Abstract
DNA methylation has been traditionally viewed as a highly stable epigenetic mark in post-mitotic cells, however, postnatal brains appear to exhibit stimulus-induced methylation changes, at least in a few identified CpG dinucleotides. How extensively the neuronal DNA methylome is regulated by neuronal activity is unknown. Using a next-generation sequencing-based method for genome-wide analysis at a single-nucleotide resolution, we quantitatively compared the CpG methylation landscape of adult mouse dentate granule neurons in vivo before and after synchronous neuronal activation. About 1.4% of 219,991 CpGs measured show rapid active demethylation or de novo methylation. Some modifications remain stable for at least 24 hours. These activity-modified CpGs exhibit a broad genomic distribution with significant enrichment in low-CpG density regions, and are associated with brain-specific genes related to neuronal plasticity. Our study implicates modification of the neuronal DNA methylome as a previously under-appreciated mechanism for activity-dependent epigenetic regulation in the adult nervous system.
Epigenetic modifications of chromatin molecules, including genomic DNA and histone proteins, play critical roles in orchestrating the transcriptome of different cell types and their developmental potentials1–3. In contrast to readily reversible histone modifications, DNA methylation has been generally regarded as a highly stable epigenetic mark in differentiated cells to ensure transcriptional gene silencing and maintain cell type identity1, 2, 4. DNA methylation, catalyzed by DNA methyltransferases (DNMTs), occurs on cytosine bases almost exclusively in CpG dinucleotides in somatic cells. Although clustered 5-methylcytosines (5mCs) are well established transcriptional repressors in gene silencing and inactivation of endogenous transposable elements5, emerging evidence suggests diverse roles of DNA methylation in various contexts, including promoting gene expression2, 6–8. In addition, 5-hydroxymethylcytosines (5hmCs) are present in the genomic DNA of mature neurons, although their potential function in gene regulation remains largely unknown4, 9.
How experience-driven transient synaptic activity leads to long-lasting modifications of neural circuits and neuronal properties is a long-standing question. Emerging evidence suggests important roles for epigenetic regulation in activity-dependent mature brain functions10–13, including synaptic plasticity13, learning and memory14, circadian rhythm15, drug addiction16, and adult neurogenesis17. Interestingly, recent studies have implicated critical roles of CpG methylation changes in neural plasticity18–21. Deletion of Dnmt1 and Dnmt3a in mouse forebrain excitatory neurons leads to deficits in neuronal morphology, synaptic plasticity, and learning and memory20. Dnmt3a is also critical for emotional behaviour and spine plasticity in adult mouse nucleus accumbens21. Recent studies have identified several specific CpGs that could be acutely modified by neuronal activity or behaviour in postnatal neural tissues17, 22–29. For example, neuronal stimulation induces Gadd45b-dependent DNA demethylation at specific promoters of Bdnf and Fgf1 in the adult mouse dentate gyrus17. However, the scope and global properties of activity-induced changes in neuronal DNA methylation remain unknown.
Here we used an improved genome-wide profiling method for analysis of the DNA methylome of dentate granule neurons in the adult mouse hippocampus in vivo, before and after synchronous neuronal activation. We identified a large number of changes in CpG methylation status in response to neuronal stimulation. Bioinformatic analysis further revealed unexpected properties of the genomic distribution of these activity-modified CpGs. Our study provides the first global view of how the DNA methylation landscape of mature neurons is rapidly modified in response to external stimuli in vivo and implicates active DNA modifications as a general mechanism for activity-dependent epigenetic regulation in the adult brain.
RESULTS
Activity-induced modification of neuronal DNA methylome
To determine the extent to which neuronal activity modifies the landscape of neuronal DNA methylation in vivo, we took a genome-wide approach to profile the DNA methylome of dentate granule cells in the adult mouse hippocampus with or without synchronous activation by electroconvulsive stimulation (ECS; Supplementary Fig. 1; See Methods), a procedure used clinically to treat patients with drug-resistant depression30. At 0 hr (E0; sham control), 4 hrs (E4) or 24 hrs (E24) after a single ECS, we purified genomic DNA from micro-dissected dentate gyrus tissues, which were highly enriched with post-mitotic NeuN+Prox1+ dentate granule neurons (Supplementary Fig. 1; ref17). We used an improved Methyl-Sensitive Cut Counting (MSCC) method7 to measure CpG methylation levels at a single-nucleotide resolution (Supplementary Fig. 2). The methylation/hydroxymethylation-sensitive restriction enzyme HpaII and its methylation-insensitive isoschizomer MspI used in the study recognize the same restriction site CCGG. Notably, locations of CCGGs largely reflect those of all CpGs in the mouse genome (Supplementary Fig. 3), allowing unbiased genome-scale analysis.
We constructed MSCC libraries and obtained a total of ~ 82 million uniquely mapped reads by next-generation sequencing (1.4 gigabase; Supplementary Table 1). The overall CpG methylation profiles of dentate granule neurons at E0, E4 and E24 are very similar (Fig. 1a) and the correlation between samples increases with sequencing depth (Supplementary Fig. 4). After excluding MSCC sites with low sequencing depth (< 30 reads), we profiled 219,991 CpGs (MSCC30+ sites), representing ~1% of all CpGs in the mouse genome. Global methylation levels were comparable across all autosomes and X chromosomes, whereas the Y chromosome was hypermethylated (Supplementary Fig. 5a). A recent study has shown the presence of 5mCs and 5hmCs in the mammalian mitochondrial genome31. However, the neuronal mitochondrial genome is virtually unmethylated (Supplementary Fig. 5a), which is consistent with early findings32. Mapping of MSCC sites to their relative location to associated genes showed a clear pattern of transcription starting site (TSS) hypomethylation and gene body methylation (Supplementary Fig. 5b), which is also similar to findings from the human methylome7, 33, 34. In addition, exon-intron junctions showed consistent methylation patterns as reported using a bisulfite sequencing-based method33 (Supplementary Fig. 5c). These results collectively confirm the validity of our MSCC profiles.
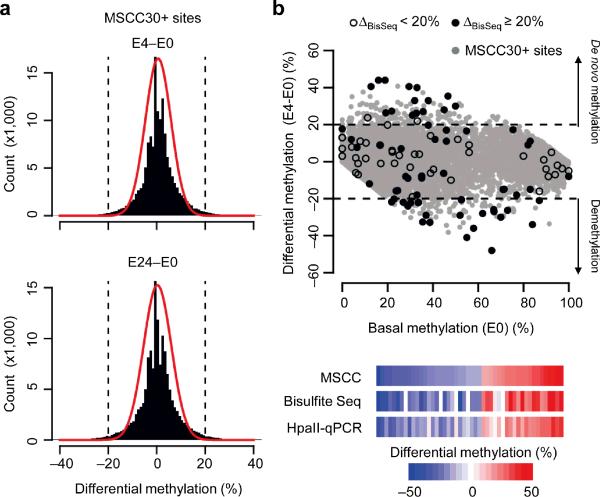
Figure 1.
Modification of DNA methylation landscape in the adult dentate gyrus by neuronal activity. (a) Comparison of CpG methylation profiles of the dentate gyrus of adult mice at different time points after a single ECS. Shown are histograms of differential CpG methylation between sham control and 4 hrs after ECS (ΔMSCC, E4-E0; top) and between sham control and 24 hrs after ECS (ΔMSCC, E24-E0; bottom). Red lines represent Gaussian distributions with the same means and variances. Dashed lines represent ± 20% cut-off. (b) A scatter plot of differential CpG methylation (E4-E0) at individual CpGs versus their basal methylation levels at E0 (MSCC estimates with 30+ reads; gray dots). Black dots indicate bisulfite analysis of selected MSCC sites for validation (solid: ΔBisSeq ≥ 20%; open: ΔBisSeq < 20%). Below are heat-maps of methylation changes detected by MSCC, bisulfite sequencing, and HpaII-qPCR, from independent biological samples.
Despite the global similarity, methylation changes were evident after neuronal activation, which outnumber those that would be predicted by a Gaussian model of measurement variation with the same mean and variance (Fig. 1a). To validate detected methylation changes, we selected a set of 116 CpGs with different levels of methylation changes (ΔMSCC; either increase or decrease) for bisulfite sequencing using independent biological samples (Fig. 1b, Supplementary Fig. 6a; Supplementary Table 2). At ΔMSCC ≥ 20%, our method achieved 94.6% specificity and 76.1% sensitivity (Supplementary Fig. 6b). We further used HpaII-digestion followed by quantitative PCR (HpaII-qPCR) to examine a set of 48 CpGs (ΔMSCC ≥ 20%) in multiple individual animals (Supplementary Table 3). Notably, the inter-individual variation of methylation levels at these CpGs was small (8 ± 4%; s.d.). There are also strong correlations for measurements between MSCC and bisulfite sequencing or HpaII-qPCR analysis (Supplementary Figs. 7 and 8). Using a cut-off of ΔMSCC ≥ 20%, 1,892 and 1,158 CpGs exhibit activity-induced de novo methylation and demethylation at E4, respectively (Supplementary Table 4). The 3,050 CpGs in total represent ~ 1.4% of all profiled CpGs and likely constitute an underestimation due to the stringent depth requirement (≥ 30 reads) and cut-off for differential methylation (≥ 20%). For example, we confirmed CpG demethylation in Nlgn3, an autism-associated gene (< 30 MSCC reads; Supplementary Fig. 6c). Thus, in contrast to the general belief that DNA methylation is a highly stable modification in terminally differentiated cells, a large number of CpGs are rapidly modified by neuronal activity in the adult brain.
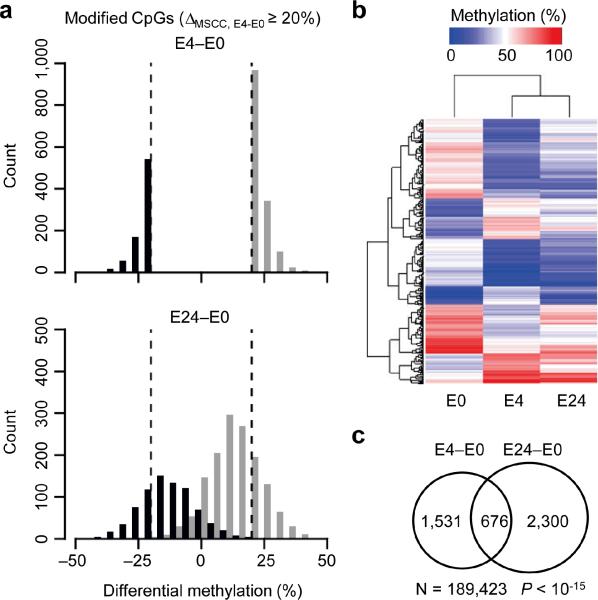
To determine the duration of activity-induced CpG modifications, we obtained the neuronal DNA methylome profile at E24 (Supplementary Table 4). While some of the methylation changes were reversed to basal levels by E24, a significant number of CpGs showed sustained changes (Fig. 2a). Unsupervised hierarchical clustering analysis further revealed that the methylation pattern of these CpGs at E24 more closely resembles E4 than E0 (Fig. 2b). Specifically, 31% of activity-modified CpGs detected at E4 remained at their modified states at E24 (Fig. 2c). Thus, unlike activity-induced immediate early gene expression, which usually lasts up to several hours, activity-induced DNA modifications are relatively long-lasting.
Figure 2.
Sustainability of activity-induced CpG modifications. (a) Histograms showing distributions of activity-modified CpGs (ΔMSCC;E4-E0 ≥ 20%) at E4 (top) and E24 (bottom). Note that de novo methylated (black) and demethylated (gray) CpGs remain well segregated from E0 at E24. (b) Unsupervised hierarchical clustering of methylation levels of top 500 MSCC sites with activity-induced modifications. Note that the E24 profile is more similar to E4 than to E0, suggesting that activity-induced CpG modifications in mature neurons in vivo are relatively sustained. (c) Venn diagram showing highly significant overlapping between activity-modified CpGs identified at E4 and E24. Note that fewer MSCC sites (N) are used in the analysis due to the additional requirement of sequencing depth of the E24 sample (P value, exact binomial test).
Properties of activity-induced CpG modifications
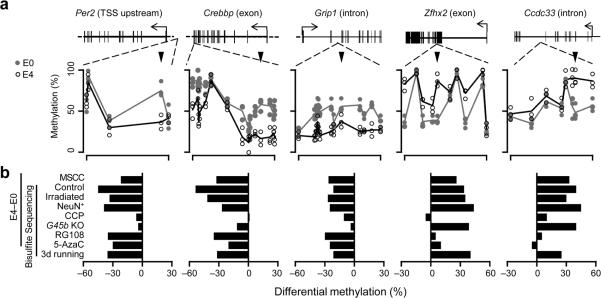
To gain mechanistic insights into activity-induced CpG modifications, we subjected animals to pharmacological, genetic and behavioural manipulations and selected representative regions (putative promoters, exons, introns) for detailed bisulfite sequencing analysis (Fig. 3). Similar to HpaII-qPCR results, inter-individual variation was small (7 ± 3 %; s.d.; n = 124), and consistent ECS-induced methylation changes were observed from multiple individual animals (Fig. 3a). Pre-treatment of animals with a highly selective NMDA receptor antagonist 3-(2-carboxypiperazin-4-yl)propyl-1-phosphonic acid (CPP) abolished ECS-induced changes (Fig. 3b), confirming that these modifications are neuronal activity-dependent. Infusion of either 5-azacytidine or RG108, two mechanistically distinct DNMT inhibitors35 previously shown to be effective in adult brains19, 21, 24, 26, 27, abolished activity-induced de novo methylation with no obvious effect on demethylation (Fig. 3b). Interestingly, Dnmt3a, but not other Dnmts, was up-regulated by ECS (Supplementary Fig. 9), suggesting its potential role in neuronal activity-induced de novo methylation. On the other hand, activity-induced demethylation was abolished in Gadd45b knockout (G45b KO) mice, consistent with recent findings on the critical role of Gadd45 family proteins in DNA demethylation36. Bisulfite sequencing of consecutive CpGs also revealed that activity-induced modifications are highly site-specific (Fig. 3a; Supplementary Fig. 6a), which may partially explain why acute methylation changes could not be readily detected by previous low-resolution profiling methods.
Figure 3.
Biological properties of activity-induced CpG methylation changes in the adult dentate gyrus. (a) Examples of bisulfite sequencing analysis of CpGs within five representative regions: Per2 (demethylated in TSS upstream region; chr1:93356934-93357200), Crebbp (demethylated in the exon; chr16:4085747-4086068), Grip1 (demethylated in the intron; chr10:119374673-119374888), Zfhx2 (de novo methylated in the exon; chr14:55691325-55691565), and Ccdc33 (de novo methylated in the intron; chr9:57897719-57898139). Shown are bisulfite sequencing results of these representative regions of interest at E0 (gray dots) and E4 (open circles) from multiple individual animals. Lines represent mean values. Arrowheads point to MSCC sites of interest. (b) Summaries of methylation changes at the MSCC sites upon different manipulations (with independent corresponding controls). CCP (10 mg/kg body weight) or saline was injected 1 hr before ECS. RG108 (1 mM), 5-azacytidine (5-AzaC; 1 mM), or saline was infused into lateral ventricles 2 days before ECS. Adult male Gadd45b knockout (G45b-KO) and wild-type (WT) littermates were used as indicated. Data represent a minimal of 20 bisulfite reads for each condition from at least two animals.
DNA demethylation can occur passively during cell division4. The adult dentate gyrus region harbours a small number of proliferating neural and glia progenitors (Supplementary Fig. 10). We carried out two experiments to directly test whether the observed DNA methylation changes are active in nature and independent of cell division. In the first experiment, we permanently inhibited cell proliferation in the adult hippocampus with targeted irradiation37 (Supplementary Fig. 10). In the second experiment, we directly purified NeuN+ post-mitotic neuronal nuclei from the dentate gyrus of normal adult mice with fluorescence-activated cell sorting (FACS), which also removed potential contribution from other cell types, such as progenitors, glia and microglia (Supplementary Fig. 11). Similar ECS-induced CpG modifications were observed in both experiments (Fig. 3b), indicating that the observed CpG modification is predominantly neuronal in its origin and independent of DNA replication.
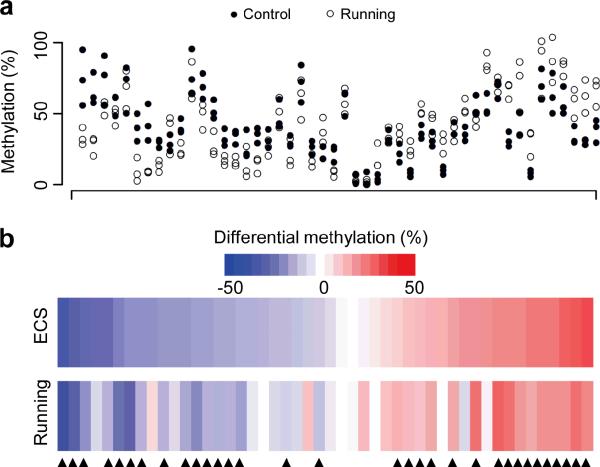
To further determine whether a physiological paradigm of neuronal stimulation could induce DNA methylation changes, we subjected adult mice to a three-day course of voluntary running and detected highly similar changes in these regions (Fig. 3b). Analysis of the same set of 48 CpGs by HpaII-qPCR analysis further showed that 67% of ECS-induced methylation changes were also observed from multiple animals after running (Fig. 4). Thus, widespread CpG methylation changes occur in post-mitotic neurons in vivo in response to both therapeutic and physiological neuronal activation.
Figure 4.
Voluntary exercise-induced CpG methylation changes in dentate granule cells of the adult mouse hippocampus. (a) Strip plot of methylation levels of CpGs from three control mice (filled circles) and three mice with running (open circles). Adult mice were housed in standard cages with or without free access to a running wheel for three days. Dentate gyrus tissues were microdissected for quantification of DNA methylation levels with HpaII-qPCR analysis for the same set of 48 CpGs as in Supplementary Fig 6a. (b) Heat maps for mean methylation changes of the set of 48 CpGs induced by a single ECS (at 4 hrs) or running (after three days). Values represent means from three sets of animals. About 67% of CpGs examined exhibit similar methylation changes 4 hrs after ECS and 3 days after running (indicated by arrowheads).
Genomic characteristics of activity-modified CpGs
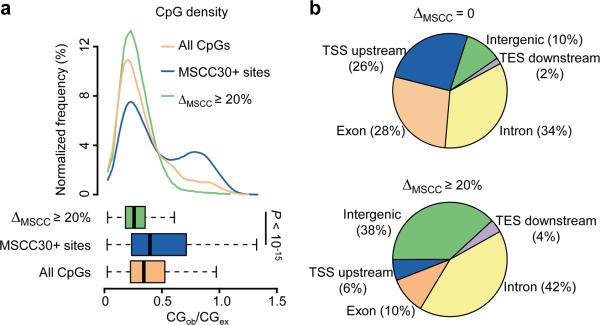
We next examined the genomic characteristics of the 3,050 MSCC hits at E4 (ΔMSCC, E4-E0 ≥ 20%). Previous DNA methylation analyses have mainly focused on CpG islands, which are highly clustered CpGs often located at gene promoter regions1, 2. Despite a modest bias of the CCGG sequence toward CpG islands, a striking exclusion of methylation changes from CpG-dense regions was observed (Fig. 5a), for both activity-induced de novo methylation and demethylation (Supplementary Fig. 12a). We further mapped MSCC sites to experimentally determined CpG islands38, and observed significant resistance of both gene-associated and intergenic CpG islands from activity-induced methylation changes (Supplementary Fig. 12b). Thus, low-density CpGs are main targets of activity-induced acute modifications in mature neurons in vivo, which is reminiscent of previously reported methylation changes during cell differentiation in vitro39.
Figure 5.
Genomic characteristics of activity-modified CpGs. (a) Enrichment of activity-induced methylation changes in regions with low CpG density. Top: distributions of CpG densities of 500 bp windows flanking activity-modified CpGs (green), all MSCC 30+ sites (blue), and all CpGs in the mouse genome (orange); Bottom: boxplots showing median and quartiles of the three distributions (P value, Student's t-test). (b) Distribution of modified CpGs in different genomic subregions. Shown are pie-charts of unchanged (top) and activity-modified CpGs (bottom) mapped to each genomic subregion. TSS upstream: within 5 kb upstream from the transcription start site (TSS); TES downstream: within 5 kb downstream from the transcription end site (TES); intergenic: over 5 kb away from any known genes.
We next mapped all MSCC hits to their nearby gene annotations. Unchanged and activity-modified CpGs exhibited significant differences in their distribution (Fig. 5b). The 3' downstream regions from the transcription end sites (TESs) are relatively depleted of MSCC sites, and minimally contribute to activity-modified CpGs (4%). Activity-modified CpGs are also under-represented in 5' upstream regions from gene TSS (putative promoters) and exonic regions, but slightly enriched in introns. Surprisingly, intergenic CpGs (≥ 5 kb away from any known genes) are most susceptible to changes by neuronal activity, comprising only 10% of CpGs with no methylation changes, but 38% of activity-modified CpGs (P < 10−15; chi-square test). Given the possibility of reactivation of DNA methylation-silenced retro-elements in intergenic regions40, we compared methylation patterns of 400 subclasses of repetitive elements in the mouse genome (Supplementary Table 5). While most of these repetitive elements were heavily methylated as expected, no significant changes were detected on a global level (Supplementary Fig. 13). It remains possible that the methylation status of an individual copy of retro-elements could be modified by neural activity.
We also cross-compared our single-base-resolution genome-wide neuronal DNA methylation map with recently reported activity-dependent transcript factor binding profiles of cultured mouse neurons41. Interestingly, although no significant enrichment for activity-induced methylation changes was observed, both intergenic and intragenic activity-regulated neuronal enhancers are significantly hypomethylated in dentate granule neurons in vivo (Supplementary Fig. 14a). Furthermore, activity-dependent transcription factor binding sites, including NPAS4 and SRF, are also hypomethylated (Supplementary Fig. 14b), revealing a common signature of DNA methylation at both constitutive and activity-dependent transcription factor binding sites in neurons.
Relationship between DNA methylation and gene expression
DNA methylation regulates gene expression in a highly context-dependent manner2. We obtained whole-genome expression profiles of dentate granule neurons at E0 and E4 using a mouse exon microarray (Supplementary Table 6). Integrated analysis comparing the relationship between genome-wide CpG methylation and gene expression with previously characterized cell types, including MSCC-profiled human B lymphocytes7 and MethylC-seq-profiled human embryonic stem cells34, revealed a distinct epigenetic pattern in the adult mouse dentate granule neurons in vivo (Supplementary Fig. 15a). Similar to other cell types, CpG methylation near TSS in dentate granule neurons is anti-correlated with the gene expression. Surprisingly, the previously observed positive correlation between gene body CpG methylation and gene expression is absent in these neurons. The anti-correlation between CpG methylation and gene expression is extended throughout the whole gene body into the 3' downstream regions from TESs. Furthermore, the robust anti-correlation was strongly dependent on low-density CpGs (Supplementary Fig. 15b), suggesting that the role of low-density CpGs in regulating gene expression may have been previously under-appreciated42.
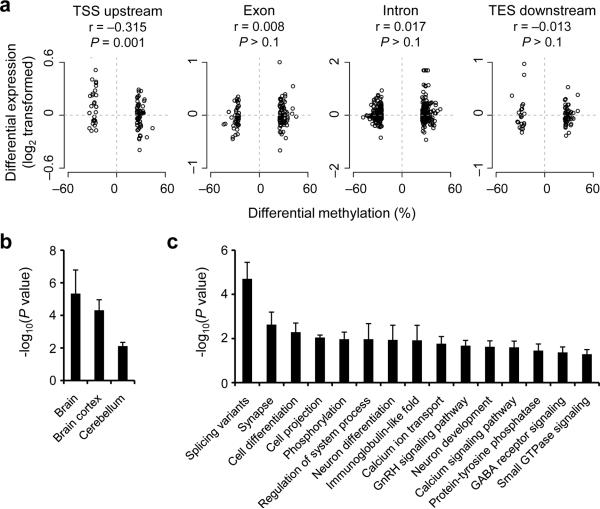
Although activity-modified CpGs are enriched in intergenic regions, 1,819 activity-modified CpGs are mapped to 1,518 genes (Supplementary Table 7). We next determined whether activity-induced changes in CpG methylation are correlated with changes in the expression of associated genes (Fig. 6a). The methylation changes located in TSS upstream regions (putative promoters) were modestly but significantly anti-correlated with changes in gene expression. We further confirmed the results using quantitative real-time PCR analysis of multiple individual animals (Supplementary Fig. 16; Supplementary Table 8). In contrast, no significant correlation was detected between methylation changes in other gene structures and gene expression changes (Fig. 6a). These results support the notion that activity-induced methylation changes may regulate gene expression in a highly context-dependent manner and may have roles other than directly regulating transcription levels. Our finding resembles the lack of correlation between active DNA modifications and gene expression changes observed during differentiation of human peripheral blood monocytes43.
Figure 6.
Correlation between changes in CpG methylation and gene expression and enrichment of activity-modified CpGs in brain-specific genes and neuronal pathways. (a) Correlation between activity-induced methylation changes of CpGs within different genomic subregions and mRNA level changes of associated genes between E4 and E0 (P values, Pearson's correlation test). (b, c) Tissue-specific expression (b) and GO analysis (c) of genes associated with activity-modified CpGs. Only non-redundant GO terms are shown in (c). To control for the gene length effect, only GO terms that show statistic significance using each of the three independent random CpG sets generated from all MSCC30+ sites are shown.
Activity-modified CpGs are enriched in brain genes
Finally, we examined the characteristics of the 1518 genes that are associated with the activity-modified 1819 CpGs. Using three randomly generated gene sets as the background to stringently control for gene length effects, the 1518 genes are significantly enriched with genes that are expressed in the brain (Fig. 6b). Activity-modified CpGs are also preferentially associated with alternative splicing variants (Fig. 6c), suggesting a potential role of DNA methylation changes in regulating alternative splicing in neurons.
Gene ontology (GO) analysis revealed significant over-representations of genes involved in synaptic function, protein phosphorylation, neuronal differentiation and the calcium signalling pathway (Fig. 6c; Supplementary Figs. 17–19; Supplementary Table 9), some of which are also enriched in activity-regulated genes at the mRNA level (Supplementary Fig. 20). Surprisingly, multiple genes encoding Notch signalling components exhibited CpG methylation and gene expression changes (Supplementary Fig. 21). Identification of the Notch signalling pathway as a novel epigenetic target of neuronal activity in mature neurons supports its emerging role in synaptic plasticity and long-term memory44.
DISCUSSION
Our study provides a striking view of how the epigenetic DNA methylation landscape of mature neurons is rapidly modified in response to external stimuli in vivo. Recent technical advances made it possible for accurate and large-scale profiling of CpG methylation status across the mammalian genome7, 33, 34, 39. In contrast to previous studies that have compared different mammalian cell types in vitro7, 33, 39 or mainly focused on CpGs within promoter regions in heterogeneous tissues45, we describe the first genome-wide, quantitative characterization of activity-induced acute CpG methylation changes for a defined mature neuronal subtype in vivo. Together with the gene expression profiles, these data sets will serve as a useful resource for the neuroscience community. The adult dentate gyrus provides a relatively homogenous population of post-mitotic neurons that can be activated synchronously in vivo and readily accessible in large quantities17. Such properties are particularly important for epigenetic analysis because each diploid cell displays only two locus-specific modifications. Our study reveals several key aspects of activity-induced epigenetic DNA modifications. First, our result provides direct evidence that extensive active DNA modifications occur in mature neurons in vivo, including DNA demethylation and de novo methylation. A significant subset of CpGs (~ 1.4%) in the genome of mature neurons is acutely modified in a site-specific fashion in response to various stimuli, including behavioural stimulation. Second, our analysis reveals novel characteristics of the genomic distribution of activity-induced DNA modifications in mature neurons in vivo (Supplementary Fig. 22). Highly clustered CpGs form CpG islands around TSSs of genes and are mostly unmethylated at the basal state. CpG-island-associated hypermethylation often occurs at tumor suppressor gene promoters in cancer46. Recent studies have also identified a subset of CpGs located near CpG islands, named CpG shores45, that are subjected to tissue-specific and/or cancer-related methylation. In contrast, activity-modified CpGs reside mostly in low CpG-density regions and in both intragenic and intergenic regions, resembling aging-associated CpG methylation changes of liver cells47. Third, our study identifies a large number of genes and pathways that are subject to modulation by neuronal activity at the level of DNA modification, including those that are not traditionally associated with mature neuronal functions, such as the Notch signalling pathway.
Our finding is supported by the continuous expression of DNMTs and putative DNA demethylation machinery genes (e.g., Gadd45, TDG, MBD4, TETs) in the adult brain3, 9, 48. Indeed, activity-induced de novo CpG methylation requires DNMT activity, consistent with recent genetic studies that identified a critical role of the persistent expression of these enzymes in activity-dependent neuronal plasticity20. We confirmed our previous finding that Gadd45b plays an essential role in activity-induced DNA demethylation17. The recent discovery of the TET family of proteins as mammalian 5mC hydroxylases has raised significant interest in the potential role of TET-catalyzed 5mC hydroxylation in active DNA demethylation4, 9. Interestingly, 5hmCs, the product of 5mC hydroxylation, are abundant in the adult dentate gyrus48. We recently show that Tet1 is required for activity-induced demethylation of Bdnf IX and Fgf1B promoters in adult dentate granule cells in vivo48. Because bisulfite sequencing and most methylation-sensitive restriction enzymes cannot distinguish 5mC and 5hmC48, new high-throughput methods to profile 5hmCs at the genome-scale with single-nucleotide resolution need to be developed.
Emerging evidence suggests that DNA modification regulates gene transcription in a highly context-dependent manner2. Densely methylated CpG islands are known to repress gene transcription1, 2, whereas methylation in adjacent “CpG shores” has been shown to be more widely and anti-correlated with tissue-specific gene expression45 (Supplementary Fig. 22). In some cases, low-density CpGs can also play a repressive role in regulatory regions49. In sharp contrast, recent epigenomic studies have suggested a novel function of gene body methylation in transcriptional activation7, 8. Furthermore, 5hmCs are shown to be intermediate products of DNA demethylation48, but may also play independent roles in regulating gene expression9. Our integrated analysis of the single base resolution DNA methylome and the whole-genome expression profile of endogenous neurons revealed a distinct pattern between gene body methylation and gene expression, compared with other previously profiled cell types7, 34. Given the high site-specificity, the broad genomic distribution, and the enrichment in intergenic and low CpG-density regions, activity-induced CpG methylation changes alone may not be sufficient for transcriptional control of their associated genes. Indeed, only CpG modifications in TSS upstream regions were modestly anti-correlated with gene expression changes. Thus, significant transcriptional control may require not only DNA methylation changes, but also the coordination with transcription factor binding and histone modifications. Neuronal activity-induced DNA modification could thus provide a permissive condition for other activity-dependent events that eventually leads to changes in the level, duration and/or isoforms of gene expression. It will be interesting to determine the potential function of both intergenic and intragenic activity-modified CpGs in the future.
The scope and specificity of activity-induced, relatively sustained DNA modifications in vivo suggest that such epigenetic regulation can serve as a general mechanism for activity-induced epigenetic control and long-lasting plasticity in neurons. Thus, in addition to histone modifications and transcription factors, epigenetic DNA modifications may expand the capacity of mature neurons for perpetuating transient stimuli into long-lasting covalent modifications in the nucleus. The MSCC method provides a cost-efficient and single-base resolution method for digital profiling of CpG methylation across the whole-genome and can be readily applied to other biological conditions. Given suggested roles of epigenetic aberrations in aging, neurological and psychiatric disorders3, 10, the inducible reversibility of epigenetic DNA methylation in the adult brain by neuronal activation and behavioural stimulation offers the possibility for novel therapeutic treatments.
METHODS
Biological samples
Adult mice (wild-type and Gadd45b knockout; 8–10 weeks old; male; C57BL/6 background) were housed in the standard facility. All animal procedures used in this study were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee. Electroconvulsive stimulation (ECS) was administered with pulses consisted of 1.0 s, 100 Hz, 18 mA stimulus of 0.3 ms delivered using the Ugo Basile ECT unit (Model 57800) as previously described17. Sham animals (E0) were similarly handled in parallel without the current delivery. Previous studies did not find any detectable damages resulted from ECS17, through assays including Western blot and immunostaining of the activated form of caspase (Caspase 3-a) and phospho-ATM (Ataxia telangiectasia mutated kinase). In one set of experiments, +3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP, 10 mg/kg body weight, i.p.) was injected 1 hr before ECS to block NMDA receptors. In another set of experiments, osmotic pumps (flow rate 1.0 μl/hr, pre-equilibrated in saline at 37°C over night; Alzet Model 1003D) were implanted to deliver one of the two structurally different DNMT inhibitors35, RG108 (1 mM; Sigma) and 5-azacytidine (1 mM; Sigma), or saline into one side of brain ventricles two days before ECS or sham treatment. Only ipsilateral dentate gyri from infusion experiments were used for DNA methylation analysis. In experiments with voluntary exercise (running) for three days, adult mice were randomly separated into two groups in standard home cages with or without free access to a mounted running wheel as described previously17.
To eliminate proliferating cells in the adult dentate gyrus, targeted irradiation at the hippocampus region was carried out as previously described37. Sham and irradiated mice were allowed to recover for five weeks before use when no difference in microglia numbers or activation was detected between sham and irradiated animals (Supplementary Fig. 10). To examine the status of DNA replication in the dentate gyrus of treated mice at use, animals were injected with BrdU (200 mg/kg body weight, i.p.) and sacrificed 2 hrs later. Coronal brain sections (40 μm thick) were prepared and processed for immunostaining using anti-BrdU antibodies (rat; 1:400; Accurate) as previously described17. Images were acquired on a Zeiss LSM 510 Live confocal system (Carl Zeiss). Stereological quantification of BrdU+ cells within the subgranular zone and granule cell layer were carried out as previously described (Supplementary Fig. 10; ref17).
Dentate gyrus tissues were rapidly micro-dissected bilaterally from adult mice unless specified; preparation was highly enriched for mature neurons as shown by immunohistology to contain ~90% NeuN+ neurons17. Each MSCC sample contained pooled DNA from two mice (four denate gyri). All validation experiments were performed using independent biological samples of bilaterally micro-dissected dentate gyri from individual animals.
In some experiments, fluorescence activated cell sorting (FACS) was used to further enrich the post-mitotic neuronal population and remove the potential contribution from glia, progenitors and other none neuronal cell types (Supplementary Fig. 11). Briefly, dentate gyrus tissues were removed from animals receiving ECS or sham treatment, flash-frozen in a mix of dry ice powder with 100% ethanol and transferred to −80°C for at least overnight. Tissue from each condition was pooled and homogenized on ice using a dounce homogenizer (Fisher) in lysis buffer containing 0.32 M sucrose, 5 mM CaCl2, 3 mM Mg(Acetate)2, 0.1 mM EDTA, 10 mM Tris-HCl pH 8.0, 1 mM DTT (Roche), and 0.1 % Triton X-100 for 1 min. Cell lysate was then loaded onto a sucrose gradient containing 1.8 mM sucrose, 3 mM Mg(Acetate)2, 10 mM Tris-HCl (pH 8.0) and 1 mM DTT, and centrifuged for 2.5 hrs at 26,000 rpm at 4°C (Beckman Coulter). The supernatant containing cell debris was aspirated and 500 μl of 1× PBS was added to the nuclear pellet. After 20 min on ice, the nuclear pellet was resuspended in PBS with gentle triteration. An immunostaining mixture containing 195 μl PBS with 0.6 μl anti-NeuN (Millipore), 1 μl R-PE (Invitrogen), and 5 μl normal goat sera was briefly vortexed, incubated for 5 min, and applied to the nuclei for 45 min in the dark. Control reactions were performed by omitting the primary antibody (negative control) and by adding 2 μg/ml 7-AAD (Sigma) as a nuclear counterstain (positive control). After incubation, stained nuclei were gently triterated and passed through a 40 μm filter into sorting tubes. Control samples were then used to determine the forward-scatter (FSC), side-scatter (SSC) nuclear profiles and background PE gating. NeuN+ nuclei from sham and ECS samples were purified using the FACSVantage SE (BD) into PBS according to FSC, SSC, and PE profiles. FACS profiles were processed using CellQuest software (BD). Lysis buffer was directly added to purified nuclei and genomic DNA was to harvested as previously described17. To verify FACS samples by immunofluorescence microscopy, the nuclear counterstain DAPI was added (2 μg/ml) to a post-sorted NeuN+ sample.
MSCC library construction, sequencing, mapping and data analysis
Genomic DNA was extracted and purified using Qiagen DNeasy Kit as previously described17. Concentrations and 260/280nm UV absorbance ratios were determined by Thermo NanoDrop2000 to ensure the sample quality. Typically, 1.5–2.5 μg genomic DNA can be obtained from each dentate gyrus. For each sample, four dentate gyri from two mice were pooled to increase the sample concentration and diminish micro-dissection variations.
Methyl-sensitive Cut Counting (MSCC)7 profiles CpG methylation status across the genome by using two restriction enzymes, the methylation-sensitive HpaII and the methylation-insensitive isoschizomer MspI. Both enzymes target the same CCGG sequence. HpaII cuts unmethylated CCGG sites, but not methylated or hydroxymethylated ones, whereas MspI cuts all CCGG sites. Sequencing read counts mapped to HpaII and MspI cutting sites provide quantitative and digital measurement of CpG methylation levels within the CCGG site. For the improved MSCC method (Supplementary Fig. 2), two libraries were constructed for each DNA sample, an HpaII library and an Inverse library, as previously described7. Briefly, two custom adaptors were synthesized by IDT. Adaptor A contains a 5' MmeI recognition site and a 5' CG overhang, while adaptor B contains a 3' NN overhang. Both adapters also contain end sequences required by Illumina library construction. For the HpaII Library, 2 μg of genomic DNA was digested with HpaII. Adaptor A was ligated to HpaII digested fragments. After digestion with MmeI, the resulting fragments were ligated to adaptor B. The final library fragments were then purified using 6% polyacrylamide gel. For the Inverse library, after DNA was digested with HpaII, the fragment ends were deactivated by treating with Antarctic Phosphatase. Then DNA was digested by MspI, purified and treated in the same manner as the HpaII library. After sequencing, the number of sequencing reads matched to a site from HpaII library came from the unmethylated sites; the number of sequencing reads matched to a site from the Inverse library came from the methylated or hydroxymethylated sites. Standard DNA with artificially introduced methylation levels was used to normalize the counts before estimating the methylation level.
For sequencing read alignment, a set of possible 17/18 bp tags was created from all CCGG sites in the mouse genome (mm9, downloaded from UCSC). Our custom Java-based software MOM was used to match sequencing reads to the set of tags50. Sequencing reads that can be mapped uniquely with up to one mismatch were tallied for each location and then used to calculate methylation level by the following formula as previously described7.
| (1) |
| (2) |
CpG sites with low counts (e.g. Inverse + HpaII < 30 counts) were excluded from further analysis.
R packages were used for all bioinformatic analysis unless specified. For CpG density calculation, 500 bp flanking sequences were fetched from mouse genome (mm9) for different subsets of MSCC sites. CpG density was calculated by the following formula:
| (3) |
where p(C) and p(G) denote the percentage of Cs and Gs in the sequence, respectively.
For functional annotation, MSCC sites were mapped to known genes using relevant tables from UCSC genome browser by in-house programs. Tissue-specificity and gene ontology analysis were done by DAVID (http://david.abcc.ncifcrf.gov). Three independent equal-sized gene sets were generated from all MSCC30+ sites as background sets to control for the gene length effect.
Bisulfite sequencing and HpaII-qPCR analysis
For bisulfite sequencing analysis, 1 μg of genomic DNA was treated with sodium bisulfite (Zymo) as previously described17. Sodium bisulfite converts unmethylated cytosines to uracils, whereas 5-methylcytosines and 5hmCs remain unconverted. The converted DNA was purified and 50 ng was used as the template in the 50 μl PCR reaction using Choice-Taq polymerase (Denville). Specific primers were designed to amplify regions of interest (listed in Supplementary Table 2). Fresh PCR products were purified (Qiagen) and cloned by TOPO-TA cloning method (Invitrogen) and sequenced. Unmethylated cytosines appear as thymines and methylated cytosines appear as cytosines. The efficiency of bisulfite conversion is confirmed by the complete conversion of non-CG cytosines to thymines in PCR-amplified sequences. We consider a difference of ≥ 20% in bisulfite sequencing as true differences in calculation of specificity and sensitivity of MSCC at different cut-offs by the following formulas:
| (4) |
| (5) |
For HpaII-based methylation-sensitive q-PCR analysis, 1 μg of genomic DNA was mock treated, or digested by HpaII or MspI for at least 4 hrs. After heat inactivation at 80°C for 20 min, the same volumes of reaction products were taken as templates for q-PCR with primers flanking the restriction sites (Supplementary Table 3). HpaII-resistant fraction was calculated as 2^(Ctundigested-CtHpaII). Less than 5% of MspI-resistant fractions confirmed the PCR specificity.
Expression profiling and bioinformatic analysis
Total RNA was extracted by TRIzol reagent (Invitrogen), and further purified by RNeasy columns (Qiagen). Processing, labelling, hydridization, and intensity acquisition was performed by Johns Hopkins Microarray Core Facility using standard protocols for Affymetrix GeneChip Mouse Exon 1.0ST Array. Two biological replicates were performed for both E0 and E4. Preprocessing of CEL files was performed using the oligo package in R. Raw intensities were log2-transformed, background-corrected, and quantile-normalized by RMA. Probe set information was downloaded from Affymetrix website. Using Refseq tables from UCSC Genome Browser, all non-exonic probesets were filtered out. Next, all exonic probesets within each Refseq transcript were averaged to obtain the gene-level expression values (Supplementary Table 6). To correlate CpG methylation and gene expression, relative distance of each MSCC site to its nearby gene(s) was calculated. The relative distance is represented by a value between − 0.5 to 1.5. Values between − 0.5 and 0 represent 5000 to 0 bp upstream from TSS. Value between 0 and 1 represent 0 to 100% along the gene body. Values between 1 and 1.5 represent 0 to 5000 bp downstream from TES. A moving window of 0.1 for relative distance was used to calculate averaged methylation levels for genes expressed at different levels, or Spearman's correlations of all sites within the window. To calculate the correlation between methylation changes and expression changes, MSCC sites of ΔMSCC ≥ 25% were used. For pathway analysis, genes with differential expression ≥ 0.5 were used.
Supplementary Material
ACKNOWLEDGEMENTS
We thank G. Church, S. Baylin, D. Ginty, and K. Christian for comments and suggestions, G. Sun for help with FACS, and W. Kim for providing some animals. This work was supported by NIH (AG024984, NS047344), McKnight, NARSAD, and Simons Foundations to H.S., by NIH (HD069184, NS048271), Johns Hopkins BSI, Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and NARSAD to G-l. M., and Lieber Institute start-up fund to Y.G.; J.U.G. was a FARMS fellow. M.H.J. was supported by a NIMH K99 award (MH090115). M.A.B. was a MSCRF fellow; M.P.B. was supported by grants from NIH to G. Church.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Accession Code. GEO accession number for microarray data: GSE30493.
REFERENCE
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Ma DK, et al. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–44. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 6.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 7.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–8. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-Hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011 doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 11.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–74. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mutagen. 2008;49:46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- 13.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1299–1440. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan JS, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 16.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–7. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller CA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–6. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–30. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–43. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–3. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 23.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 24.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 25.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–9. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–3. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 28.Murgatroyd C, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–66. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 29.McGowan PO, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisanby SH. Electroconvulsive therapy for depression. N Engl J Med. 2007;357:1939–45. doi: 10.1056/NEJMct075234. [DOI] [PubMed] [Google Scholar]
- 31.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–5. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nass MM. Differential methylation of mitochondrial and nuclear DNA in cultured mouse, hamster and virus-transformed hamster cells. In vivo and in vitro methylation. J Mol Biol. 1973;80:155–75. doi: 10.1016/0022-2836(73)90239-8. [DOI] [PubMed] [Google Scholar]
- 33.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–31. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 36.Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–31. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford EC, et al. Localized CT-Guided Irradiation Inhibits Neurogenesis in Specific Regions of the Adult Mouse Brain. Radiat Res. 2011;175:774–83. doi: 10.1667/RR2214.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Illingworth RS, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 41.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 43.Klug M, et al. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–55. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 45.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 47.Thompson RF, et al. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9:506–18. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–90. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- 50.Eaves HL, Gao Y. MOM: maximum oligonucleotide mapping. Bioinformatics. 2009;25:969–70. doi: 10.1093/bioinformatics/btp092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.