Abstract
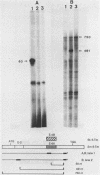
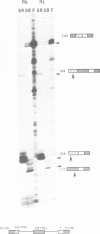
A subgenomic fragment of the chicken beta tropomyosin gene which contains two alternative exons flanked by common exons was isolated and placed under the control of the SV 40 early promoter. This construction was subsequently used to transfect quail myoblasts together with a Neomycin resistance gene, and to isolate stable transfectants. mRNAs were isolated before and after differentiation and analyzed using a modification of the primer extension method. We show that myoblasts accumulate transcripts which contain the non muscle specific exon joined to the common exons while myotubes accumulate transcripts containing the muscle specific exon. These results, therefore demonstrate that such a subgenomic fragment contains all the necessary information to direct a correct developmentally regulated mutually exclusive splicing. They also strongly suggest that trans acting factors must be involved in the switch of the splicing pattern which takes place during the transition from myoblasts to myotubes. The same regulation cannot be faithfully reproduced during transient expression, since no difference in the use of exons 6A/6B is observed during differentiation and two aberrant minor splicing products are obtained which contain or lack both exons. We suggest that failure of exon 6A to splice to exon 6B is due to the existence of some structural constraints which lower the efficiency with which the intron between them is excised.
Full text
PDF

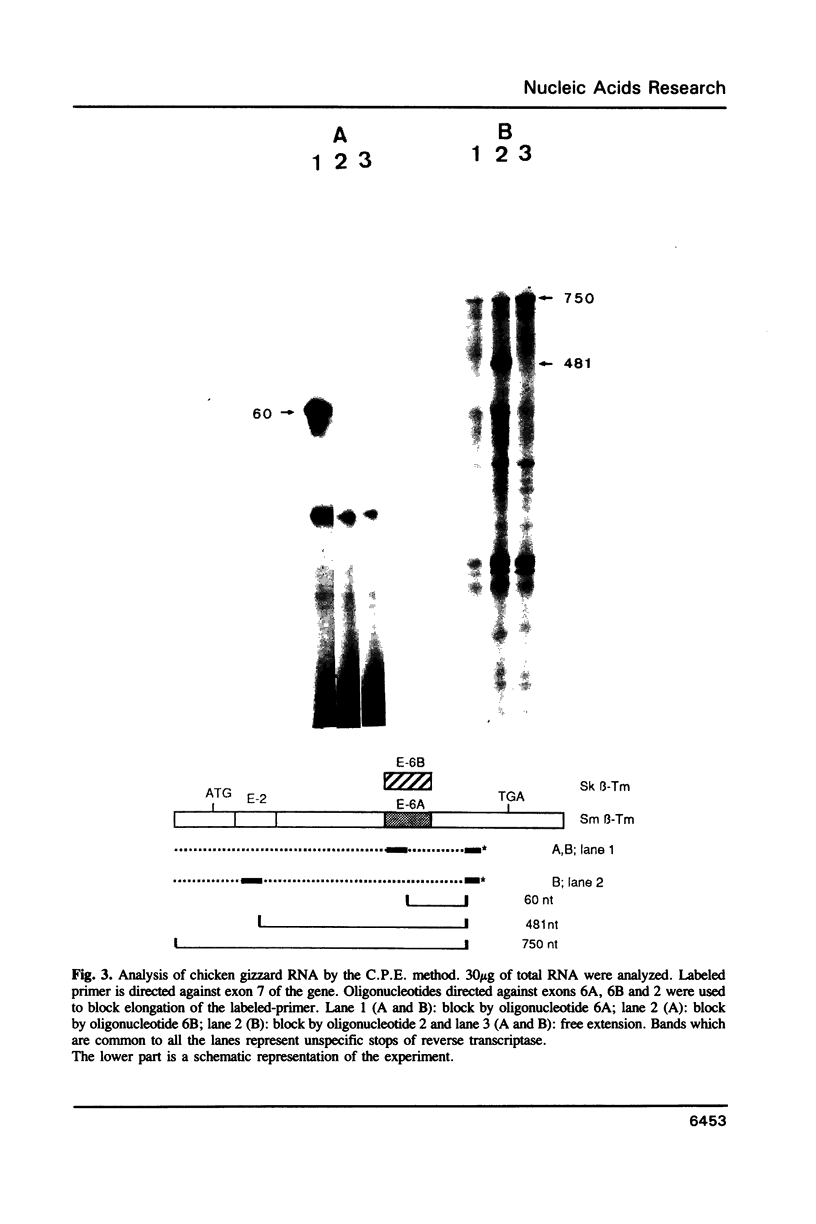
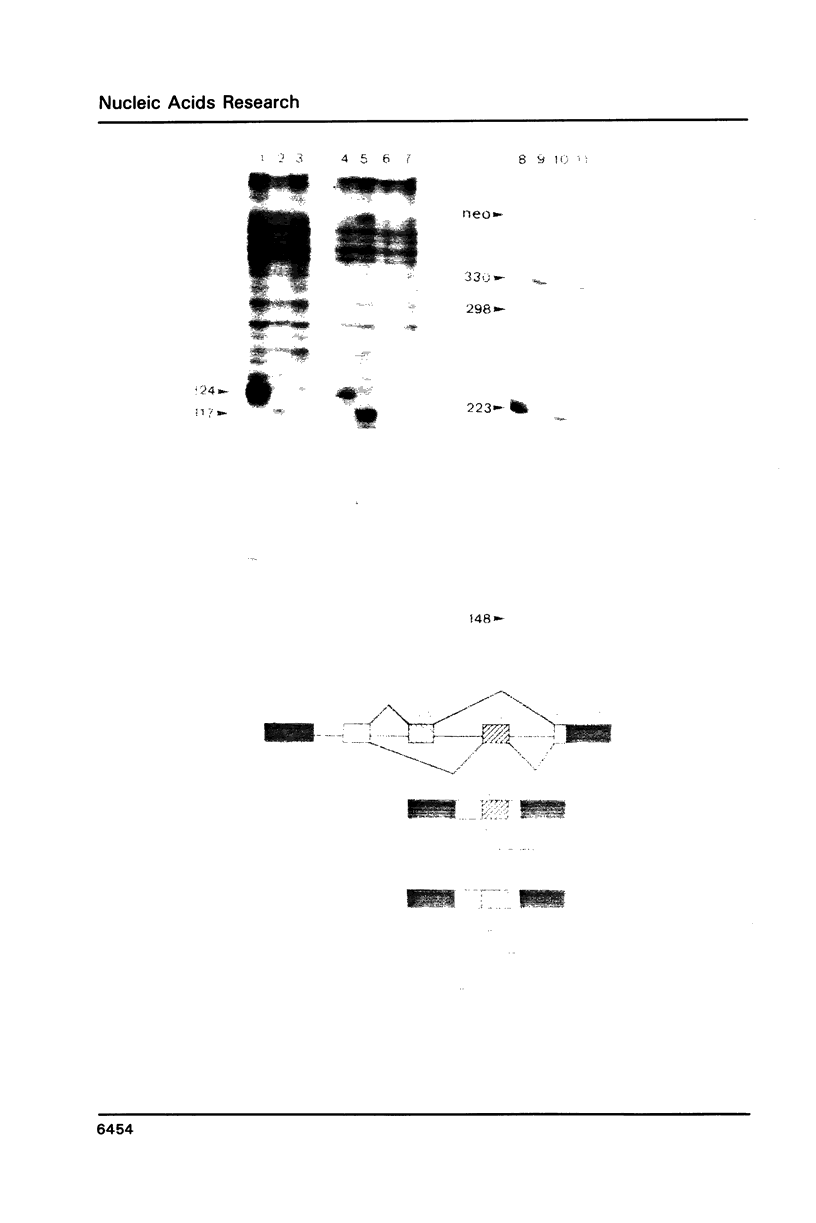


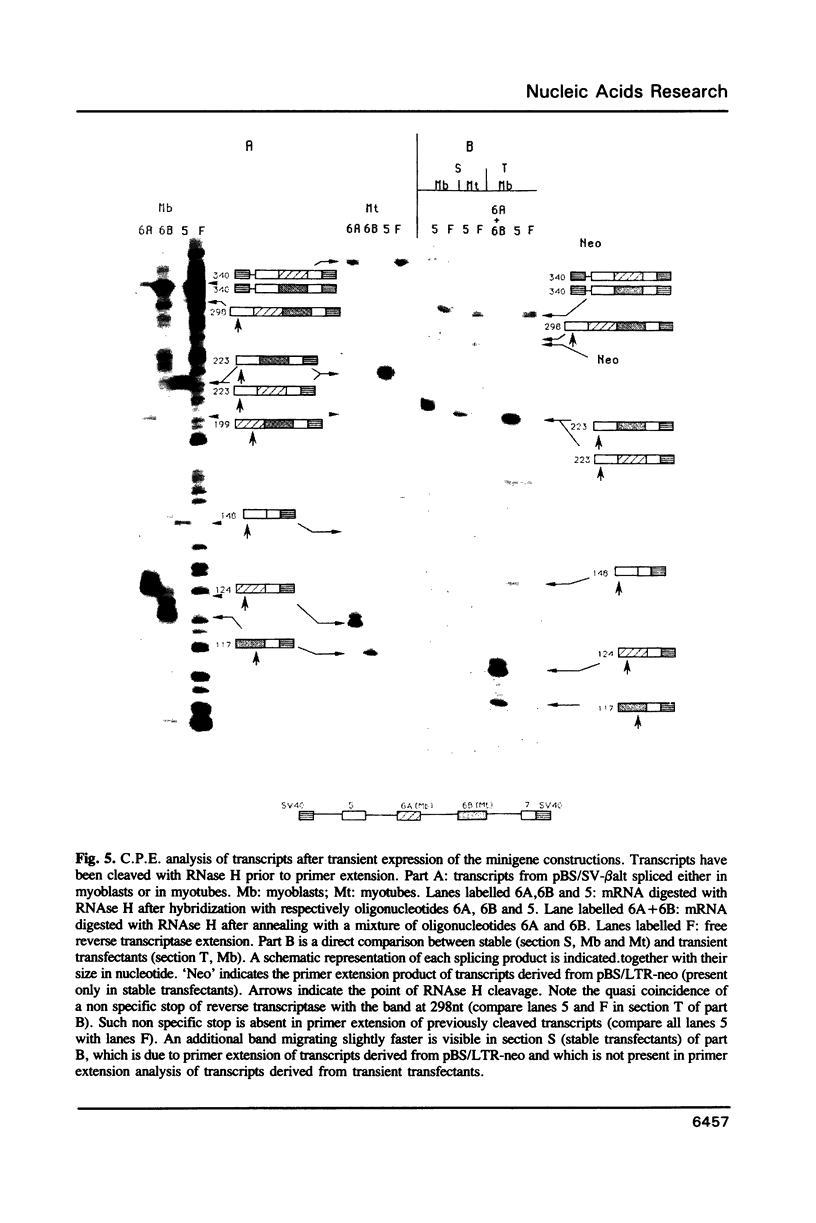





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Gallego M. E., Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–242. doi: 10.1146/annurev.cb.03.110187.001231. [DOI] [PubMed] [Google Scholar]
- Barone M. V., Henchcliffe C., Baralle F. E., Paolella G. Cell type specific trans-acting factors are involved in alternative splicing of human fibronectin pre-mRNA. EMBO J. 1989 Apr;8(4):1079–1085. doi: 10.1002/j.1460-2075.1989.tb03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradac J. A., Gruber C. E., Forry-Schaudies S., Hughes S. H. Isolation and characterization of related cDNA clones encoding skeletal muscle beta-tropomyosin and a low-molecular-weight nonmuscle tropomyosin isoform. Mol Cell Biol. 1989 Jan;9(1):185–192. doi: 10.1128/mcb.9.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nguyen H. T., Medford R. M., Destree A. T., Mahdavi V., Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985 May;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Erster S. H., Finn L. A., Frendewey D. A., Helfman D. M. Use of RNase H and primer extension to analyze RNA splicing. Nucleic Acids Res. 1988 Jul 11;16(13):5999–6014. doi: 10.1093/nar/16.13.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M., Finn L. A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988 Dec;2(12A):1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
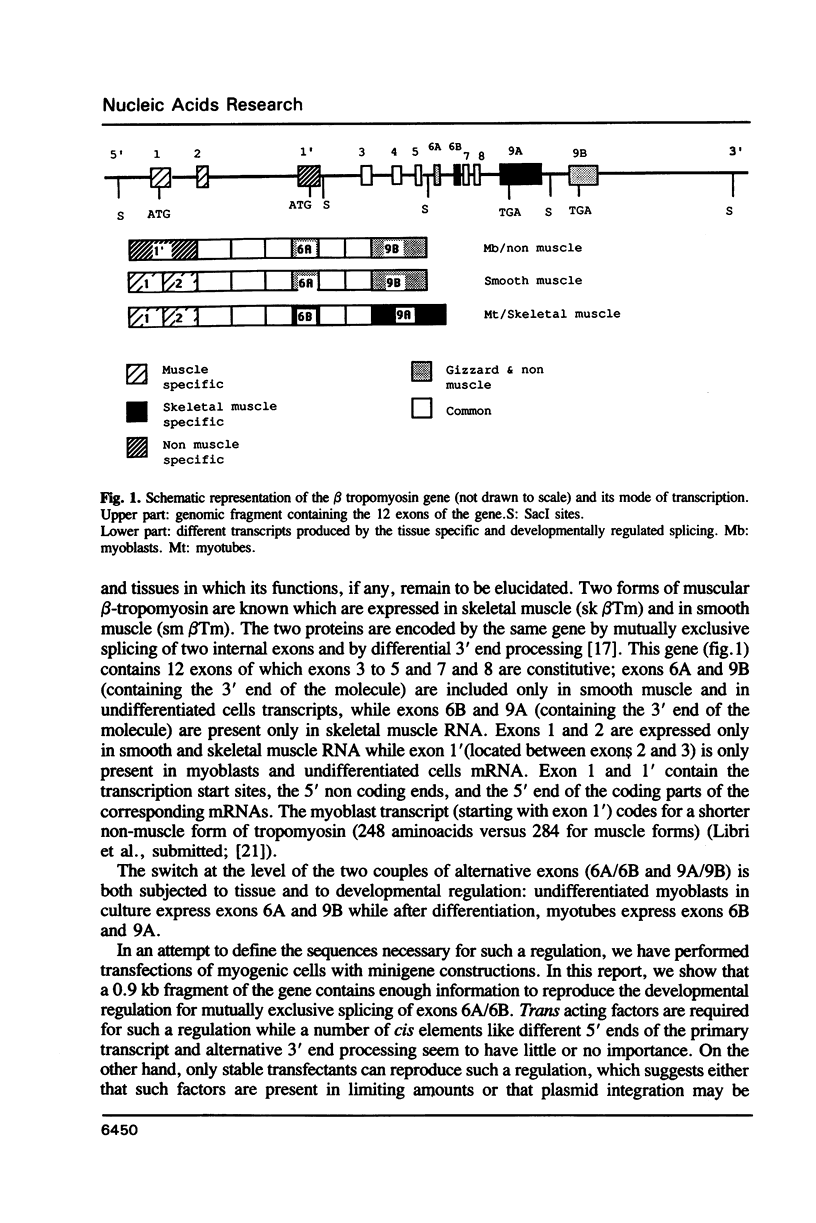
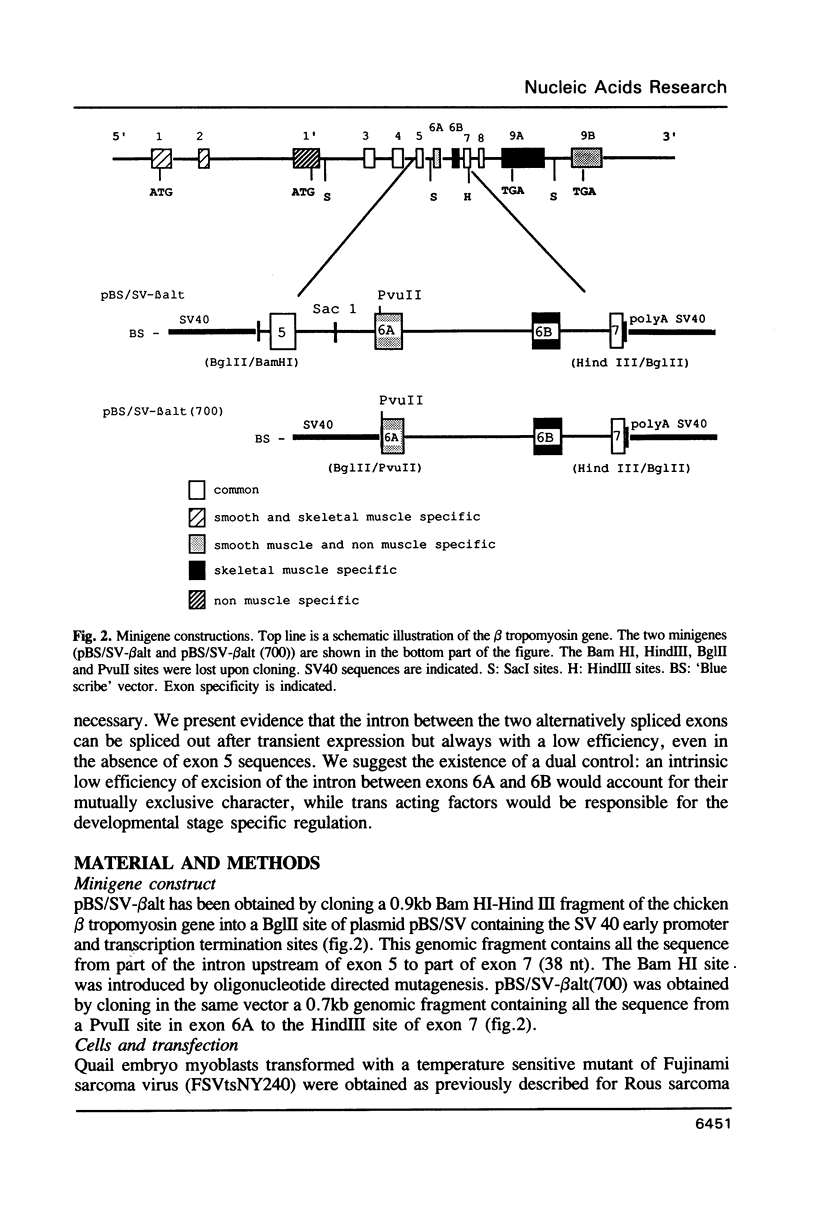
- Libri D., Lemonnier M., Meinnel T., Fiszman M. Y. A single gene codes for the beta subunits of smooth and skeletal muscle tropomyosin in the chicken. J Biol Chem. 1989 Feb 15;264(5):2935–2944. [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Fiszman M. Y. A new muscle phenotype is expressed by subcultured quail myoblasts isolated from future fast and slow muscles. J Biol Chem. 1983 Mar 25;258(6):3883–3888. [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Opazo N., Nadal-Ginard B. Alpha-tropomyosin gene organization. Alternative splicing of duplicated isotype-specific exons accounts for the production of smooth and striated muscle isoforms. J Biol Chem. 1987 Apr 5;262(10):4755–4765. [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]