Abstract
Infections of the vaginal tract result from perturbations in the complex interactions between the microbiome and the host vaginal ecosystem. Recent data have linked specific vaginal microbes and urogenital infection with pre-term birth. Here we discuss how next generation sequencing-based approaches to study the vaginal microbiome will be important for defining what constitutes an imbalance of the microbiome and the associated host conditions that lead to subsequent infection and disease states. These studies will provide clinicians reliable diagnostic tools and treatments for women who are at increased risk for vaginal infections, preterm birth, HIV and other sexually acquired diseases, and will provide opportunities for intervention.
Microbial infections of the vaginal tract
Urogenital infections [e.g. bacterial vaginosis (BV), urinary tract infection, and vaginitis], afflict over 1 billion women each year [1-5]. These infections are associated with a large percentage of cases of vulvitis, cervicitis, and pelvic infections. BV, for example, is a complex condition thought to occur as a result of a shift in the microbial community from the normally dominant Lactobacillus species to a polymicrobial community, with concentrations of other aerobes and anaerobes being elevated 100- and 1000-fold, respectively [6, 7]. BV affects 10-15% of women of reproductive age and is associated with genital tract infections and pregnancy complications, including pelvic inflammatory disease, premature rupture of membranes, intrauterine growth restriction, intrauterine fetal demise, chorioamnionitis, endometritis, pre-term labor and delivery, postpartum infection, ectopic pregnancy and tubal factor infertility [8-11]. The cost of BV-related pregnancy complications in the United States alone is nearly $1 billion annually [3]. Similar statistics can be found for fungal vaginitis [12]. Approximately three out of four women experience at least one episode of vaginal candidiasis, while 20% of healthy women are asymptomatically colonized with Candida [13]. Recent studies have shown that the vaginal tract contains a remarkably complex microbial community and that defining a healthy vaginal environment is a considerable challenge. The emerging methods for studying the microbiome provide an opportunity to define a healthy vaginal microbiome, and link vaginal infections to a microbial pathogen or community structure. In this review we discuss the potential for the application of metagenomic approaches to vaginal diseases for improving women’s health.
Urogenital infections and pathophysiological complications
We are just beginning to understand the interactions and relationships between the abundant, complex and dynamic microbial populations (microbiome) of the vagina and womens’health [7, 14, 15]. Despite increasing efforts and awareness, we currently know very little about how the microbiome is affected by age, menstrual cycle, genetic background or other factors in the daily lives of women, including pregnancy [16-21]. Although the final outcome of a microbial infection likely depends on many factors, the host’s genetic make-up is thought to play a major role [22, 23]. For example, studies have indicated that black women are more likely to have BV than are non-Hispanic white women and that known risk factors do not explain the observed racial disparities [20, 24, 25]. Very little is known about the factors that contribute to these differences. Other metabolic processes that correlate with the presence of BV are important to consider. The malodor that is characteristic of BV has been attributed to the presence of amine-containing metabolites, and several diagnostic tests are based on the detection of these metabolites [26-34]. However, amines and amine-producing microbes have been found in asymptomatic women as well [35-41], bringing into question the association between BV and amine production and whether microbes present in a healthy vagina also produce amine-containing metabolites that may cause malodor.
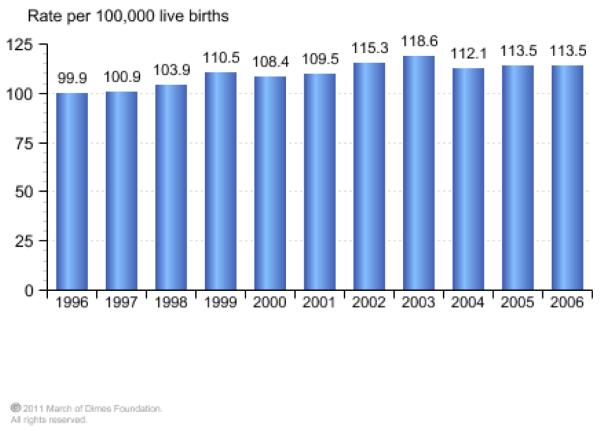
In addition to the complications already discussed, urogenital infections, such a BV, are associated with the costly problem ($26.2 billion annually in societal costs), of pre-term delivery. For much of the 20th century, pre-term birth, defined as birth before 37 weeks of gestation, was viewed as an unpredictable and inevitable fact of life. About 13 million premature births occur each year, and this is a direct cause of nearly 30% of all neonatal deaths (1 million each year)[42]. In the United States prematurity ranks second for causes of infant mortality behind birth defects (Figure 1A). The world’s rate of pre-term birth has also risen consistently over the past couple of decades. For example, the rate of singleton pre-term births has increased by 14% since 1990, and now accounts for 11.1% of deliveries worldwide. In 2006, pre-term births constituted 12.8% of live births in the United States, an increase of 20% since 1990, and with this there has been a corresponding increase in infant deaths related to prematurity (Figure 1B).
Figure.
A) Prematurity was the second leading cause of infant death in the United States in 2006 and accounted for 31.3% of all infant deaths. B) Infant deaths due to prematurity per 100,000 live births continues to be a significant problem in the United States and even increased from 1996-2006. (www.marchofdimes.com/peristats)
How is the vaginal tract microbiome associated with pre-term birth?
In most cases of pre-term labor and delivery, intrauterine infection is not clinically apparent. This may be because many hospital laboratories still use the traditional culturing methods for detecting bacterial infections [43, 44]. As uncultivated species may not be detected without the use of molecular approaches, it is possible that uncultivated vaginal species associated with preterm birth may also be discovered through analysis of the vaginal microbiome. Histologic evidence of inflammation in the decidua, fetal membranes, or umbilical cord is relatively common, and microbiologic evidence suggests that infection may contribute to approximately 25% of pre-term births, with bacterial colonization rates as high as 79% for birth at 23 weeks of gestation, declining to 11% at 31 to 34 weeks[45-47]. A recent meta-analysis study concluded that BV more than doubled the risk of preterm delivery in asymptomatic patients and of preterm labor in patients with symptoms, as well as significantly increased the risk of late miscarriages and maternal infection [48]. Moreover, there are specific associations between various vaginal microbes linked to BV (Mycoplasma hominis, Gardnerella vaginalis and Atopobium vaginae, alone or together) and pre-term birth [49-52]. Changes in the vaginal microbiome have also been associated with another known risk factor for pre-term delivery, shortening of the cervix in pregnancy [53]. A conundrum exists, however, as treatment for BV has not been shown to decrease rates of pre-term delivery [54]. While some microbes can be cultured from the conceptus when suitable methods are used, pregnant women at risk for pre-term birth due to vaginal colonization with these organisms receive little or no benefit from prophylactic treatment to eradicate the organisms. The question remains, then, what is the relationship between vaginal microbes and preterm deliveries and other perturbations in the vaginal ecosystem that lead to disease states?
Understanding the vaginal tract microbiome in the metagenomic era
Our understanding of this relationship between the vaginal tract microbiome and human disease is now poised to change - A report by the National Research Council supporting a Global Metagenomics Initiative states that metagenomics “presents the greatest opportunity to revolutionize understanding of the microbial world” [55]. The term metagenomics was first used in the late 1990s, and was defined as the genomic analysis of microorganisms by direct extraction and cloning of DNA from an assemblage of microorganisms without the necessity for culturing. The availability of “next generation” sequencing technologies have made it such that a cloning step is no longer essential for metagenomic projects and one can now rapidly, and with unprecedented depth, define the total microbial community or microbiome (the number and relative abundance of microbial species present), and their metagenomes (the microbial functional content). The application of these technological advances will provide an understanding of the overall composition and physiology of the vaginal microbiome, and will provide objective analytical and biomolecular tools to assess and improve women’s health. Ultimately, we believe this work will revolutionize the diagnosis and treatment of vaginal diseases, including pre-term birth, and thus will have a significant overall impact on women’s health.
Metagenomics holds significant promise for increasing our understanding of many microbial diseases associated with the human body, especially those considered to be polymicrobial in origin. Indeed, the landscape of microbial ecology as it relates to human disease has been altered forever by the advent of metagenomics and has led to the National Institutes of Health’s (NIH) Human Microbiome Project (HMP). The HMP has the overall goal of demonstrating the role the human microbiome plays in human health and disease by promoting a combined assessment of microbial population structure with an assessment of community function. Ultimately, the goal of the HMP is to provide a reference database of genome and metagenome sequences and an estimation of the microbial community structure at various body sites in healthy individuals. This will establish if there is a “core microbiome” at each body site indicative of a healthy status, and will provide the baseline community structure databases that will enable application of Koch’s Postulates to previously intractable human diseases. That is, the correlation of microbial community structure or microbiome, rather than a single microorganism, to healthy and diseased states.
Initial studies are beginning to define the microbial, metabolic and immunologic components of the vaginal ecosystem and the effects of demographic variables on these parameters. Despite this progress, several large gaps exist in our knowledge about the vaginal ecosystem including the role of factors such as host genetics[56, 57], environmental exposure[58] and health status[59] on the abundance and interactions of individual microbial species, related taxa, and groups of distantly related microorganisms. For example, it is commonly believed that the vaginal microbiome is an indicator of health, whereas women with an altered composition of vaginal microbes are more likely to exhibit symptomatic infections and increased risk of sexually acquired diseases or pregnancy complications [60-63]. Culture-dependent identification of vaginal microbes has provided only an incomplete understanding of the vaginal microbiome [16, 64], and a profound knowledge gap remains regarding the linkage between the vaginal microbiome and pre-term birth. For example, until recently there was a common belief that the vaginal microbiome include only a few species, predominantly from the genus Lactobacillus, the so-called “good” bacteria. Vaginal disease has been simplistically modeled to result from a decrease in the percentage of lactobacilli and concomitant overgrowth by other bacterial species such as Gardnerella vaginalis. However, it is clear from genomic analysis of G. vaginalis[65, 66] and Lacobacillus iners[67] that it is the virulence potential, both genomic and metabolic, of the strain of that is crucial, as BV isolates have numerous gene set and metabolic potentials that increase their pathogenic potential over non-BV isolates. Thus, the mere presence or absence of these bacterial species is not indicative of the disease state, but more importantly it is the metabolic potential harbored in these bacterial species that defines a commensal from a pathogenic strain.
Metagenomics and computational biology approaches are providing the biological, technological, and reference genome resources that we believe will enable significant advances in our understanding of health and disease and address the issues of metabolic potentials, not bacterial strains as indicators or mediators of disease. Whenever these techniques have been applied, whether to study environmental sites or commensal communities of humans and animals, the apparent diversity of microbes has been astounding. From the few such studies that have been conducted to survey the human vaginal microbiome, the number of species present has exceeded, by orders of magnitude, the diversity determined using classic culture-based methods [36, 38, 39, 41, 68]. Not only is there is an incredible diversity of microorganisms present within individual women, but the vaginal populations among different healthy women are highly varied [36, 38, 39, 41, 69]. Our recently published studies which showed that the vaginal microbiome is heterogeneous within an individual support this observation [70], but this is just the beginning. Although two recent studies. have examined racial differences and host genotype in vaginal microbial communities [69] [25], the impact of other variables that might contribute to microbiome composition and diversity such as age, race/ethnicity, other demographics, medications, medical conditions, sexual practices, or hormone status (normal menstrual cycle, birth control pills, menopause, pregnancy) is currently unknown. There is thus a critical need for additional high-throughput genomics based approaches to characterize the vaginal microbiome.
Concluding remarks
We believe that application of high-throughput genomic technologies to survey the bacterial, fungal, phage and viral microbiomes in combination with host genetics [71] and immunologic status, particularly as they relate to urogenital infections and pre-term birth, iskeenly needed. Such a multifaceted approach is the only way we can hope to understand how a microbiome evolves and adapts in the context of complex factors, including the host, and the varying contributions of polygenic traits on the abundance and interactions of individual microbial species, related taxa, and groups of distantly related organisms. Microbial and gene predictions (annotation and construction of genomes based on microbial reference genomes from the HMP) could be used to test the hypothesis that particular microbial taxa or gene(s) are predictors of increased risk and incidence of pre-term birth. We propose that microbial taxa or genetic markers will lead to personalized (genomic) medical diagnostics capable of being point of care predictors of risk for certain clinical outcomes that profoundly impact and improve women’s health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reid G, Bruce AW. Urogenital infections in women: can probiotics help? Postgrad Med J. 2003;79:428–432. doi: 10.1136/pmj.79.934.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van der Veen F, Fransen L. Drugs for STD management in developing countries: choice, procurement, cost, and financing. Sex Transm Infect. 1998;74(Suppl 1):S166–174. [PubMed] [Google Scholar]
- 3.Oleen-Burkey MA, Hiller SL. Pregnancy complications associated with bacterial vaginosis and their estimated costs. Infect Dis Obstet Gynecol. 1995;3:149. doi: 10.1155/S1064744995000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darroch JE, Frost JJ. Women’s interest in vaginal microbicides. Fam Plann Perspect. 1999;31:16–23. [PubMed] [Google Scholar]
- 5.Fathalla MF. Vaginal microbicides: a priority need for women’s health. Curr Womens Health Rep. 2003;3:263–264. [PubMed] [Google Scholar]
- 6.Spiegel CA, et al. Anaerobic bacteria in nonspecific vaginitis. N Engl J Med. 1980;303:601–607. doi: 10.1056/NEJM198009113031102. [DOI] [PubMed] [Google Scholar]
- 7.Hill GB. The microbiology of bacterial vaginosis. Am J Obstet Gynecol. 1993;169:450–454. doi: 10.1016/0002-9378(93)90339-k. [DOI] [PubMed] [Google Scholar]
- 8.Reid G, Heinemann C. The role of the microflora in bacterial vaginosis. In: Tannock GW, editor. Medical Importance of the Normal Microflora. Kluwer, Ltd.; 1998. pp. 477–487. [Google Scholar]
- 9.McGregor JA, French JI. Bacterial vaginosis in pregnancy. Obstet Gynecol Surv. 2000;55:S1–19. doi: 10.1097/00006254-200005001-00001. [DOI] [PubMed] [Google Scholar]
- 10.Flynn CA, et al. Bacterial vaginosis in pregnancy and the risk of prematurity: a meta-analysis. J Fam Pract. 1999;48:885–892. [PubMed] [Google Scholar]
- 11.Donders GG, et al. Relationship of bacterial vaginosis and mycoplasmas to the risk of spontaneous abortion. Am J Obstet Gynecol. 2000;183:431–437. doi: 10.1067/mob.2000.105738. [DOI] [PubMed] [Google Scholar]
- 12.Fidel PL., Jr. History and new insights into host defense against vaginal candidiasis. Trends Microbiol. 2004;12:220–227. doi: 10.1016/j.tim.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Sobel JD, et al. Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am J Obstet Gynecol. 1998;178:203–211. doi: 10.1016/s0002-9378(98)80001-x. [DOI] [PubMed] [Google Scholar]
- 14.Johansson M, Lycke NY. Immunology of the human genital tract. Curr Opin Infect Dis. 2003;16:43–49. doi: 10.1097/00001432-200302000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Koumans EH, Kendrick JS. Preventing adverse sequelae of bacterial vaginosis: a public health program and research agenda. Sex Transm Dis. 2001;28:292–297. doi: 10.1097/00007435-200105000-00011. [DOI] [PubMed] [Google Scholar]
- 16.Weir E. Bacterial vaginosis: more questions than answers. Can. Med. Assoc. J. 2004;171:448. doi: 10.1503/cmaj.1041174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witkin SS, et al. Individual immunity and susceptibility to female genital tract infection. Am J Obstet Gynecol. 2000;183:252–256. doi: 10.1067/mob.2000.104258. [DOI] [PubMed] [Google Scholar]
- 18.Eschenbach DA, et al. Influence of the normal menstrual cycle on vaginal tissue, discharge, and microflora. Clin Infect Dis. 2000;30:901–907. doi: 10.1086/313818. [DOI] [PubMed] [Google Scholar]
- 19.Cauci S, et al. Prevalence of bacterial vaginosis and vaginal flora changes in peri- and postmenopausal women. J Clin Microbiol. 2002;40:2147–2152. doi: 10.1128/JCM.40.6.2147-2152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Royce RA, et al. Race/ethnicity, vaginal flora patterns, and pH during pregnancy. Sex Transm Dis. 1999;26:96–102. doi: 10.1097/00007435-199902000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Sweet RL. Gynecologic conditions and bacterial vaginosis: implications for the non-pregnant patient. Infect Dis Obstet Gynecol. 2000;8:184–190. doi: 10.1155/S1064744900000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehmet N, et al. Serum and gastric fluid levels of cytokines and nitrates in gastric diseases infected with Helicobacter pylori. New Microbiol. 2004;27:139–148. [PubMed] [Google Scholar]
- 23.Gerber S, et al. Interleukin-1beta gene polymorphism in women with vulvar vestibulitis syndrome. Eur J Obstet Gynecol Reprod Biol. 2003;107:74–77. doi: 10.1016/s0301-2115(02)00276-2. [DOI] [PubMed] [Google Scholar]
- 24.Ness RB, et al. Can known risk factors explain racial differences in the occurrence of bacterial vaginosis? J Natl Med Assoc. 2003;95:201–212. [PMC free article] [PubMed] [Google Scholar]
- 25.Ryckman KK, et al. Predicting Risk of Bacterial Vaginosis: The Role of Race, Smoking and Corticotropin-Releasing Hormone Related Genes. Mol Hum Reprod. 2009;15:131–137. doi: 10.1093/molehr/gan081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaim W, et al. New technology for diagnosis of bacterial vaginosis. Eur J Obstet Gynecol Reprod Biol. 2003;111:83–87. doi: 10.1016/s0301-2115(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 27.Cook RL, et al. Clinical, microbiological, and biochemical factors in recurrent bacterial vaginosis. J Clin Microbiol. 1992;30:870–877. doi: 10.1128/jcm.30.4.870-877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Famularo G, et al. Microecology, bacterial vaginosis and probiotics: perspectives for bacteriotherapy. Med Hypotheses. 2001;56:421–430. doi: 10.1054/mehy.2000.1195. [DOI] [PubMed] [Google Scholar]
- 29.Jones BM, et al. The determination of amines in the vaginal secretions of women in health and disease. Int J STD AIDS. 1994;5:52–55. doi: 10.1177/095646249400500112. [DOI] [PubMed] [Google Scholar]
- 30.O’Dowd TC, et al. Evaluation of a rapid diagnostic test for bacterial vaginosis. Br J Obstet Gynaecol. 1996;103:366–370. doi: 10.1111/j.1471-0528.1996.tb09743.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues AG, et al. Is the lack of concurrence of bacterial vaginosis and vaginal candidosis explained by the presence of bacterial amines? Am J Obstet Gynecol. 1999;181:367–370. doi: 10.1016/s0002-9378(99)70563-6. [DOI] [PubMed] [Google Scholar]
- 32.Wolrath H, et al. Trimethylamine content in vaginal secretion and its relation to bacterial vaginosis. Apmis. 2002;110:819–824. doi: 10.1034/j.1600-0463.2002.1101108.x. [DOI] [PubMed] [Google Scholar]
- 33.Wolrath H, et al. Analysis of bacterial vaginosis-related amines in vaginal fluid by gas chromatography and mass spectrometry. J Clin Microbiol. 2001;39:4026–4031. doi: 10.1128/JCM.39.11.4026-4031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolrath H, et al. Trimethylamine and trimethylamine oxide levels in normal women and women with bacterial vaginosis reflect a local metabolism in vaginal secretion as compared to urine. Apmis. 2005;113:513–516. doi: 10.1111/j.1600-0463.2005.apm_175.x. [DOI] [PubMed] [Google Scholar]
- 35.Fredricks DN, et al. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 36.Hyman RW, et al. Microbes on the human vaginal epithelium. Proc Natl Acad Sci U S A. 2005;102:7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota T, et al. Detection and identification of amines in bacterial vaginosis. J Obstet Gynaecol. 1995;21:51–55. doi: 10.1111/j.1447-0756.1995.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 38.Verhelst R, et al. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 2004;4:16–26. doi: 10.1186/1471-2180-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verstraelen H, et al. Culture-independent analysis of vaginal microflora: the unrecognized association of Atopobium vaginae with bacterial vaginosis. Am J Obstet Gynecol. 2004;191:1130–1132. doi: 10.1016/j.ajog.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Witkin SS, et al. Bacterial flora of the female genital tract: function and immune regulation. Best Pract. Res. Clin. Obstet. Gynaecol. 2007 doi: 10.1016/j.bpobgyn.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, et al. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 42.Lawn JE, et al. Estimating the causes of 4 million neonatal deaths in the year 2000. International Journal of Epidemiology. 2006;35:706–718. doi: 10.1093/ije/dyl043. [DOI] [PubMed] [Google Scholar]
- 43.Han YW, et al. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han YW, et al. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J Clin Microbiol. 2006;44:1475–1483. doi: 10.1128/JCM.44.4.1475-1483.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbs RS, et al. Quantitative Bacteriology of Amniotic-Fluid from Women with Clinical Intra-Amniotic Infection at Term. Journal of Infectious Diseases. 1982;145:1–8. doi: 10.1093/infdis/145.1.1. [DOI] [PubMed] [Google Scholar]
- 46.DiGiulio DB, et al. Microbial Prevalence, Diversity and Abundance in Amniotic Fluid During Preterm Labor: A Molecular and Culture-Based Investigation. Plos One. 2008;3 doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watts DH, et al. The Association of Occult Amniotic-Fluid Infection with Gestational-Age and Neonatal Outcome among Women in Preterm Labor. Obstetrics and Gynecology. 1992;79:351–357. doi: 10.1097/00006250-199203000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Leitich H, Kiss H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol. 2007;21:375–390. doi: 10.1016/j.bpobgyn.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Muglia LJ, Katz M. The enigma of spontaneous preterm birth. N Engl J Med. 362:529–535. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 50.Donders GG, et al. Association between abnormal vaginal flora and cervical length as risk factors for preterm birth. Ultrasound Obstet Gynecol. 2010 doi: 10.1002/uog.7568. [DOI] [PubMed] [Google Scholar]
- 51.Menard JP, et al. High vaginal concentrations of Atopobium vaginae and Gardnerella vaginalis in women undergoing preterm labor. Obstet Gynecol. 115:134–140. doi: 10.1097/AOG.0b013e3181c391d7. [DOI] [PubMed] [Google Scholar]
- 52.Srinivasan U, et al. Vaginal and oral microbes, host genotype and preterm birth. Med Hypotheses. 2009;73:963–975. doi: 10.1016/j.mehy.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donders GG, et al. Association between abnormal vaginal flora and cervical length as risk factors for preterm birth. Ultrasound Obstet Gynecol. 2010 doi: 10.1002/uog.7568. [DOI] [PubMed] [Google Scholar]
- 54.Andrews WW, et al. Randomized clinical trial of metronidazole plus erythromycin to prevent spontaneous preterm delivery in fetal fibronectin-positive women. Obstet Gynecol. 2003;101:847–855. doi: 10.1016/s0029-7844(03)00172-8. [DOI] [PubMed] [Google Scholar]
- 55.Committee on Metagenomics: Challenges and Functional Applications, N.R.C. THE NEW SCIENCE OF METAGENOMICS, Revealing the Secrets of Our Microbial Planet. The National Acadamies Press; 2007. [PubMed] [Google Scholar]
- 56.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ochman H, et al. Evolutionary Relationships of Wild Hominids Recapitulated by Gut Microbial Communities. Plos Biology. 2010;8 doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zella GC, et al. Distinct microbiome in pouchitis compared to healthy pouches in ulcerative colitis and familial adenomatous polyposis. Inflamm Bowel Dis. doi: 10.1002/ibd.21460. [DOI] [PubMed] [Google Scholar]
- 60.Genc MR, et al. Relationship between a toll-like receptor-4 gene polymorphism, bacterial vaginosis-related flora and vaginal cytokine responses in pregnant women. Eur J Obstet Gynecol Reprod Biol. 2004;116:152–156. doi: 10.1016/j.ejogrb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 61.Martin HL, et al. Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis. 1999;180:1863–1868. doi: 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 62.Redondo-Lopez V, et al. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev Infect Dis. 1990;12:856–872. doi: 10.1093/clinids/12.5.856. [DOI] [PubMed] [Google Scholar]
- 63.Schwebke JR. Role of vaginal flora as a barrier to HIV acquisition. Curr Infect Dis Rep. 2001;3:152–155. doi: 10.1007/s11908-996-0040-6. [DOI] [PubMed] [Google Scholar]
- 64.Taylor-Robinson D. The future of bacterial vaginosis-related research. Int J Gynaecol Obstet. 1999;67(Suppl 1):S35–38. doi: 10.1016/s0020-7292(99)00137-x. [DOI] [PubMed] [Google Scholar]
- 65.Harwich MD, Jr., et al. Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genomics. 11:375. doi: 10.1186/1471-2164-11-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeoman CJ, et al. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. Plos One. 5:e12411. doi: 10.1371/journal.pone.0012411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macklaim JM, et al. At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc Natl Acad Sci U S A. 108(Suppl 1):4688–4695. doi: 10.1073/pnas.1000086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hummelen R, et al. Deep Sequencing of the Vaginal Microbiota of Women with HIV. Plos One. 2010;5 doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou X, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. The ISME Journal. 2007;1:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 70.Kim TK, et al. Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol. 2009;47:1181–1189. doi: 10.1128/JCM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adams KM, Eschenbach DA. The genetic contribution towards preterm delivery. Semin Fetal Neonatal Med. 2004;9:445–452. doi: 10.1016/j.siny.2004.04.001. [DOI] [PubMed] [Google Scholar]