SUMMARY
Recent studies have documented genome-wide binding patterns of transcriptional regulators and their associated epigenetic marks in hematopoietic cell lineages. In order to determine how epigenetic marks are established and maintained during developmental progression, we have generated long-term cultures of hematopoietic progenitors by enforcing the expression of the E-protein antagonist Id2. Hematopoietic progenitors that express Id2 are multipotent and readily differentiate upon withdrawal of Id2 expression into committed B lineage cells, thus indicating a causative role for E2A (Tcf3) in promoting the B cell fate. Genome-wide analyses revealed that a substantial fraction of lymphoid and myeloid enhancers are pre-marked by the poised or active enhancer mark H3K4Me1 in multipotent progenitors. Thus, in hematopoietic progenitors, multilineage priming of enhancer elements precedes commitment to the lymphoid or myeloid cell lineages.
INTRODUCTION
In the adult bone marrow, long-term hematopoietic stem cells (LT-HSCs) have the ability to both self-renew and reconstitute the entire immune system for the life of the organism (Spangrude et al., 1988). LT-HSCs have the ability to differentiate into short-term HSCs (ST-HSCs), which give rise to multipotent progenitors (MPPs) and then differentiate into the lymphoid-primed multipotent progenitors (LMPPs) (Adolfsson et al., 2005). LMPPs differentiate into common lymphoid progenitors (CLPs) or granulocyte-macrophage progenitors (GMPs) which give rise to either a lymphoid or myeloid restricted pathway, respectively (Inlay et al., 2009; Kondo et al., 1997).
During the past decade transcription factors have been identified that play critical roles in early B cell development. These include the E2A proteins, EBF1, FOXO1 and PAX5 (Bain et al., 1994; Zhuang et al., 1994; (Urbánek et al., 1994) Lin et al. 1995; Dengler et al., 2008; Amin et al., 2008(Treiber et al., 2010). The E2A proteins are members of the E-protein transcription factor family that also include HEB, E2-2 and E2A. The E-protein transcription factors are characterized by the presence of a helix-loop-helix (HLH) dimerization domain and a DNA binding region. The E2A locus encodes for two proteins, E12 and E47, which arise from differential splicing of exons that encode for their DNA binding and dimerization domains. E2A proteins were originally identified based on their abilities to bind the immunoglobulin kappa chain enhancer, and have since been implicated at multiple stages of lymphoid and B cell development (Bain et al., 1994; Beck et al., 2009; Dias et al., 2008; Murre et al., 1989; Semerad et al., 2009; Zhuang et al., 2004). E-proteins are antagonized by the Id proteins, which contain an HLH motif but lack a DNA binding region (Benezra et al., 1990). In mice deficient for E2A, B-cell development is blocked at the CLP cell stage (Bain et al., 1997; Bain et al., 1994; Zhuang et al., 1994). A similar block in B cell development is observed in EBF-, FOXO1- and PAX5-deficient mice (Dengler et al., 2008; Lin and Grosschedl, 1995; Nutt et al., 1997).
When cultured in B-cell supportive conditions in vitro, E2A-deficient bone marrow cells self-renew indefinitely without losing multipotent differentiation potential (Ikawa et al., 2004). B-cell differentiation in E2A deficient cells can be rescued by forced EBF expression, suggesting that the E2A proteins act upstream of EBF in B-cell development (Hagman and Lukin, 2006; Lazorchak et al., 2005; Seet et al., 2004). Recent studies have directly linked these and other transcriptional regulators into a common framework (Ghisletti et al. 2010; Heinz et al., 2010; Treiber et al., 2010; Lin et al., 2010; Natoli, 2010). E2A initiates a program of B-lineage specific gene expression by inducing the expression of Ebf1, Bcl11a, Irf4, Irf8 and Foxo1 (Lin et al., 2010). E2A, EBF1, FOXO1, IRF4 and IRF8 act in concert to activate the expression of the Pax5 locus (Decker et al., 2009; Lin et al., 2010). E2A, EBF, Foxo1 as well as Pax5 then act to induce the expression of a large subset of genes including signaling components, survival factors and regulators that modulate cell cycle progression (Lin et al., 2010; McManus et al. 2011).
Here, we have generated a cytokine-dependent cell line, named Id2-HPC, which allowed for the expansion of a progenitor cell line that maintains multipotency both in vivo and in vitro. We then harnessed the differentiation potential of these cells to monitor how enhancer repertoires are established at the very earliest stages of B and myeloid cell development. We found that a subset of poised enhancer elements are marked at the uncommitted progenitor cell stage. However, H3K4me1 marks at a subset of enhancers were elevated during developmental progression, resulting in evolving active enhancer repertoires that we propose orchestrate the myeloid and B cell fates.
RESULTS
Self-Renewal Activity in Hematopoietic Progenitors
Previous experiments demonstrated that multipotent progenitor cells can self-renew in vitro in the absence of E2A and retain pluripotency in vivo (Ikawa et al., 2004). To determine if overexpression of Id proteins would effectively inhibit E2A and thus allow in vitro expansion of multipotent progenitor cells, we created lentiviral vectors utilizing a Tet-Off promoter that expresses human ID2 (hId2) and GFP in the absence of doxycycline (Figure S1A).
Lineage depleted bone marrow from CD45.1 congenic C57/Bl6 mice was infected with lentivirus carrying Id2 (TetOff_hId2) or empty vector control (TetOff_Empty). The transduced cells were cultured in the presence of cytokines IL-3, IL-6, WEHI supernatant and SCF. Initially, both the control (C-HPCs) and hId2 infected cells (Id2-HPCs), rapidly expanded. However, upon culture for four weeks, C-HPCs had ceased growing while the Id2-HPCs continued to divide (data not shown). Id2-HPCs were then cultured in IL-7, Flt3L, and SCF on sub-confluent S17 feeder cells. In these conditions, the cells continued to expand over time. The Id2-HPC cells overexpressed hId2 mRNA, which could be robustly turned off with the addition of doxycycline to the culture media (Figure 1A).
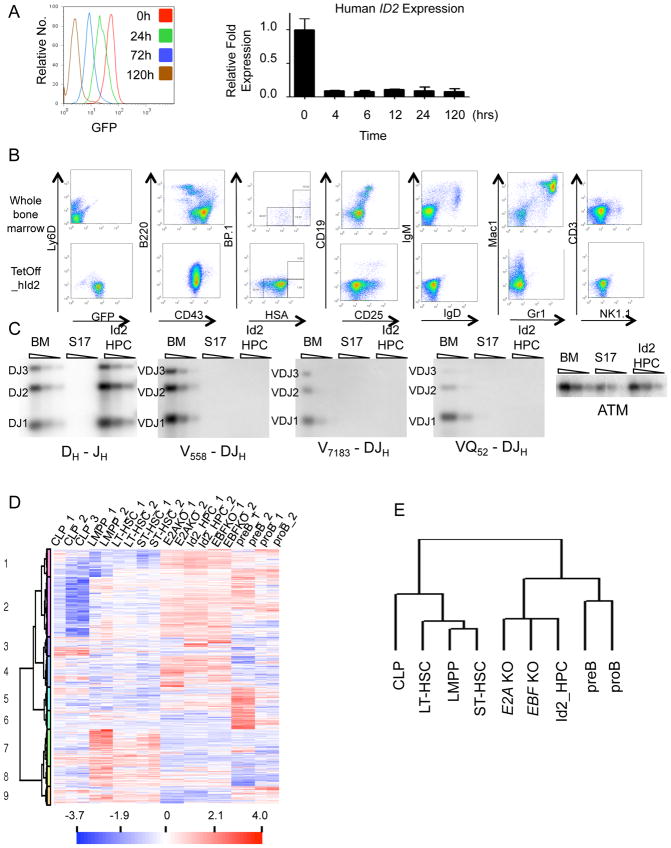
Figure 1. Establishment of a Long-Term Culture of Multipotent Hematopoietic Progenitors.
(A) Rapid and robust downregulation of GFP and human ID2 after doxycycline addition in vitro. FACS plots of GFP (left panel) at 0, 24, 72 and 120 hour post-doxycycline addition are shown. Real-time PCR analysis of human ID2 expression at 0, 4, 6, 12, 24 and 120-hour post-doxycycline addition.
(B) Phenotypic analysis of Id2-HPC cells. TetOff_hId2 infected cells were cultured on S17 feeder cells with IL-7, SCF and Flt3L. Cells were analyzed by FACS for the expression of B220, Ly6D, CD43, CD25, CD19, Mac1, Gr1, CD3 and NK1.1 after 3 months in culture. The upper panel shows staining from wild type bone marrow as controls and the lower panel shows staining from expanded Id2-HPCs in the absence of doxycycline.
(C) IgH gene rearrangement analysis of Id2-HPCs. DNA was isolated from wild type bone marrow, S17 feeder cells, and TetOff_hId2 cells and analyzed by Southern Blot for the presence of IgH DJ and V-DJ rearrangements. ATM was used as a loading control.
(D) Microarray analysis of LT-HSCs, ST-HSCs, LMPPs, CLPs, pre-B cells, pro-B cells, Tcf3−/− cell line, Ebf1−/− cell line, and Id2-HPC cell line. Duplicate genes have been removed. The color scale is shown below the diagram.
(E) Vertical clustering of cell types from microarray experiment in Figure 1D.
Id2-HPCs did not express markers characteristic for T cell and NK cell lineages, and expressed intermediate amounts of B220, high abundance of CD43, and were Ly6D, IgM and IgD negative (Figure 1B). A small percentage of the cells (1–5%) expressed CD11b, CD19 or CD25, but the majority of cells remained lineage-negative even after being cultured in vitro for relatively long periods of time. Id2-HPCs carried DHJH joints, but lacked VH-DHJH rearrangements, suggesting that they represent cells arrested at the CLP or pre-pro-B cell stage (Busslinger, 2004) (Figure 1C).
To further characterize Id2-HPCs, we performed microarray analysis (GSE30859) using RNA derived from freshly isolated LT-HSCs, ST-HSCs, CLPs, LMPPs, pre-B cells, pro-B cells, as well as cultured E2A deficient cells, EBF deficient cells and Id2-HPCs. Nine expression patterns were identified from the 11,367 changed genes (Figure 1D). Genes encoding for proteins required for B-cell development such as Foxo1, Pou2af1, IgL, Ly6d, and Vpreb1 showed equivalent expression in the Id2-HPCs relative to HSCs and LMPPs, but lower expression as compared to pro- and pre-B cells. In contrast, genes that are expressed in pluripotent progenitors such as Cd34, kit, Tie1, Slamf1, and Tal1 were upregulated in HSCs and CLPs when compared to Id2-HPCs. Vertical clustering of expression patterns showed that Id2-HPCs resemble the transcription signatures of Tcf3(E2A)−/− deficient and Ebf1−/− cells (Figure 1E). Taken together, these data indicate that forced Id2-expression in hematopoietic progenitors promotes the self-renewal of cells with a similar phenotype as observed in Tfc3−/− and Ebf1−/− pre-pro-B cells.
Id2-HPCs are Multipotent
To determine if Id2-HPCs maintain pluripotency and repopulating ability in vivo, cells were injected into irradiated CD45.2 recipients in a competitive repopulating assay. Because in the absence of E2A, B cells do not develop beyond the pre-pro-B cell stage and alternate lineage development is perturbed, we administered doxycycline via food pellets to recipient animals to turn off Id2 expression and thus allow restoration of E-protein activity (Bain et al., 1994). The mice received doxycycline food pellets 24 hours before injection and throughout the remainder of the study. Recipient mice were lethally irradiated 24 hours before tail vein injection of Id2-HPCs (CD45.1) mixed (1:1) with freshly harvested CD45.2 bone marrow cells. At 6 week post-transplantation, Id2-HPCs had successfully reconstituted the bone marrow, thymus, and spleen of irradiated recipients to varying degrees, albeit at reduced ability compared to wild type bone marrow (Figure 2 and Figure S1). The large majority of the CD45.1 cells were GFP negative, showing the robust sensitivity of the tetracycline transactivator to doxycycline in vivo (Figure 2).
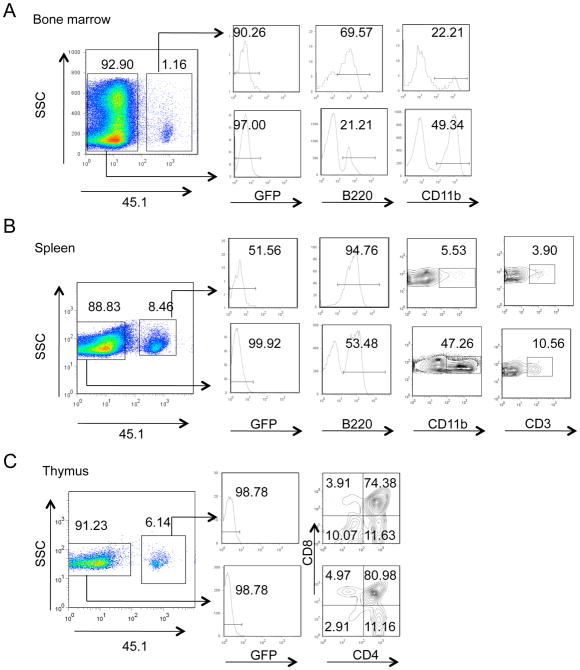
Figure 2. Id2-HPCs Reconstitute Multiple Lineages in vivo and in vitro.
Id2-HPCs can reconstitute multiple lineages in irradiated recipients. A competitive reconstitution assay was used to assess the ability of Id2-HPCs to reconstitute irradiated recipients. One million Id2-HPCs and one million freshly isolated wild type bone marrow cells were injected into lethally irradiated recipients receiving doxycycline feed. Reconstitution was analyzed 6 weeks later. The data is representative of two experiments.
(A) Id2-HPCs successfully reconstitute the bone marrow of recipients. Bone marrow cells were analyzed by FACS analysis for expression of GFP, B220 and CD11b.
(B) Id2-HPCs successfully reconstitute the spleen of recipients. Bone marrow cells were analyzed by FACS analysis for expression of GFP, B220, CD11b and CD3.
(C) Id2-HPCs successfully reconstitute the thymus of recipients. Bone marrow cells were analyzed by FACS analysis for expression of GFP, CD4 and CD8.
In the bone marrow, Id2-HPCs reconstituted the B and myeloid compartments successfully (Figure 2A). There were increased numbers of CD45.1 B220+ cells and decreased numbers of CD45.1 CD11b+ cells relative to their CD45.2 counterparts, suggesting a propensity for Id2-HPCs to commit to the B cell lineage (Figure 2A). The Id2-HPCs also successfully migrated to the peripheral lymphoid organs (Figure 2B and 2C). IgM expression was readily detectable on approximately 20% of CD45.1 splenocytes, demonstrating that Id2-HPCs have the ability to successfully progress beyond the pre-pro-B cell stage (data not shown). Id2-HPCs also reconstituted the T-cell and myeloid compartments in the spleen (Figure 2B). In the thymus, Id2-HPCs reconstituted the double negative, double positive, and single positive populations (Figure 2C). Thus, Id2-HPCs can be grown long-term in culture and have the ability to reconstitute multiple hematopoietic cell lineages in vivo.
In vitro Differentiation of Id2-HPCs into Myeloid and B Cell Lineages
The transplantation experiments revealed that Id2-HPCs possessed multilineage potential. To exclude the possibility that the multipotency of Id2-HPCs was caused by a heterogeneous populations of lineage restricted cells, single cell cultures were initiated to determine the clonal properties of Id2-HPCs. Individual cells from the highest 4%, lowest 4%, or intermediate 10% of GFP mean fluorescence were sorted onto pre-seeded S17 cells in 96-well plates and cultured as described above. Clones from each of the conditions (Low GFP= L, Intermediate GFP=I, High GFP= H) were expanded, and one clone from each group was chosen for further analysis (Figure S2A). To examine their myeloid lineage potential three Id2-HPC monoclonal cell lines (L1, I2 and H1) were cultured in the presence of IL3, Flt3-L, GM-CSF and M-CSF. Within 48-hours after initiation of the myeloid culture, Id2-HPCs expressed significant levels of CD11b (Figure 3A, Figure S2B and S2C). Interestingly, the mean fluorescence intensity of B220 was increased in the myeloid cultures in all three clones, similar to that seen in other studies (Xie et al., 2004). Additionally, Id2 expression was higher in the myeloid cultures than in progenitor cells prior to differentiating conditions (Figure 3D). Id2-HPCs possess similar myeloid potential as the Tcf3−/− cell line (Figure S1B). These data indicate that cells expressing high amounts of Id2 are favored to commit to the myeloid cell lineage.
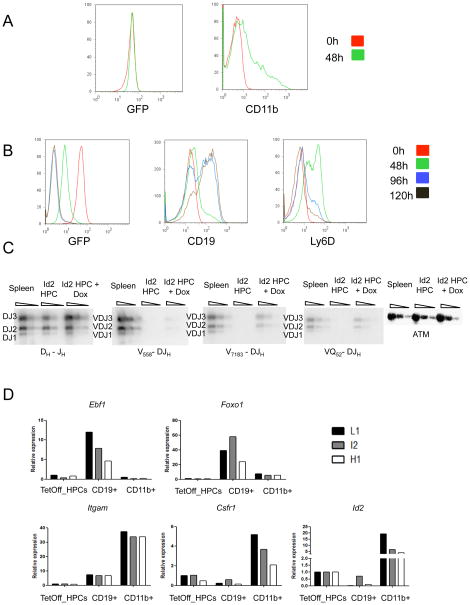
Figure 3. In vitro Differentiation Potential of Id2-HPCs.
(A) In vitro differentiation of Id2-HPCs into myeloid cells. Cells were cultured in IL-3, Flt3L, GMCSF and MCSF in the presence or absence of doxycycline for 2 days in vitro and analyzed by FACS at day 0 and day 2 for CD11b.
(B) In vitro differentiation of Id2-HPCs to pro-B cells. Cells were cultured in IL-7, SCF in the presence or absence of doxycycline for 6 days in vitro and analyzed at day 0, day 2, day 4 and day 5 for GFP, CD19, and Ly6D expression.
(C) IgH gene rearrangement in differentiated Id2-HPCs. DNA was isolated from wild type bone marrow, day 0 Id2-HPCs and day 8 plus doxycycline Id2-HPCs and analyzed by Southern blotting for IgH DJ and V-DJ rearrangements. ATM was used as a loading control.
(D) Gene expression analysis of lineage specific transcripts in undifferentiated Id2-HPCs, and CD19+ or CD11b+ differentiated Id2-HPCs. RNA was isolated from monoclonal Id2-HPCs at the undifferentiated state or the CD19+ or CD11b+ differentiated state, and transcript levels were analyzed by real-time PCR analysis.
To promote differentiation of Id2-HPCs into the B cell lineage, cells were cultured in the presence of S17 cells, IL-7 and SCF either with or without doxycycline. Id2-HPCs that were cultured in the absence of doxycycline continued to divide but maintained the pre-pro-B cell phenotype. Id2-HPCs that were grown in the presence of doxycycline rapidly downregulated GFP and showed a transient upregulation of the recently defined marker for B-lineage primed progenitors LY6D (Inlay et al., 2009; Mansson et al., 2008). This was followed by a robust increase in the levels of CD19 expression (Figure 3B, and Figure S2B and S2C). Differentiation into committed B-lineage progenitors was also supported by the presence of substantial levels of VH-DHJH joints (Figure 3C). Quantitative-PCR analysis confirmed upregulation of the myeloid specific genes, including Itgam and Csfr1 upon culture conditions that favor myeloid differentiation, and the upregulation of B-cell specific genes, including Ebf1 and Foxo1, in conditions that favor B-cell differentiation (Figure 3D). We note, however, that cultured Id2-HPCs do not retain their B cell developmental potential indefinitely. Taken together, these observations indicate that forced expression of Id2 in hematopoietic progenitors permits the establishment of a long-term, but not indefinite, culture of multipotent progenitors.
Gene Expression Signatures in B and Myeloid Cells Derived from Multipotent Progenitors
To compare the expression signatures from differentiated B- and myeloid-cells derived from hematopoietic progenitors that express Id2, Id2-HPCs were differentiated either into CD19+ B cells or CD11b+ myeloid cells. RNA was isolated from the cultures and analyzed by microarray gene expression analysis. We hierarchically clustered the expression of genes with a two-fold or more change in absolute expression levels between any two groups (Figure 4A). Several patterns emerged: genes whose transcripts became elevated in differentiated B cells, genes whose transcripts declined in differentiated myeloid cells, genes whose transcript levels were elevated in undifferentiated cells and genes that were activated upon myeloid differentiation (Figure 4A).
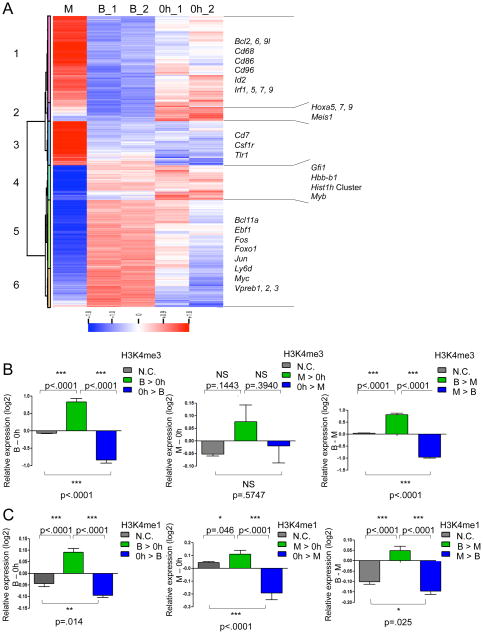
Figure 4. Establishment of Promoter and Enhancer Marks in Differentiating B and Myeloid Cells.
(A) Multipotent progenitors carrying Id2 were differentiated into either CD19+ B cells (upon exposure to doxycycline) or CD11b+ myeloid cells. Gene expression patterns of multipotent progenitors and differentiated progeny were analyzed by microarray.
(B) Changes in gene expression levels correlate to changes in proximal H3K4me3 mapped reads between HPCs, B and myeloid cells. Changes in H3K4me3 were directly compared to gene expression levels. Expression from genes with two-fold more mapped reads (tags) in one lineage versus another lineage are indicated in green (B > 0h or M > 0h) or blue (0h > B or 0h > M), and peaks with less than a two-fold change in mapped reads (tags) are shown in grey (N.C. refers to No Change). 0h refers multipotent Id2-HPC cells. M refers to myeloid cells as characterized by CD11b expression that were differentiated from Id2-HPCs. B refers to B cells as characterized by CD19 expression that were differentiated from Id2-HPCs. P-values are indicated. N.S. refers to not significant.
(C) Changes in gene expression levels correlate to changes in distal H3K4me1 mapped reads between HPCs, B cells and myeloid cells.
As expected, Ebf1, Foxo1, Bcl11a, and Vpreb transcript levels transcript levels were substantially elevated upon differentiation into the B cell lineage (Figure 4A). Interestingly, the pattern of Ly6d expression also falls into this cluster, supporting previous data implicating Ly6d as a marker that specifies the B cell fate (Figure 4A) (Inlay et al., 2009; Mansson et al., 2010). Myeloid specific genes including Cd68 and Csf1r were readily activated in culture conditions that favor myeloid development (Figure 4A; Cluster 1 and 3). A group of genes was also identified whose transcript levels declined upon differentiating into either the B or the myeloid cell lineage (Clusters 2 and 4). Interestingly, cluster 2 includes genes that are characteristic of the hematopoietic stem cell signature, including Meis1 as well as several members of the Hoxa family (Figure 4A). Collectively these data indicate that Id2-HPCs in differentiating conditions activate lineage-specific programs of gene expression and repress transcription of genes associated with the HSC fate as well as alternate differentiated cell lineages.
Establishment of Promoter and Enhancer Marks in Differentiating B and Myeloid Cells
The gene expression patterns uncovered by microarray analysis in differentiating multipotent progenitors (Id2-HPCs) indicate genome-wide changes in transcriptional activity. We next asked the question when and how these changes are initiated and established. Recent studies have demonstrated a tight correlation between transcriptionally active promoters and H3K4 trimethylation, whereas H3K4 monomethylation has been associated with enhancer activity (Pokholok et al., 2005; Heintzman et al., 2007). To determine whether the changes in gene expression patterns upon differentiation correlate with the presence of H3K4me3 as well as H3K4me1, cell lysates were derived from Id2-HPCs and differentiated into CD11b+ and CD19+ cells. Lysates were immunoprecipitated with antibodies directed against H3K4me1 and H3K4me3 and analyzed by ChIP-Sequencing (Barski et al., 2007). Over 15,000 regions were identified that showed H3K4me3 in all three cell types (Figure S3A). The number of H3K4me1 islands varied between the cell types (Figure S3A). As expected, the majority of the H3K4me3 peaks were promoter proximal, while most of the H3K4me1 peaks were promoter distal (Figure S3B). ChIP-Seq analyses for H3K4me1 were performed in duplicate using biological replicates. We note that the patterns of H3K4me1 were very similar between the duplicates (Figures S3, S4 and S6).
To determine how the presence of epigenetic marks relates to lineage specific programs of gene expression, changes in the abundance of H3K4me3 and H3K4me1 were directly compared to relative levels of gene expression (Figure 4B). As a threshold we used a two-fold change in the number of normalized reads mapped to promoter regions associated with one lineage versus another. We observed a tight correlation between changes in H3K4me3 and gene expression levels upon comparing multipotent progenitors (Id2-HPCs) to differentiated CD19+ B cells (Figure 4B, left panel). Surprisingly, there was no substantial distinction between Id2-HPCs and myeloid cells (Figure 4B; middle panel). Changes in the levels of H3K4me1 in regions adjacent to genes correlated very well with the dynamics of gene expression abundance for both myeloid and B-lineage cells when compared to multipotent progenitors (Figure 4C; left and middle panel). Thus, during the transition from the multipotent progenitor to the differentiated cell stage, changes in H3K4 monomethylation are tightly associated with the activation of either a B- or myeloid-specific program of gene expression.
Islands of H3K4me1 Mark Lineage-Specific Enhancers in Multipotent Progenitors
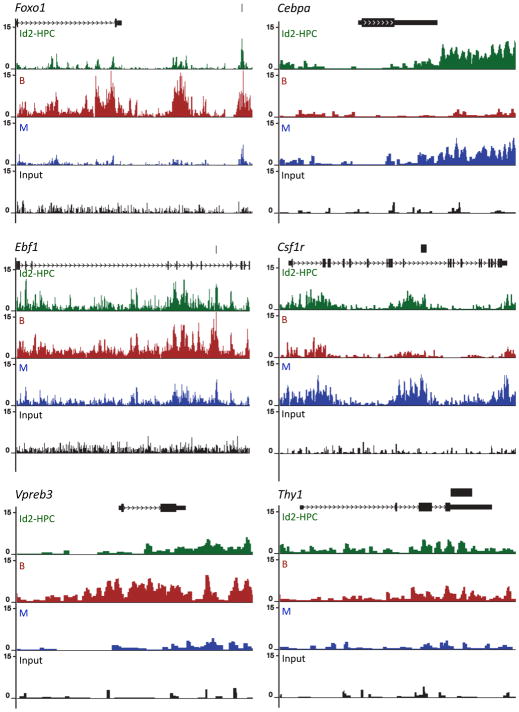
To explore the possibility that enhancer regions are primed in Id2-HPCs prior to differentiation, we examined H3K4me1 enrichment for a subset of cell-type specific genes including the B-lineage specific genes Foxo1, Ebf1 and Vpreb3. We also analyzed myeloid specific loci such as Csfr1 and Cepba, and we examined the patterns of H3K4me1 in genes that are repressed upon specification into either the B or myeloid cell lineages, such as Thy1 (Figure 5). We note that the patterns were normalized to a total number of ten million tags. H3K4me1 islands are already present in Id2-HPCs prior to differentiation, but are markedly elevated in CD19 positive cells at the Foxo1, Ebf1 and Vpreb3 loci, whose expression is activated upon commitment to the B-lineage but not upon commitment to the myeloid lineage (Figure 5). Similarly, the relative abundance of H3K4 monomethylation is elevated in loci Cebpa and Csfr1, whose expression increases in myeloid cells but declines upon differentiating into the B-lineage (Figure 5). Correspondingly, abundance of H3K4 monomethylation at the T cell specific Thy1 locus decreases upon differentiation into either the B- or myeloid cell lineage (Figure 5). Potential enhancer elements in the Foxo1, Ebf1, Thy1 and Csfr1 loci were cloned by PCR and inserted upstream of a basal promoter in a modified pGL3 vector and transfected, with control renilla luciferase, into the pro-B cell line 22D6, the double positive T cell line 166, and T-cell line A12. Luciferase activity was quantitated after 24 hours. As expected, regions in the Foxo1 and Ebf1 loci that gained H3K4 monomethylation upon differentiation to the B-cell state had greater luciferase activity in the B cell line as opposed to the T cell lines (Figure S5). This is consistent with previous observations (Lin et al., 2010) In contrast, sites in the Thy1 locus that lose H3K4 monomethylation upon differentiation into B cells showed slightly higher levels of enhancer activity in the T cell lines (Figure S5). Taken together, these findings indicate that in multipotent progenitors (Id2-HPCs), H3K4 monomethylation patterns mark a wide spectrum of enhancers associated with lineage restricted genes that become further activated or repressed in a lineage restricted manner.
Figure 5. The Enhancer Repertoires of Multipotent Progenitors are Primed for Activation.
Multipotent progenitors (Id2-HPC) and differentiated progeny (B and M) were analyzed for active enhancer repertoires. Distributions of H3K4me1 in multipotent progenitors as well as differentiated progeny are shown across Foxo1 (chr3: 52,356,303-52,540,000), Ebf1 (chr11: 44,460,539-44,849,952), Vpreb3 (chr10: 75,390,000–75,394,700), Cebpa (chr7: 34,825,439-34,833,310), Csf1r (chr18: 61,229,941-61,257,506), and Thy1 (chr9: 43,793,835-43,800,231) genomic regions. Patterns were normalized against ten million tags. The transcript on top of each graph is shown as in the UCSC Browser. Numbers indicate the number of normalized mapped reads (tags) observed. Black bars on top of transcripts indicate regions cloned for luciferase studies.
Establishment of Lineage-Specific Enhancer Repertoires
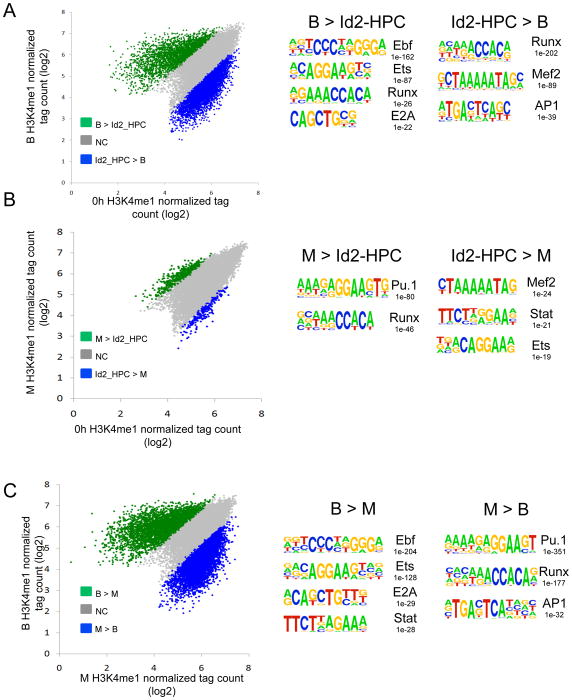
The data described above raise the question as to whether changes in the levels of H3K4 monomethylation in the B versus myeloid cell lineages correlate with the presence of distinct cis-regulatory codes. To address this question, we examined H3K4me1 associated sites for enriched DNA sequences. First, we created scatter plots comparing the abundance of H3K4me1 in multipotent (Id2-HPCs) and differentiated cells (Figure 6). Next H3K4me1 regions specifically associated with multipotent progenitors and differentiated cells were examined for the presence of enriched cis-regulatory elements, using a de novo motif finder algorithm, HOMER (Heinz et al. 2010). The ranking of such associated sequences was based on their enrichment as compared to background genomic DNA sequences.
Figure 6. Establishment of Lineage-Specific Enhancer Repertoires.
(A) Analysis of changes in H3K4me1 mapped reads (tags) between hematopoietic multipotent progenitors (Id2-HPCs) and CD19+ B cells. B cells were derived from Id2-HPCs upon in vitro differentiation. Scatter plot displays the number of H3K4me1 mapped reads across H3K4me1 marked regions in Id2-HPCs versus CD19+ B cells. Distributions of H3K4me1 mapped reads showing a two-fold or greater increase in mapped reads in Id2-HPCs versus differentiated CD19+ B-cells are shown in blue, and peaks with a two-fold or greater increase in mapped reads in CD19+ B cells are indicated in green. Numbers reflect P-values. Cis-regulatory elements associated with H3K4me1 regions were identified by computational analysis (HOMER).
(B) Analysis of changes in H3K4me1 mapped reads (tags) between hematopoietic multipotent progenitors (Id2-HPC) and myeloid cells. Myeloid cells were derived from Id2-HPCs upon in vitro differentiation. Scatter plot displays the number of H3K4me1 mapped reads across H3K4me1 marked regions in Id2-HPCs versus CD11b+ myeloid cells. Distributions of H3K4me1 mapped reads showing a two-fold or greater increase in mapped reads in Id2-HPCs versus differentiated CD11b+ myeloid cells are shown in blue, and peaks with a two-fold or greater increase in mapped reads in CD11b+ myeloid cells are indicated in green. Numbers reflect P-values. Cis-regulatory elements associated with H3K4me1 regions were identified by computational analysis (HOMER).
(C) Analysis of changes in H3K4me1 mapped reads (tags) between myeloid and CD19+ B cells. Myeloid and B cells were derived from Id2-HPCs upon in vitro differentiation. Scatter plot displays the number of H3K4me1 mapped reads across H3K4me1 marked regions in CD11b+ myeloid versus CD19+ B cells. Distributions of H3K4me1 mapped reads showing a two-fold or greater increase in mapped reads in CD11b+ myeloid cells versus differentiated CD19+ B-cells are shown in blue, and peaks with a two-fold or greater increase in mapped reads in CD19+ B cells are indicated in green. Numbers reflect P-values. Cis-regulatory elements associated with H3K4me1 regions were identified by computational analysis (HOMER).
Ebf, Ets, Runx and E2A were ranked among the top-scoring motifs associated with putative enhancers in cells with at least a two-fold increase in H3K4me1 mapped reads in B cells as compared to multipotent progenitors (Id2-HPCs) (Figure 6A). In multipotent progenitors (Id2-HPCs) compared to B cells, H3K4me1 marked regions were enriched for Runx, Mef2 and AP-1 consensus binding sites (Figure 6A). When comparing myeloid cells to Id2-HPCs and B cells, H3K4me1 peaks associated the myeloid lineage were significantly enriched for the Pu.1, Runx and AP-1 consensus binding sites (Figure 6B and 6C). Alternatively, Mef2, Stat and Ets consensus binding sites were primarily enriched in Id2-HPCs compared to myeloid cells, while B cell specific H3K4me1 sites were strongly enriched for Ebf, Ets and E2A consensus binding sites (Figure 6C). Thus, lineage specific enhancer repertoires, as characterized by H3K4me1, are established upon developing from multipotent progenitors into committed differentiated B and myeloid cells.
Evolving Global Enhancer Repertoires in Developing B cells
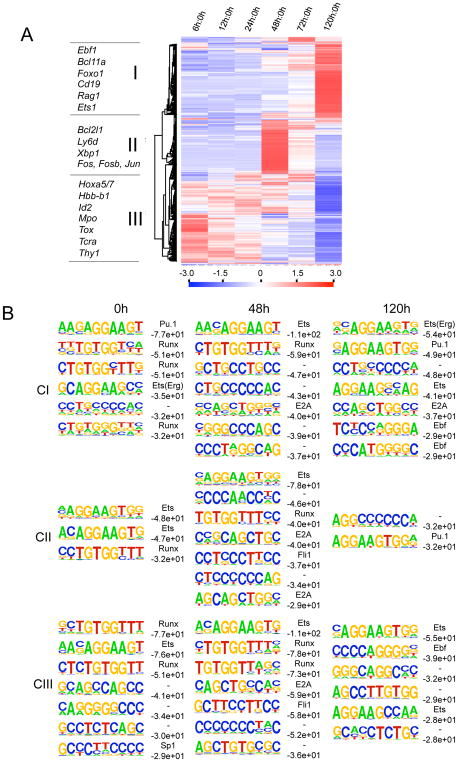
The development of a cell line that gives rise to differentiated progeny allows for the analysis of intermediate steps, not just the endpoints. We thus used the Id2-HPCs cells to analyze intermediate changes in transcript levels from the multipotent HPC to the committed pro-B stage. To this end, Id2-HPCs were differentiated into CD19+ pro-B cells and isolated RNA at different time-points. We then analyzed patterns of gene expression by microarray analysis and performed hierarchical clustering of genes that had a two-fold or greater change in expression at any given time-point. From this analysis we observed three main patterns: transcript levels that were increased (Cluster I), transcript levels that were transiently elevated (Cluster II) and transcript levels that declined (Cluster III) upon developmental progression (Figure 7A). Genes associate with elevated transcript abundance include Ebf1, Foxo1 and Rag1. Genes linked with a decline in transcript levels include those involved with alternate cell lineage programs of gene expression, such as Hbb-b1, Tox and Tcra, as well as loci potentially involved in the maintenance of the HSC phenotype, including members of the Hoxa family (Figure 7A, Table S1). Interestingly, a group of genes associated with cell growth, including Jun, Fos and Xbp1, showed a transient pattern of expression, possibly reflecting a stage in B cell development linked to cellular expansion (Figure 7A). Taken together, these data identify clusters of genes whose expression patterns are coordinately activated or silenced during developmental progression.
Figure 7. Evolving Enhancer Repertoires in Developing Multipotent Progenitors.
(A) Multipotent progenitors (Id2-HPCs) were differentiated into pro-B cells over a 5-day timecourse, and RNA was taking at 7 timepoints for microarray analysis. Expression was normalized to day 0. Cluster I refers to genes activated 120 hours post differentiation. Cluster II refers to genes activated 48 hours post differentiation. Cluster III refers to genes repressed 120 hours post differentiation.
(B) Enriched regulatory motifs at distal H3K4me1 sites during early B cell differentiation. ChIP-Seq was performed on multipotent progenitors (Id2-HPC) at 0-, 48-, and 120-hours after induction of B cell differentiation. De novo motif finding was performed to determine transcription factor binding motifs associated with active enhancers of genes that are upregulated (CI), intermediately upregulated (C2), and downregulated (C3). The known motif names are indicated to the left of the de novo motifs. Log P-values of the de novo motifs are indicated below of the names of the motifs. “-”refers to unknown motif.
The data described above show that the very early stages of B cell development are associated with a dynamic pattern of gene expression signatures. Specifically, we identified enhancers by the presence of H3K4me1 at 0-hours, 48-hours and 120-hours post differentiation (Figures S3, S4 and S6). The data was obtained from two independent experiments generated for all three time-points. To accomplish this, the reads from the ChIP-Seq analyses were merged. Next, H3K4me1 islands with FDR (False Discovery Rate) less than 0.001 were selected from the merged files, and then were analyzed for enriched transcription factor binding motifs within the three clusters at 0-hours, 48-hours and 120-hours post differentiation using HOMER (Figure 7B).
As aforementioned, Cluster I represents loci whose expression is activated during the developmental progression from the multipotent (0h) to the committed B cell stage (120h). At the multipotent HPC stage, putative enhancers modulating the expression of Cluster I loci frequently contain binding motifs for Pu.1, Runx and a potential novel participant named Erg. Upon developmental progression to the committed B cell stage, initially E2A and ultimately Ebf binding sites were frequently associated with active enhancers, consistent with the induction of a B-lineage program of gene expression (Figure 7B: CI-120h).
Cluster II represents loci whose transcript levels transiently are elevated but decline prior to the commitment cell stage (Figure 7: CII). The cistromic elements associated with this subset of genes was distinct from that of cluster I. Analysis of this cluster showed a substantial enrichment for Runx and Ets binding sites (Figure 7B; CII). Two days past induction of differentiation, novel enhancer repertories were identified that included E2A and a potential novel cis-regulatory element, Fli-1, (Figure 7B; CII-48h). Five days post-differentiation, the enhancer repertoire again was altered as characterized by the singular presence of Pu.1 binding sites (Figure 7B: CII-48h).
Cluster III represents loci whose transcript levels decline upon developmental maturation from the multipotent HPC to the committed B cell (Figure 7B; CIII). They include genes whose expression is associated with alternate lineages and HSC cell identity, including Hoxa5, Hoxa7, Hbb-b1, Ptcra and Thy1. Interestingly, upon developing into committed B cells, H3K4me1 islands in these loci were substantially enriched for E2A and Fli binding sites (Figure 7B: CIII-120h). Five days post-induction, the enhancer repertoires were changed as demonstrated by the lack of E2A and Fli1 binding sites and the presence of Ebf consensus cis-regulatory elements (Figure 7B: CIII-120h). Thus, it appears that these combinations of regulatory elements suppress rather than activate the expression of genes associated with alternate cell lineages in committed B cells. In sum, we describe the differentiation of multipotent HPCs into committed B-lineage cells in terms of evolving enhancer repertoires that involve both known regulators such as E2A, EBF and PU.1 as well as potential participants such as Erg and Fli1.
DISCUSSION
Recent genome-wide studies have provided insight into how transcriptional regulators act in concert with others to specify a developmental stage (Ghisletti et al., 2010; Heinz et al., 2010; Treiber et al., 2010; Lin et al., 2010; (Natoli, 2010). We are now faced with the fundamental question as to how such networks are established during the developmental progression from the multipotent HPC to a committed lymphoid or myeloid cell stage. Here we describe experimental and computational strategies to address this question.
Previous studies have demonstrated that long-term cultures of E2A-deficient progenitors remain pluripotent both in vitro and in vivo (Ikawa et al., 2004). A limitation of these cells is that E2A-deficient cells cannot be differentiated into committed B cells since the E2A proteins are required for the initiation of the B cell pathway. Additionally, forced E47 expression has until now, not permitted rescue of the developmental block observed in E2A-deficient pre-pro-B cells. Here we have developed a strategy that overcomes this obstacle. Briefly, we have generated a long-term culture of HPCs by enforcing the expression of the E-protein inhibitor Id2 (Rivera and Murre, 2001). Id2-HPCs are multipotent both in vitro and in vivo (Ikawa et al., 2004). However, unlike E2A-deficient progenitors, Id2-HPCs have the ability to differentiate into the B cell lineage upon downregulation of Id2. These studies indicate that it is the activity of E2A that plays a causative role in B cell specification. It is the accumulation of E2A activity in multipotent progenitors that permits B cells to begin their developmental journey.
A tight correlation between enhancers associated with transcriptionally active genes and H3K4 monomethylation has recently been established (Heintzman et al., 2007). We demonstrate that multipotent HPCs lineage restricted enhancer elements are already primed as demonstrated by the presence of H3K4 methylation prior to differentiation. As expected, upon commitment to the B- or myeloid cell lineage, we observed an increase in H3K4 mono- and trimethylation across genomic regions associated with a B- or myeloid lineage programs of gene expression, respectively. In addition, the degree of H3K4 methylation declined across genomic regions linked with the alternate programs of gene expression. Taken together, these data confirmed the previous notion that H3K4me1 regions are associated with transcriptional poised or active regions. These findings are in line with the concept of multilineage priming, previously proposed to be involved in the maintenance of multipotency in HSCs (Hu et al., 1997).
The simplest explanation of multi-lineage priming is that low level of expression of several lineage-determining transcription factors at the precursor stage “primes” genomic regions characteristic of different hematopoietic lineages. Therefore, it is likely that, in progenitor cells, these pre-marked enhancers are already associated, possibly at low levels, with the very same transcription factors that will bind these regions in fully differentiated cells. It is very well possible that pre-lineage priming involves E2A. E2A proteins are expressed in HSCs and act to maintain the HSC pool (Dias et al., 2008; Semerad et al., 2009; Yang et al., 2008). Thus, it may very well be that E2A functions in progenitor population to pre-mark lymphoid enhancer repertoires. This raises the question as to why this pre-marking generates a low degree of H3K4 monomethylation in multipotent progenitors relative to lineage-committed cells. The level of E2A at these developmental stages is similar to that of pro-B cells. Thus, it does not seem likely that the expression of E2A in HSCs versus pro-B cells generates differences in the degree of H3K4 monomethylation. Although still to be proven, it is conceivable that E2A associates with different partners including SCL/TAL1 in HSCs. The SCL proteins are well known to heterodimerize with E2A. Thus, we suggest that the E2A proteins could function in HSCs to promote the pre-marking of a lymphoid specific enhancer repertoire while in committed B cells, they act as homodimers to elevate H3K4me1 abundance across poised enhancer regions and thus induce a B-lineage program of gene expression.
The establishment of committed lymphoid cells from mulitpotent HPCs involves multiple intermediate stages. How are such intermediate stages established? Developmental stages can be characterized in terms of gene expression signatures. Gene expression signatures are in turn primarily established by enhancer repertoires. Here, we have used genome-wide and computational approaches to describe the evolving enhancer repertoires that underpin early B cell development. These repertoires include E2A, Ebf, Ets, Runx and Pu.1, as well as Erg and Fli1 consensus binding sites. Erg and Fli1 are particularly intriguing since they have previously been suggested, based on their expression patterns, to play critical roles in early B cell development (Rivera et al., 1993).
In sum, we have generated long-term cultures of HPCs that are multipotent and self-renew indefinitely in vitro. Remarkably, the distribution of motifs across enhancer repertoires does not change dramatically during developmental progression. However, we note that a fraction of the enhancer repertoires has changed upon becoming committed to the B cell lineage. This is particularly apparent by the increased frequency of Ebf motifs at 120 hours relative to 0 hour. Finally, using directed differentiation, gene expression microarrays and identification of enhancer repertoires by H3K4me1, we describe the early stages of B cell development in terms of evolving global enhancer and silencer repertoires. In principle, this analysis can be applied to any developmental setting including β-selection, positive selection, receptor editing and beyond.
EXPERIMENTAL PROCEDURES
Viral Vectors and Virus Production
Human ID2 was amplified from 293T cDNA with the addition of BamH1 sites at both 3′ and 5′ ends, and then cloned into pRRL.sin-18.PPT.PGK.MCS.IRES.GFP.pre, kindly provided by Dr. Irving Weissman (Stanford University). The hId2_IRES-GFP expression cassette was excised with AgeI and BsrG1, replacing the eGFP gene in pBob_TA1_R2_corrected120803, kindly provided by Dr. Inder Verma (Salk Institute). hId2 was then cut out of the resulting vector to create an empty vector control. These vectors were named TetOff_hId2 and TetOff_Empty, respectively.
Transduction and Cell Culture
CD45.1 congenic animals (8–12 weeks of age) were injected with 250 mg/kg of 5-fluorouracil 4 days before bone marrow was harvested and lineage depleted by auto-MACS. Purified lineage negative cells were cultured overnight in expansion media (DMEM + 15% FBS in the presence of 10 ng/mL IL-3, 10 ng/ml IL-6, 1:200 SCF and 1:20 WEHI). After 12–18 hours in culture, cells were pelleted and resuspended in fresh expansion media plus TetOff_hId2 or TetOff_Empty lentivirus in the presence of 4 ug/mL polybrene and the above cytokines. Cells were spin infected twice for 1.5 hours at 2500 rpm, 30° C, with a 12–18 hour rest in between spin infections. After the second spin, cells were resuspended in fresh expansion media and cultured for 4–5 weeks. Cells were then cultured in IMDM + 10% FCS, 2% PSG and 2 μL β-me on subconfluent S17 feeder cells in the presence of 1:100 IL-7, 1:100 Flt3L and 1:200 SCF. S17 feeder cells were maintained in α-MEM + 10% FBS and 2% PSG. Animal studies were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego.
In Vitro Differentiation
TetOff_hId2 expanded cells were depleted of small (<1–5%) numbers of CD19, CD25 and CD11b positive cells by auto-MACS. For myeloid differentiation, cells were cultured for up to 6 days in IMDM + 10% FCS and β-me in the presence of IL3, Flt3L, GMCSF and MCSF and in the presence or absence of 1 ug/mL doxycycline. To promote B-cell differentiation cells were cultured for up to 10 days in IMDM + 10% FCS and β-me on S17 feeder cells in the presence of 1:100 IL-7 and 1:200 SCF in the presence or absence of 1 ug/mL doxycycline, or alternatively, in α-MEM in the presence of cytokines and the Tst-4 stromal cell line (kind gift from Dr. Ikawa). Fresh doxcycline and cytokines were added every two days. E2A −/− cells were cultured as previously described (Ikawa et al., 2004)
Microarray Profiling
All RNA was prepared using RNeasy Columns (Qiagen). Gene expression profiling depicted in Figure 1 was performed by hybridization to the Affymetrix MOE430 2.0 gene expression array according to the manufacturer’s instructions. Gene expression profiling depicted in Figures 1, 4 and 7 was performed by amplification and hybridization to the Illumina Mouse WG-6 v1.1 and v2 gene expression arrays according to the manufacturer’s instructions. Differentially expressed gens were determined using the Limma package from BioConductor. A Benjamini-Hochberg-adjusted p-value cut-off of 0.05 and a fold-change threshold of 2. Hierarchical clustering was performed on this set of genes. The distance metric used was (1 - Pearson correlation coefficient), and the ‘average’ (unweighted pair-group average method, UPGMA) method was used for agglomeration. Clusters were obtained by empirically cutting the tree. The microarray data was deposited in GEO database (GSE30859).
ChIP-Sequencing
The chromatin immunoprecipitation (ChIP) protocol was essentially the same as previously described (Agata et al., 2007; Lin et al., 2010). Anti-H3K4me1 (ab8895) and anti-H3K4me3 (07–473) were purchased from Abcam and Millipore, respectively. Anti H3K4me1 ChIP-Seq was performed in duplicate. Peaks form duplicate experiments were merged to identify putative peaks. Those peaks positions and each of the individual experiments were used to score the peak and to ensure significance relative to the input. The R package “edgeR” was used to perform the significance calculations. The ChIP_Seq data was deposited in GEO database (GSE30859).
Data Analysis
Clusters of tags across the analyzed genomes were identified based on the significantly enrichment of tags relative to background as well as local tag counts. Data was analyzed using HOMER software (http://biowhat.ucsd.edu/homer/). Bound sites were identified using HOMER. ChIP-Seq tag counts were normalized between experiments to a total of 107 mapped tags. The peak region was required to show 4-fold more tags (as compared to total tag count) than controls. Additionally, DNA bound elements were required to exhibit at least two-fold more tags within a 1 kb region versus flanking 10 kbp DNA regions. The threshold for the number of tags generating an interacting site was determined for a false discovery rate of 0.001. Furthermore, peaks were required to exhibit at least 4-fold more tags (normalized versus total number) versus input control samples. To avoid identifying DNA elements that contain genomic duplications or non-localized occupancy a four-fold increase in the number of tags relative to tags located within immediate genomic proximity was chosen as a threshold. The detailed description of the heat map generation was found in http://biowhat.ucsd.edu/homer/. Briefly, a peak file of the first biological replicate of the top 5000 high confidence peaks that had at least four-fold more tags within a 1 kb region versus flanking 10 kbp DNA regions. Then, the sequences within 3000 bp upstream and downstream of the peak centers of each replicate were clustered by the average linkage method using Cluster 3.0. The heat map was visualized by Java Tree View.
Replicates for ChIP-Seq of H3K4me1 in Id2-HPCs, Id2-HPCs induced to differentiate towards the B cell lineage for two and five days, and myeloid cells were merged into one file each in a single “META”-experiment. EdgeR was used to calculate the false discovery rates to determine the difference between the merged files and input. Motifs associated with transcription factor occupancies were identified using HOMER software (Benner et al., in preparation; Heintz et al. 2010). To analyze H3K4me1 islands for enriched motifs, DNA sequences (plus or minus 500 bp centered across H3K4me1 peaks) were compared to 50,000 randomly identified genomic fragments of similar size and matched for the proportion of GC content.
Supplementary Material
HIGHLIGHTS.
Long-term culture of multipotent hematopoietic progenitors
The E2A proteins play a causative role in initiating B cell development
Enhancers are primed for lineage plasticity in multipotent progenitors by H3K4me1
Acknowledgments
We thank G. Hardiman, C. Ludka, J. Sprague, M. Harabaglia, and Z. Ye for help with Solexa DNA Sequencing. We are grateful to J. Sprague for microarray analysis. We thank the members of the Murre laboratory for comments on the manuscript. E.M. was supported by a training grant from the NIH. Y.C.L. was supported by the Ruth L. Kirschstein Research Service Award (1F32CA130276). The studies were supported by grants from the NSF (NSF IIS-0803937) to T.I. and from the NIH to C.B. (P01DK074868), C.K.G. (CA52599) and C.M. (CA054198-20) and BIOGEM DK063491 to the University of California, San Diego Core Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LAM, et al. Identification of Flt3+ Lympho-Myeloid Stem Cells Lacking Erythro-Megakaryocytic Potential: A Revised Road Map for Adult Blood Lineage Commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T Cell Receptor [beta] Gene Rearrangements and Allelic Exclusion by the Helix-Loop-Helix Protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag E, te Riele H, Voland J, Sharp L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Maandag ECR, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Busslinger M. Transcriptional Control of Early B Cell Development1. Annual Review of Immunology. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Månsson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Current Opinion in Immunology. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes & Development. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Wright LYT, Murre C. Long-Term Cultured E2A-Deficient Hematopoietic Progenitor Cells Are Pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes & Development. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco AC, Stegmann APA, Heemskerk MHM, Couwenberg F, Bakker AQ, Weijer K, Spits H. Genetic modification of human B-cell development: B-cell development is inhibited by the dominant negative helix loop helix factor Id3. Blood. 1999;94:2637–2646. [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of Clonogenic Common Lymphoid Progenitors in Mouse Bone Marrow. Nat Immunol. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lazorchak A, Jones ME, Zhuang Y. New insights into E-protein function in lymphocyte development. Trends in Immunology. 2005;26:334–338. doi: 10.1016/j.it.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Lin HH, Grosschedl R. Failure of B-Cell Differentiation in Mice Lacking the Transcription Factor Ebf. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SEW, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- McManus S, Ebert A, Slavagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Natoli G. Maintaining Cell Identity through Global Control of Genomic Organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Urbánek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes & Development. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- Rivera R, Murre C. The regulation and function of the Id proteins in lymphocyte development. Oncogene. 2001;20:8308–8316. doi: 10.1038/sj.onc.1205091. [DOI] [PubMed] [Google Scholar]
- Rivera RR, Stuiver MH, Steenbergen R, Murre C. Ets proteins: new factors that regulate immunoglobulin heavy-chain gene expression. Mol Cell Biol. 1993;13:7163–7169. doi: 10.1128/mcb.13.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL. Early B Cell Factor Promotes B Lymphopoiesis with Reduced Interleukin 7 Responsiveness in the Absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proceedings of the National Academy of Sciences. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G, Heimfeld S, Weissman I. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R. Early B Cell Factor 1 Regulates B Cell Gene Networks by Activation, Repression, and Transcription-Independent Poising of Chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Urbánek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, Graf T. Stepwise Reprogramming of B Cells into Macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 Controls the Developmental Integrity and Cell Cycle Quiescence of Multipotential Hematopoietic Progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Jackson A, Pan L, Shen K, Dai M. Regulation of E2A gene expression in B-lymphocyte development. Molecular Immunology. 2004;40:1165–1177. doi: 10.1016/j.molimm.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.