, but was absent in control immuno-precipitation from hiw-null larval brains (Fig. 1c). Furthermore, the Hiw-Rae1 interaction remained unaffected in Fsn mutant brains, indicating that Fsn is not required for the binding of Rae1 to Hiw. Although Fsn was not required for the Hiw-Rae1 interaction, Rae1 was detected in Fsn-immunoprecipitated complexes (Supplementary Fig. 1), further supporting the existence of a Rae1/Hiw/Fsn protein complex in vivo. Taken together, our data suggest that Rae1 associates with the Hiw complex in post-mitotic neurons.
Rae1 is required to restrain synaptic terminal growth
The strong association of Rae1 and Hiw in neurons led us to postulate that Rae1 may work with Hiw during synaptic development as either a positive or negative regulator of the Hiw/Fsn ligase complex. To assess the function of Rae1 in synaptic development, we carried out a Rae1 loss-of-function study. We first generated Rae1 imprecise excision alleles by mobilizing a transposable P-element (P{GawB}NP3499) located ~400 bp upstream of the first exon of the Rae1 locus. After screening over 800 excision events, we recovered two independent Rae1 imprecise excision mutants that survived to the second instar larval stage, Rae1EX28 and Rae1EXB12. The mutant larvae started wandering out of food with a body size close to second instar. They then continued to wander for several days without a further increase in their body size and eventually died without pupating. PCR of the genomic DNA extracted from Rae1EX28 and Rae1EXB12 mutants revealed that deletions of the Rae1 locus in both mutants encompassed almost the entire Rae1 promoter region and a small region of the 3′ untranslated region (UTR) of the neighboring gene pirk (Fig. 2a). Western blot analysis showed that Rae1 protein levels were reduced to less than 10% of the wild-type level in Rae1EX28, Rae1EX28/Df(2R)5764 and Rae1EXB12/Df(2R)5764 mutants (Fig. 2b,c). Given that Rae1EX28 mutants had a very similar level of residual Rae1 protein to Rae1EX28/Df(2R)5764 mutants and carried a smaller lesion on the neighboring gene pirk, we focused our analysis on the Rae1EX28 mutants.
Figure 2.
Generation of Rae1 mutant alleles. (a) The gene structure of Rae1. The black-arrowed boxes indicate exons. A transposable element, P{GawB}NP3499, located ~400 bp upstream of the first exon of Rae1 was used to generate imprecise excision mutants of Rae1. PCR analysis indicates that ~900 bp and a ~1.5-k bp DNA fragment in the Rae1 promoter region are missing in Rae1EX28 and Rae1EXB12, respectively. (b) Western blot analysis on Rae1 protein in the brain lysate of wild-type, Df(2R)5764/+, Rae1EX28/+, or late second instar Rae1EX28, Rae1EX28/Df(2R)5764(/Df) and Rae1EXB12 /Df(2R)5764 larvae. β-tubulin was used as the loading control. Full-length blots are presented in Supplementary Figure 9. (c) Quantification of Rae1 protein levels in the Rae1 heterozygous and homozygous mutant brains. Rae1 protein levels were first normalized to β-tubulin protein levels and are presented as percentage of wild-type level. *P < 0.05, **P < 0.001. Error bars denote s.e.m.
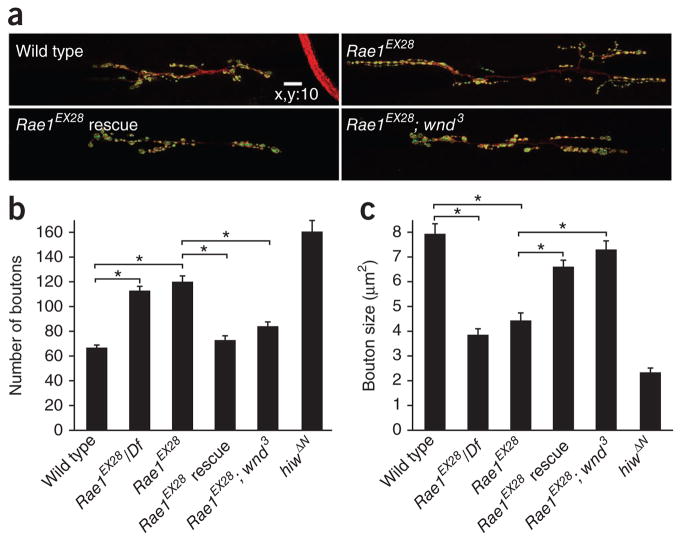
Morphological analysis of the NMJs from Rae1EX28 and Rae1EX28/Df(2R)5764 larvae revealed a morphological defect that was similar to that seen in the hiw mutant NMJ: synaptic terminal overgrowth and decreased bouton size (Fig. 3a). We quantified this phenotype at the NMJ of muscle 6/7 on segment A3, although all phenotypes were qualitatively similar at all of the other type I NMJs. The average muscle sizes of all of the quantified NMJs were similar (muscle 6 on segment A3 (×1,000 μm2): wild type, 27.1 ± 1.9; Rae1EX28/Df(2R)5764, 23.4 ± 1.4; Rae1EX28, 26.9 ± 0.9; Rae1EX28 neuronal rescue, 25.6 ± 1.2, Rae1EX28 and wnd suppression, 23.2 ± 0.8; hiwΔN, 27.3 ± 2.0). However, the number of boutons was increased by almost twofold (120 ± 5 versus 67 ± 5 in wild type) and bouton size was decreased by ~50% (4.4 ± 0.4 μm2 versus 7.9 ± 0.5 μm2 in wild type) in homozygous Rae1EX28 mutants compared with wild-type larvae with similar body size (Fig. 3). The Rae1EX28/Df(2R)5764 mutant’s phenotype was very similar to that of the homozygous Rae1EX28 mutant, suggesting that Rae1EX28 is a strong loss-of-function allele. These findings suggest that Rae1 acts to restrain synaptic growth at the NMJ.
Figure 3.
Rae1 is required to restrain synaptic terminal growth at the NMJ. (a) Representative confocal images of segment A3 muscle 6/7 synapses stained for both DVGLUT (green) and FasII (red), in late 2nd/early 3rd instar wild-type, Rae1EX28, Rae1EX28 presynaptic rescue (Rae1EX28; elav-Gal4/UAS–NTAP-Rae1) and Rae1EX28 wallenda suppression (Rae1EX28; wnd3) larvae. Scale bar represents 10 μm. (b,c) Quantification of bouton number (b) and size (c) in wild-type, Rae1EX28/Df, Rae1EX28, Rae1EX28 rescue, Rae1EX28; wnd3 suppression and hiwΔN larval NMJs (segment A3 muscle 6/7; n = 20, 21, 18, 20, 20 and 11, respectively, for bouton number; n = 271, 465, 548, 726, 486 and 826, respectively, for bouton size). There was no significant difference in either bouton number or bouton size between Rae1EX28 and Rae1EX28/Df (P > 0.1 for both comparisons). *P < 0.001. Error bars denote s.e.m.
On the basis of the Rae1 mutant phenotype and the association between Rae1 and Hiw, we predicted that Rae1 functions in the presynaptic motoneuron to restrain the synaptic growth at NMJs, as both Hiw and Fsn regulate synaptic development on the presynaptic side19,21. To test this idea, we expressed the UAS-TAP-Rae1 transgene under the control of elav-Gal4, a pan-neuronal Gal4 driver, in the Rae1 mutant flies. We found that both the increased bouton number and the reduced bouton size were restored to almost wild-type levels by neural expression of Rae1 in these larvae (Fig. 3). These data indicate that the synaptic phenotype caused by Rae1EX28 mutation is a result of the loss of Rae1, but not pirk, and that Rae1 is required in the presynaptic motoneurons to control the growth of synapse.
Rae1 genetically interacts with hiw
Our biochemical data and Rae1 loss-of-function analysis strongly suggest that Rae1 and Hiw function in the same complex to restrain synaptic overgrowth. To further test this hypothesis, we analyzed genetic interactions between Rae1 and hiw. We reasoned that if Rae1 and Hiw function together in a complex, then partial loss-of-function of Rae1 should enhance the intermediate synaptic terminal overgrowth caused by the hiw hypomorphic mutant, hiwND512,3. A partial loss-of-function of Rae1, Rae1 heterozygosity (Rae1EX28/+), resulted in a ~50% reduction of Rae1 protein (Fig. 2) and a significant increase in the number of boutons on synapses on muscle 6/7 (P < 0.001; Supplementary Fig. 2), but did not change the number of boutons on muscle 4 (Fig. 4). However, when we combined hiwND51 with Rae1 heterozygosity, we observed a marked enhancement of NMJ overgrowth on muscle 4 (Fig. 4). The dominant enhancement of a weak hiw allele by the heterozygous Rae1 mutant is consistent with the hypothesis that the two genes directly interact in the same genetic pathway. Consistent with this model, we found that a null allele of hiw, hiwΔN, was not further enhanced by Rae1 heterozygosity (Fig. 4). Together, our data provide strong genetic evidence that Rae1 and hiw collaborate to restrain synaptic terminal growth.
Figure 4.
Rae1 genetically interacts with hiw to restrain synaptic terminal growth. (a) Representative confocal images of muscle 4 synapses stained for both DVGLUT (green) and FasII (red) in wild-type, Rae1EX28/+, hiwND51, hiwND51; Rae1EX28/+, hiwΔN and hiwΔN; Rae1EX28/+ larvae. Scale bar represents 10 μm. (b) Quantification of bouton number at muscle 4 NMJs in wild-type, Rae1EX28/+, hiwND51, hiwND51; Rae1EX28/+, hiwΔN and hiwΔN; Rae1EX28/+ larvae (n = 27, 23, 42, 46, 23 and 25 cells, respectively). There was a significant increase in the number of boutons formed in hiwND51; Rae1EX28/+ compared with hiwND51 3rd instar larval NMJs (*P < 0.001). The number of boutons in the hiwΔN; Rae1EX28/+ double mutant was not significantly different from that in hiwΔN (**P > 0.5), demonstrating a lack of enhancement of the hiwΔN phenotype by Rae1EX28/+. Error bars denote s.e.m.
The Hiw/Fsn E3 ligase complex regulates synaptic terminal growth by downregulating Wnd23. Loss of wnd function suppressed the morphological defects in either hiw or Fsn mutant NMJs. Given that we found that Rae1 interacts with Hiw to restrain synaptic growth, one would expect that Rae1 should also work through Wnd in this process. To test this hypothesis, we assessed the ability of wnd to suppress the morphological defects caused by Rae1EX28. We found that wnd suppressed both the increased bouton number (84.1 ± 3.9 in wnd suppression versus 119.8 ± 5.2 in Rae1EX28) and decreased bouton size (7.2 ± 0.3 μm2 in wnd suppression versus 4.3 ± 0.3 μm2 in Rae1EX28) seen in the Rae1 mutant at the NMJ (Fig. 3). Collectively, our data indicate that Rae1 is required in motoneurons and works together with hiw, upstream of wnd, to restrain synaptic growth at the Drosophila NMJ.
A structure and function analysis of Hiw functional domains
Hiw and its homologs are huge proteins with multiple conserved domains. The identification of Hiw binding partners, such as Medea32, Fsn21 and Rae1, and their functional relevance to Hiw activity suggest that different Hiw functional domains may interact with different cofactors and together modulate Hiw’s action. To study the functional requirement of these conserved domains and to develop tools for dissecting domain-specific interactions, we performed a systematic structure and function analysis on Hiw. We previously found that a full-length hiw transgene rescues the hiw mutant phenotype when expressed in neurons and that a similar transgene with point mutations in its RING-finger domain fails to do so, indicating that the E3 ligase domain is essential for Hiw function19. We then generated additional truncated hiw transgenes lacking the RCC1 domain, the PHR repeats, the C-terminal half of the Hiw protein or the N-terminal half of the Hiw protein. In addition, we generated a transgene encoding the 1,000 amino-acid fragment of the Hiw C terminus containing the E3 ubiquitin ligase domain (Fig. 5a). We next confirmed the effective expression levels of all of the hiw mutant transgenes in larval brains by western blot, and found that all of the Hiw deletion mutants were expressed with expected molecular weights at levels close to or higher than the wild type hiw transgene (Fig. 5b). When expressed in neurons in hiw mutants, only the full-length hiw transgene completely rescued the hiw phenotype19; none of the truncated hiw transgenes were able to rescue the synaptic terminal overgrowth phenotype (Fig. 5a and Supplementary Fig. 3). We next examined Wnd protein levels in these larval brains, we found a consistent correlation between elevated Wnd protein levels and the inability of the mutated hiw transgenes to rescue hiw morphological phenotype, in contrast with the fully restored Wnd protein levels and synaptic terminal size that were observed during neuronal expression of the hiw full-length transgene (Supplementary Figs. 3 and 4). The inability of the hiw mutant transgenes to restore Wnd protein level and to rescue the hiw morphological phenotype is unlikely to be a result of insufficient or excessive expression levels of these transgenes because most of the presented mutant hiw trans-genes were expressed at higher levels than the wild-type hiw transgene and because we analyzed multiple insertion lines of each transgene that had similar expression levels as the wild-type hiw transgene and found that none of them rescued the hiw overgrowth phenotype (data not shown). Thus, the results of our structure and function analysis on Hiw suggest that an intact Hiw protein with all of the functional domains is required for the proper control of Wnd protein expression and presynaptic arborization at the fly NMJ.
Figure 5.
A structure and function analysis of Hiw functional domains. (a) Schematic presentation of NTAP-tagged (NT-) and HM-tagged (HM-) hiw transgenes encoding full-length, mutated or truncated Hiw proteins. Hiw functional domains are marked with colored boxes. The positions of amino-acid substitution in NT-HiwΔRING and the added amino acid residue necessary to maintain the reading frame in NT-Hiw-RCC1 and NT-Hiw-PHR are indicated. The right column summarizes the ability of the given hiw transgene to rescue the synaptic overgrowth phenotype in hiw mutants, to cause dominant negative overgrowth phenotype when expressed in wild-type background and to bind to Rae1. (b) Western blot analysis of fly brain lysate using antibody to TAP (PAP, Sigma) or Myc revealed the expression of wild-type and mutant hiw transgenes in predicted size. (c) Representative confocal images of muscle 4 synapses stained for DVGLUT (green) and FasII (red) in wild-type (+/hiwND8; +/+; GS-elav-Gal4/+), NT-Hiw overexpression (+/hiwND8; UAS-NT-hiw/+; GS-elav-Gal4/+), NT-Hiw-NT overexpression (+/hiwND8; UAS-NT-hiw-NT/+; GS-elav-Gal4/+), NT-Hiw-CT1000 overexpression (+/hiwND8; +/+; GS-elav-Gal4/UAS-NT-hiw-CT1000), NT-Hiw-PHR overexpression (+/hiwND8; +/+; GS-elav-Gal4/UAS-NT-hiw-PHR), HM-Hiw-HindIII overexpression (+/hiwND8; UAS-HW-hiw-HindIII/+; GS-elav-Gal4/+), NT-Hiw-RCC1 overexpression (NT-hiw-RCC1/hiwND8; +/+; GS-elav-Gal4/+), or NT-Hiw-CT overexpression (+/hiwND8; +/+; GS-elav-Gal4/UAS-NT-hiw-CT) 3rd larval NMJs. Overexpression is indicated by an up arrow. Scale bar represents 10 μm. (d) Quantification of the number of boutons at muscle 4 NMJs in wild-type larvae or larvae overexpressing wild-type or mutant hiw transgenes (n = 23, 24, 26, 27, 26, 21, 25 and 27, respectively; *P < 0.001). Error bars denote s.e.m.
The inability of all the truncated Hiw proteins to rescue the hiw morphology may be a result of disruptions of the normal Hiw function or a gross alteration in protein folding, stability and/or localization. Although none of the mutant transgenes rescued the hiw morphology, some did act in a dominant-negative manner. When we used a strong neuronal Gal4 driver, GS-elav-Gal4, to express each of the hiw trans-genes in a hiw heterozygous background (hiwND8/+, no overgrowth phenotype), four hiw transgenes resulted in synaptic terminal overgrowth. Among them, hiw-phr overexpression resulted in a mild overgrowth, whereas Hiw-RCC1 and Hiw-HindIII overexpression resulted in an intermediate phenotype, and Hiw-CT overexpression resulted in notable hiw-like synaptic terminal overgrowth (Fig. 5c,d). The dominant-negative effects likely arise in relation to hiw, as, when expressed in a wild-type background using the same driver, each of the four hiw transgenes resulted in a milder overgrowth phenotype than that observed in the hiw heterozygous background (data not shown). These data indicate that the four truncated Hiw proteins can act as dominant-negative mutants, likely by sequestering important Hiw cofactors. Collectively, our data suggest that all of the Hiw functional domains are necessary for the ability of Hiw to restrain synaptic terminal growth. The fact that each individual domain is necessary for Hiw action suggests that these domains coordinate with each other, potentially by binding to different factors, and work together to modulate the activity of a Hiw complex.
Rae1 interacts with a Hiw C-terminal domain
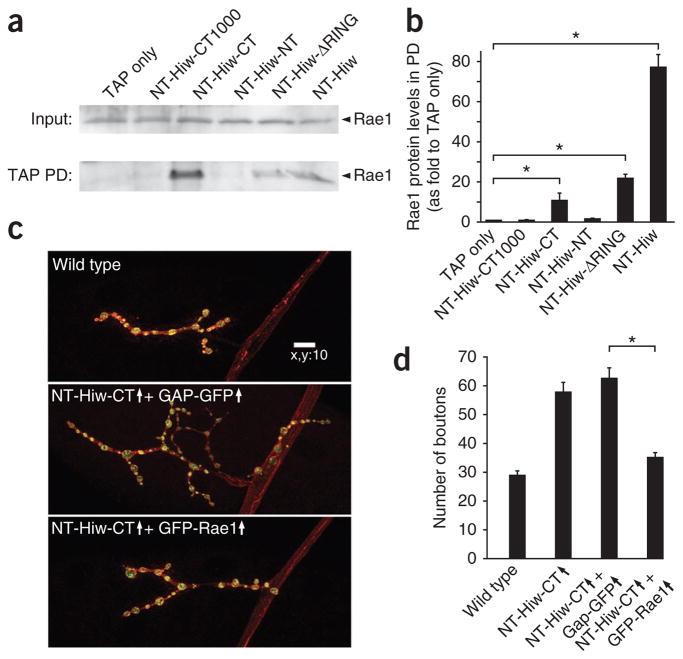
Except for Hiw-HindIII, the Hiw transgenes were tagged with TAP at their amino-terminus (Fig. 5a), allowing us to map the domain(s) of Hiw that mediate its interaction with Rae1. To do this, we first expressed Hiw deletion transgenes in neurons and performed the first step of TAP pull-down experiments with 3rd instar larval brain lysates. We used immunoblotting to determine whether Rae1 co-precipitates with different Hiw truncated proteins. All of the transgenic proteins, except for Hiw N-terminal half (Hiw-NT) and Hiw very C terminus (Hiw-CT1000), strongly associated with Rae1, whereas Hiw-NT completely and Hiw-CT1000 nearly completely lost their ability to pull-down Rae1 (Fig. 6a,b and Supplementary Figs. 1 and 5). These results suggest that the domain required for the Hiw-Rae1 interaction resides primarily in the C-terminal portion of Hiw outside of the Hiw-CT1000 region (amino acids 2,885 to 4,082). Our data are consistent with a detailed structure and function analysis (B. Grill, L. Chen, E.D. Tulgren, S.T. Baker, W. Bienvenut et al., personal communication), which mapped the Rae1-binding motif on Hiw to the corresponding protein fragment, amino acids 2,932–2,959.
Figure 6.
Rae1 interacts with a fragment in the Hiw C-terminal region, and coexpression of Rae1 with NT-Hiw-CT suppresses the NT-Hiw-CT–induced dominant-negative overgrowth phenotype. (a) Indicated NTAP-tagged hiw transgenes (described in Fig. 5) and a TAP only transgene were expressed in neurons under the control of the BG380-Gal4 driver. Larval brain lysates from each sample were subject to IgG pulldown. Both the inputs and the IgG pulldown complexes were analyzed by western blot using antibody to Rae1. Rae1 was present in all of the pulldown complexes except for those from TAP only, NT-Hiw-CT1000 and NT-Hiw-NT. Full-length blots are presented in Supplementary Figure 10. (b) Quantification of the interaction between various Hiw transgenic proteins and Rae1. Rae1 intensities in TAP pulldown blots were normalized to both the intensities and molecular weights of the corresponding TAP pulldown proteins. (c) Representative confocal images of muscle 4 synapses stained for both DVGLUT (green) and FasII (red) in wild-type (BG380/Y;+/+; +/+), NT-Hiw-CT and GAP-GFP (BG380/Y; UAS-GAP-GFP/+; UAS-NT-hiw-CT/+), and NT-Hiw-CT and Rae1 (BG380/Y; +/+; UAS-NT-hiw-CT/UAS-GFP-Rae1) larvae. Scale bar represents 10 μm. (d) Quantification of number of boutons at muscle 4 NMJs in wild-type, NT-Hiw-CT overexpression (BG380/Y; +/+; UAS-NT-hiw-CT/+), NT-Hiw-CT and GAP-GFP, and NT-Hiw-CT and Rae1 (n = 24, 27, 32 and 26, respectively) larvae. Error bars denote s.e.m.; *P < 0.001.
Notably, neuronal expression of Hiw-CT, which contains the Rae1-binding domain, had prominent dominant-negative effects on synaptic terminal growth at the NMJs (Fig. 5c,d). On the basis of the results of our domain-mapping study, we postulated that the dominant-negative effect of Hiw-CT is partly caused by its ability to sequester Rae1 and consequently prevent the formation of functional Hiw-Rae1 complexes. We further predicted that increasing Rae1 expression would suppress the dominant-negative effects of Hiw-CT. To test this hypothesis, we coexpressed Hiw-CT and Rae1 or a control GAP-GFP protein, a fusion of the myristoylation sequence from GAP43 to GFP that targets GFP to the cell membrane, in neurons. We found that the synaptic overgrowth phenotypes caused by Hiw-CT expression were completely suppressed by coexpression of Rae1, but not by GAP-GFP (Fig. 6c,d). These data confirm that Rae1 is a cofactor that is required for the function of the Hiw complex, which is diluted out by this dominant-negative Hiw fragment. This notion is also supported by the fact that all of the mutant hiw transgenes that contain the Rae1-binding region exhibited various levels of dominant-negative effect when overexpressed in the wild-type background (Fig. 5a).
Rae1 promotes Hiw protein abundance
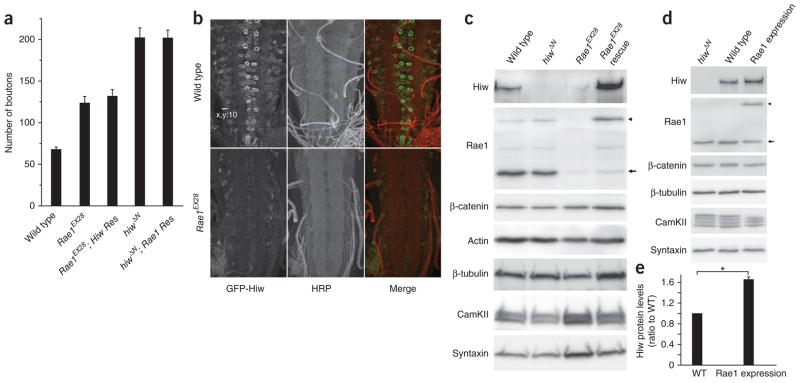
To understand how Rae1 positively regulates Hiw function at the molecular level, we first tested whether neuronal expression of Hiw can rescue the Rae1 morphological phenotype and vice versa. We found that neither GFP-Hiw nor N-terminal TAP-tagged Rae1 can rescue the mutant phenotype of the other (Fig. 7a), suggesting that they are required for each other and the Hiw-Rae1 interaction is essential for a functional E3 ubiquitin ligase complex. To exclude the possibility that this inability to rescue is caused by insufficient expression levels of either genes, we examined the expression levels of both transgenes in wild-type and mutant larval CNS. Although we detected no substantial difference in the expression levels of elav-Gal4–driven Rae1 in hiw mutant and wild-type larval CNS (data not shown), elav-Gal4–driven GFP-Hiw expression in Rae1 homozygous mutant brains was markedly diminished compared with the broad neuronal cell body expression of GFP-Hiw in Rae1 heterozygous mutant brains (Supplementary Fig. 6). To confirm that transgenic GFP-Hiw protein is downregulated in Rae1 mutant brains, we analyzed the expression level of BG380-Gal4–driven GFP-Hiw protein in Rae1EX28 and wild-type larval brains by immunocytochemistry. Again, we observed a marked reduction of GFP-Hiw expression in Rae1EX28 brains compared with wild-type brains (Fig. 7b). These data suggest that Rae1 may function to regulate the abundance and stability of Hiw.
Figure 7.
Rae1 promotes Hiw abundance. (a) Neuronal expression of neither Hiw nor Rae1 rescues the synaptic terminal overgrowth phenotype caused by the loss of function of the other gene. Quantification of the number of boutons in wild-type, Rae1EX28, Rae1EX28; Hiw rescue (Res) (Rae1EX28; elav-Gal4/UAS-GFP-hiw), hiwΔN and hiwΔN; Rae1 rescue (hiwΔN; elav-Gal4/UAS-NTAP-Rae1) larval NMJs (segment A3 muscle 6/7; n = 12, 12, 12, 7 and 11, respectively). (b) Representative confocal images of wild-type (BG380-Gal4/+; +/+; UAS-GFP-hiw/+) or Rae1 mutant (BG380-Gal4/+; Rae1EX28; UAS-GFP-hiw/+) ventral ganglions with UAS-GFP-hiw trangene expressed in neurons. The larvae were stained with antibodies to horseradish peroxidase (HRP, red) and GFP (green). Scale bar represents 10 μm. (c) Western blots of total proteins extracted from wild-type, hiwΔN, Rae1EX28 and Rae1EX28 rescue (Rae1EX28; elav-Gal4/UAS-NTAP-Rae1) larval brains probed with indicated antibodies. Full-length blots are presented in Supplementary Figure 11. (d) Western blot analysis on total proteins extracted from hiwΔN, wild-type (C155-Gal4/+) and Rae1 expression (C155-Gal4/+; UAS-NTAP-Rae1/+) larval brains. Full-length blots are presented in Supplementary Figure 12. (e) Quantification of Hiw protein levels in the wild-type and the Rae1 expression larval brains (*P < 0.001, based on seven independent experiments). Arrows indicate a 38-kDa endogenous Rae1 protein and the arrowheads indicate the 58-kDa transgenic NTAP-Rae1 protein. Error bars denote s.e.m.
To test this hypothesis, we examined whether the endogenous Hiw protein is also reduced in Rae1 mutants. We assayed Hiw protein levels in the brains of wild-type, hiwΔN, Rae1EX28 and Rae1EX28 larvae with neuronal rescue by a Rae1 transgene. We extracted total proteins from dissected larval brains using a lysis buffer that contains urea, SDS, and a comprehensive panel of protease and proteosome inhibitors to ensure the maximal preservation of total neuronal proteins in the lysates. Hiw immunoblot of these brain lysates revealed that endogenous Hiw protein in Rae1 mutant brain lysate was markedly reduced compared with that in wild-type brains (Fig. 7c). The reduction of Hiw protein level seemed to depend on the dose of Rae1, as the Rae1 heterozygous mutants also showed a mild decrease in Hiw expression (Supplementary Fig. 7). The neuronal expression of the NT-Rae1 transgene completely restored Hiw protein level (Fig. 7c), consistent with its ability to rescue the synaptic terminal overgrowth phenotype at the Rae1 mutant NMJs (Fig. 3). In contrast with the markedly reduced protein level of Hiw that we observed in Rae1 mutant brain lysate, Rae1 protein level was not altered in hiw mutants (Figs. 1c and 7c,d and Supplementary Fig. 7), indicating that Rae1 is not a Hiw ubiquitination target. We next tested whether neuronal overexpression of transgenic Rae1 protein is sufficient to elevate endogenous Hiw protein level in wild-type background. We consistently detected a ~70% increase in the level of endogenous Hiw protein when we expressed transgenic NTAP-Rae1 protein in wild-type brains at about the same level as endogenous Rae1 (driven by a pan-neuronal driver C155-Gal4; Fig. 7d,e). These data indicate that Rae1 is both necessary and sufficient to promote Hiw protein abundance.
Given the established role of Rae1 in regulating mRNA export in yeast27, the observed effect of Rae1 on Hiw protein levels could be a result of a general defect in mRNA export when Rae1 is mutated. If this were true, we would predict a gross change in protein levels for a broad range of molecules. However, none of the neuronal proteins that we tested, Futsch, CamKII, β-tubulin, Syntaxin and the neural form of β-catenin, showed decreased protein levels in Rae1 mutant or increased protein levels in Rae1-expression brain lysate (Fig. 7c,d and Supplementary Fig. 7). Hence, both the reduction of protein level in the Rae1 mutant brains and the increase of protein level in Rae1-expression brains are selective to Hiw. Rae1’s regulation of Hiw protein abundance is therefore unlikely mediated by general defects in mRNA transportation. This observation is also consistent with the previous finding that knockdown of Rae1 activity by RNA interference in cultured Drosophila cells does not lead to mRNA transport defects29.
The direct association of Rae1 to Hiw complex strongly suggests that Rae1 regulates the abundance of Hiw at the post-translational level. However, given the broad functions of Rae1 homologs in other systems, Rae1 could also regulate hiw transcription or hiw mRNA stability. To test these possibilities, we examined hiw mRNA levels in wild-type and Rae1 mutant brains using quantitative reverse-transcription PCR (RT-PCR). We found that there was no significant change in hiw transcript levels in Rae1 mutant brains compared to wild-type brains (ΔCt ratio of specific transcript to 18S rRNA internal control transcript: Rae1EX28 17.36 ± 0.58, versus wild type 17.48 ± 0.19; P > 0.5; see Online Methods). These data therefore exclude the possibility that Rae1 regulates hiw transcription or mRNA stability. In addition, Rae1 mutants were unable to sustain a comparable level of the transgenic GFP-Hiw protein expressed via the UAS or Gal4 systems in larval CNS (Fig. 7b). The UAS–GFP-hiw transgene contains the full-length Hiw coding sequence without introns and UTRs. Given that RNA-binding proteins often target 3′ UTRs to regulate mRNA stability, degradation, nuclear export, subcellular localization and translation efficiency33,34, it is unlikely that Rae1 regulates the transgenic hiw mRNA via these mechanisms. Thus, Rae1 controls endogenous Hiw protein expression via post-translational regulation. Altogether, these findings strongly suggest that Rae1 regulates Hiw protein expression through direct protein-protein interactions with the Hiw E3 ligase complex.
Rae1 suppresses autophagy-mediated degradation of Hiw
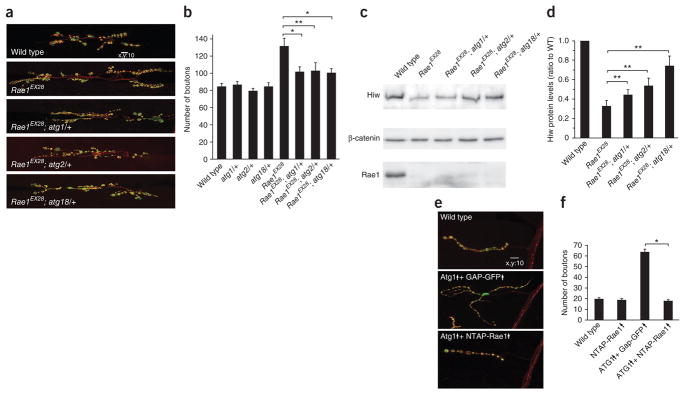
How does Rae1-Hiw interaction promote the abundance of Hiw protein? How does it interact with known mechanisms that regulate Hiw protein level? Autophagy regulates synaptic development by promoting the degradation of Hiw protein25. It is thus possible that Rae1-Hiw interaction may protect Hiw from being degraded by the autophagy pathway. If this were true, we would expect that reducing autophagic activity in Rae1 mutant should suppress both the reduction of Hiw protein level and the associated synaptic terminal overgrowth. We tested this possibility by crippling autophagic activity via removal of one copy of three different essential autophagy genes35, atg1, atg2 or atg18, respectively, in the Rae1 mutant background. We found mutations of these essential autophagy genes can each dominantly suppress the Rae1 mutant morphological phenotype (Fig. 8a,b), while heterozygous mutants of atg1, atg2 or atg18 alone have no synaptic overgrowth phenotype (Fig. 8b). Furthermore, this suppressions correlates with Hiw protein levels; there was a significant increase in the expression of Hiw protein in Rae1 mutant brains when one copy of atg1, atg2 or atg18 was removed (P < 0.05; Fig. 8c,d). These data indicate that Hiw is rapidly degraded via autophagy in the absence of Rae1.
Figure 8.
Rae1-Hiw interaction prevents autophagy-mediated degradation of Hiw protein. (a) Representative confocal images of segment A3 muscle 6/7 synapses stained for both DVGLUT (green) and FasII (red), in wild-type, Rae1EX28, Rae1EX28; atg1/+, Rae1EX28; atg2/+ and Rae1EX28; atg18/+ wandering larvae. Scale bar represents 10 μm. (b) Quantification of the number of boutons in wild-type, atg1/+, atg2/+, atg18/+, Rae1EX28, Rae1EX28; atg1/+, Rae1EX28; atg2/+ and Rae1EX28; atg18/+ larval NMJs (segment A3 muscle 6/7; n = 12, 12, 12, 12, 12, 16, 12 and 21, respectively). (c) Western blot analysis on total proteins extracted from wild-type, Rae1EX28, Rae1EX28; atg1/+, Rae1EX28; atg2/+ and Rae1EX28; atg18/+ larval brains. The neural form of β-catenin (82 kDa, see Online Methods) was used as an internal control of neuronal proteins. Full-length blots are presented in Supplementary Figure 13. (d) Quantification of Hiw protein levels in wild-type, Rae1EX28, Rae1EX28; atg1/+, Rae1EX28; atg2/+ and Rae1EX28; atg18/+ larval brains (based on six independent experiments). (e) Representative confocal images of muscle 4 synapses stained for both DVGLUT (green) and FasII (red) in wandering larvae of wild-type (C155-Gal4/+), Atg1 Gap-GFP (C155-Gal4/+; UAS-Gap-GFP/+; UAS-Atg16B/+), and Atg1 NTAP-Rae1 (C155-Gal4/+; UAS-NTAP-Rae1/+; UAS-Atg1/+). Scale bar represents 10 μm. (f) Quantification of the number of boutons at muscle 4 synapses in wild-type, Rae1 expression (C155-Gal4/+; UAS-NTAP-Rae1/+), Atg1 Gap-GFP and Atg1 NTAP-Rae1 wandering larvae (segment A2–4, n = 28, 27, 33 and 34, respectively). Error bars denote s.e.m. *P < 0.001, **P < 0.05.
Neuronal expression of Atg1 induces high levels of autophagy and causes synaptic terminal overgrowth by downregulating Hiw25. If the interaction between Rae1 and Hiw protects autophagy-mediated degradation of Hiw, coexpression of Rae1 with Atg1 should suppress Atg1-induced overgrowth. We coexpressed NTAP-Rae1 with Atg1 and found that the Atg1-induced NMJ overgrowth was fully suppressed (Fig. 8e,f), This suppression is not a result of a simple dilution of Gal4, as coexpression of GAP-GFP had no effect on Atg1-induced NMJ overgrowth. Together, these findings indicate that an important function of Rae1 in the Hiw E3 ligase complex is to prevent autophagy-mediated degradation of Hiw.
DISCUSSION
Molecular pathways mediated by Hiw and its orthologs are important for neural development and regeneration. By identifying and characterizing the Hiw-interacting protein Rae1, we identified a previously unknown function of Rae1 in the post-mitotic neuron: regulating synaptic development by binding to Hiw and protecting Hiw from autophagic degradation.
Rae1 associates and functions with Hiw in neurons
We provide three pieces of evidence that Rae1 is a bona fide Hiw-binding protein. First, Rae1 and Hiw were co-purified from adult head homogenate in reciprocal TAPs using either Hiw or Rae1 as the tagged proteins, respectively. Second, endogenous Rae1 co-immunoprecipitated with Hiw in fly larval brains. Finally, coexpressing Rae1 suppressed the dominant-negative effects caused by overexpression of the truncated Hiw protein (Hiw-CT) that contains the Rae1-binding region.
In addition to Rae1, a number of other molecules have been identified that bind Hiw and its orthologs. These factors include the fly medea/SMAD (a component of the TGF-β signaling pathway32), adenylate cyclase (an enzyme controlling cAMP levels15,36), Tuberin (a component of the TSC complex that regulates TOR signaling37), GLO-4 (a positive regulator of a Rab GTPase pathway38) and FSN-1/Fsn/Fbox45 (an F-box protein20–22). Of these interactions, only the Fsn-PHR and Rae1-PHR (our results and those of B. Grill, L. Chen, E.D. Tulgren, S.T. Baker, W. Bienvenut et al., personal communication) interactions have been shown to be conserved across species. Studies of the PHR-FSN interaction have defined a common mechanism in the action of PHR proteins, RING E3 ligases working in an SCF-like ubiquitin ligase complex20–22. Similarly, the PHR-Rae1 interaction may reveal a previously unknown and conserved mechanism that positively regulates the action of PHR proteins in post-mitotic neurons.
Drosophila nucleoporin (Nup98) is a binding partner of Rae1 involved in many biological processes, such as facilitating mRNA export and the Rae1-APC interaction27,28. Our Rae1 TAP experiment also pulled down Nup98 with high confidence (Fig. 1b and Supplementary Table 2), suggesting that Nup98 strongly associates with Rae1 in Drosophila neurons. However, Nup98 was not detected in the TAP-Hiw pulldown complex. This observation suggests that the interaction between Hiw and Rae1 may not require Nup98 and that Rae1 and Nup98 may carry out functions in neurons that are independent of Hiw.
We investigated the biological relevance of the Hiw-Rae1 interaction in post-mitotic neurons. Rae1 has multiple important functions in mitotic cells, but its role in post-mitotic neurons is unknown. Our loss of function analysis of Rae1 at the Drosophila larval NMJ revealed that Rae1 functions in post-mitotic motor neurons to restrain synaptic growth. Rae1 is unlikely to be a ubiquitin target of the Hiw E3 ubiqutin ligase complex, as Rae1 functions in the same direction as hiw, and Rae1 protein expression was unchanged in hiw null mutants compared with wild-type animals. Instead, our data suggest that Rae1 works as a positive regulator to facilitate the function of the Hiw E3 ligase complex. This notion is supported by the similar loss of function phenotype in Rae1 and hiw null mutants, the strong genetic interactions between Rae1 and hiw, and the ability of wnd, a known Hiw downstream target, to suppress Rae1 phenotypes. Thus, our biochemical data, together with our genetic analysis, suggest that Rae1 acts in the post-mitotic neurons to restrain the growth of synaptic terminals through its interaction with the Hiw E3 ubiquitin ligase complex.
The role of Rae1 in stabilizing Hiw protein
The Rae1-Hiw protein interaction could promote the function of Hiw ubiquitin ligase by regulating Hiw’s localization, enzymatic activity or substrate specificity, or protein stability. Our results favor the last possibility, that Rae1 regulates Hiw protein abundance and stability. We found that Hiw protein levels were markedly reduced in the Rae1EX28 mutant brains and substantially increased in the brains of flies expressing Rae1, and that both the reduction and increase were selective for Hiw, as the levels of many other neuronal proteins were unaltered. Thus, an important function of Rae1 in the post-mitotic neuron is to dynamically regulate Hiw protein expression. To further understand the molecular basis of how Rae1 promotes Hiw protein abundance, we have excluded the possibilities that reduction of Hiw protein in the Rae1 mutants could be due to defects in mRNA export in general, or reduced hiw transcription or mRNA stability. It is also unlikely that Rae1 controls the translation of hiw, as the promoter and 3′ UTR region of hiw were unnecessary for Rae1’s regulation of Hiw abundance. These data, together with the finding that Rae1 physically associates with Hiw, strongly suggest that Rae1 promotes Hiw protein stability via its association with Hiw and prevents Hiw’s degradation.
Protein clearance and quality control in eukaryotic cells are executed by two major pathways, ubiquitin-proteosome–mediated degradation and autophagy. The former is generally anticipated to be a more specific targeting mechanism than the latter. However, recent studies have suggested that selective autophagy can be achieved through a number of autophagic adaptor proteins, although the molecular mechanisms remain to be elucidated39. Autophagy was recently implicated in the regulation of Hiw protein levels in Drosophila25, but it is unclear whether this regulation is selective and how the selectivity is achieved. We found, both genetically and biochemically, that the association of Rae1 and Hiw prevents Hiw from being degraded by autophagy. Our results suggest a mechanism for regulating selective autophagy: specific factor(s) can bind to the target protein and prevent it from being selected for autophagy-mediated degradation.
Although our data do not exclude a functional requirement of Rae1 for Hiw activity and localization, they clearly indicate that Rae1 is essential for controlling Hiw protein levels during development. Rae1, a microtubule-binding protein with multiple cellular functions, is of great interest as a regulator of synaptic development. Our data suggest that decreasing and increasing Rae1 from its basal protein levels drives corresponding changes in Hiw protein levels. It is intriguing to speculate that Rae1 functions as a molecular switch in response to either environmental cues or microtubule networks to rapidly and precisely regulate Hiw protein levels during neural development and regeneration.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureneuroscience/.
Supplementary Material
Acknowledgments
We are particularly grateful to D. Sitterlin for providing the antibody to Rae1. We would like to thank the A.J. Siteman Cancer Center at the Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Missouri, and the Department of Physiology at the Louisiana State University Health Sciences Center in New Orleans for the use of their proteomic core facilities. We are grateful to T. Neufeld and the Bloomington Stock Center for fly stocks, and the Developmental Studies Hybridoma Bank for antibodies. We also thank B. Grill and Y. Jin for helpful comments. This work is supported by grants from the US National Institutes of Health (NS070962 to C.W. and DA020812 to A.D.).
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
AUTHOR CONTRIBUTIONS
C.W. and A.D. initiated the project and conducted the Hiw structure and function analysis. C.W. directed the rest of the studies. X.T. and C.W. conceived and designed the experiments. X.T., J.L. and C.W. performed the experiments. V.V. contributed to the characterization of the Rae1 mutant phenotype. C.W. and X.T. wrote the manuscript with input from A.D.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Po MD, Hwang C, Zhen M. PHRs: bridging axon guidance, outgrowth and synapse development. Curr Opin Neurobiol. 2010;20:100–107. doi: 10.1016/j.conb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 2.DiAntonio A, et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 3.Wan HI, et al. Highwire regulates synaptic growth in Drosophila. Neuron. 2000;26:313–329. doi: 10.1016/s0896-6273(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 4.Uthaman SB, Godenschwege TA, Murphey RK. A mechanism distinct from highwire for the Drosophila ubiquitin conjugase bendless in synaptic growth and maturation. J Neurosci. 2008;28:8615–8623. doi: 10.1523/JNEUROSCI.2990-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaefer AM, Hadwiger GD, Nonet ML. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 2000;26:345–356. doi: 10.1016/s0896-6273(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhen M, Huang X, Bamber B, Jin Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 2000;26:331–343. doi: 10.1016/s0896-6273(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 7.Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess RW, et al. Evidence for a conserved function in synapse formation reveals Phr1 as a candidate gene for respiratory failure in newborn mice. Mol Cell Biol. 2004;24:1096–1105. doi: 10.1128/MCB.24.3.1096-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza J, et al. Formation of the retinotectal projection requires Esrom, an ortholog of PAM (protein associated with Myc) Development. 2005;132:247–256. doi: 10.1242/dev.01578. [DOI] [PubMed] [Google Scholar]
- 10.Culican SM, Bloom AJ, Weiner JA, DiAntonio A. Phr1 regulates retinogeniculate targeting independent of activity and ephrin-A signaling. Mol Cell Neurosci. 2009;41:304–312. doi: 10.1016/j.mcn.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewcock JW, Genoud N, Lettieri K, Pfaff SL. The ubiquitin ligase Phr1 regulates axon outgrowth through modulation of microtubule dynamics. Neuron. 2007;56:604–620. doi: 10.1016/j.neuron.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong X, et al. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholich K, Pierre S, Patel TB. Protein associated with Myc (PAM) is a potent inhibitor of adenylyl cyclases. J Biol Chem. 2001;276:47583–47589. doi: 10.1074/jbc.M107816200. [DOI] [PubMed] [Google Scholar]
- 16.Guo Q, Xie J, Dang CV, Liu ET, Bishop JM. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc Natl Acad Sci USA. 1998;95:9172–9177. doi: 10.1073/pnas.95.16.9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbarini N, Delpire E. The RCC1 domain of protein associated with Myc (PAM) interacts with and regulates KCC2. Cell Physiol Biochem. 2008;22:31–44. doi: 10.1159/000149781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abrams B, Grill B, Huang X, Jin Y. Cellular and molecular determinants targeting the Caenorhabditis elegans PHR protein RPM-1 to perisynaptic regions. Dev Dyn. 2008;237:630–639. doi: 10.1002/dvdy.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Wairkar YP, Collins CA, DiAntonio A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J Neurosci. 2005;25:9557–9566. doi: 10.1523/JNEUROSCI.2532-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 21.Wu C, Daniels RW, DiAntonio A. DFsn collaborates with Highwire to down-regulate the Wallenda/DLK kinase and restrain synaptic terminal growth. Neural Dev. 2007;2:16. doi: 10.1186/1749-8104-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saiga T, et al. Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol Cell Biol. 2009;29:3529–3543. doi: 10.1128/MCB.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins CA, Wairkar YP, Johnson SL, DiAntonio A. Highwire restrains synaptic growth by attenuating a MAP kinase signal. Neuron. 2006;51:57–69. doi: 10.1016/j.neuron.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Nakata K, et al. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 2005;120:407–420. doi: 10.1016/j.cell.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–79. doi: 10.1083/jcb.200907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JA, et al. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeganathan KB, Malureanu L, van Deursen JM. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 29.Sitterlin D. Characterization of the Drosophila Rae1 protein as a G1 phase regulator of the cell cycle. Gene. 2004;326:107–116. doi: 10.1016/j.gene.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Kraemer D, Dresbach T, Drenckhahn D. Mrnp41 (Rae1p) associates with microtubules in HeLa cells and in neurons. Eur J Cell Biol. 2001;80:733–740. doi: 10.1078/0171-9335-00216. [DOI] [PubMed] [Google Scholar]
- 31.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 32.McCabe BD, et al. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004;41:891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- 33.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 34.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 35.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierre SC, Hausler J, Birod K, Geisslinger G, Scholich K. PAM mediates sustained inhibition of cAMP signaling by sphingosine-1-phosphate. EMBO J. 2004;23:3031–3040. doi: 10.1038/sj.emboj.7600321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murthy V, et al. Pam and its ortholog highwire interact with and may negatively regulate the TSC1. TSC2 complex J Biol Chem. 2004;279:1351–1358. doi: 10.1074/jbc.M310208200. [DOI] [PubMed] [Google Scholar]
- 38.Grill B, et al. C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1. Neuron. 2007;55:587–601. doi: 10.1016/j.neuron.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.