Abstract
The 5q- syndrome is a subtype of myelodysplastic syndrome (MDS) with a defined clinical phenotype associated with heterozygous deletions of Chromosome 5q. While no genes have been identified that undergo recurrent homozygous inactivation, functional studies have revealed individual genes that contribute to the clinical phenotype of MDS through haploinsufficient gene expression. Heterozygous loss of the RPS14 gene on 5q leads to activation of p53 in the erythroid lineage and the macrocytic anemia characteristic of the 5q- syndrome. The megakaryocytic and platelet phenotype of the 5q- syndrome has been attributed to heterozygous deletion of miR145 and miR146a. Murine models have implicated heterozygous loss of APC, EGR1, DIAPH1, and NPM1 in the pathophysiology of del(5q) MDS. These findings indicate that the phenotype of MDS patients with deletions of Chromosome 5q is due to haploinsufficiency of multiple genes.
Introduction
Among the diverse clinical presentations of myelodysplastic syndromes (MDS), a subset of patients has isolated deletion of Chromosome 5q and a distinctive set of features. This hematologic phenotype, termed the 5q- syndrome, includes a severe macrocytic anemia, a normal or elevated platelet count with hypolobated micromegakaryocytes, a normal or slightly decreased neutrophil count, and a low rate of progression of acute myeloid leukemia relative to other types of MDS.1–6 The association of a clinical phenotype with a chromosomal deletion has provided a scientific opportunity to attribute individual clinical features with deletion of particular genes.
Deletions of Chromosome 5q in MDS are somatically acquired, heterozygous, and encompass many genes.7 Heterozygous chromosomal deletions in cancer often highlight the locus of a tumor suppressor gene that undergoes homozygous inactivation, but in other cases, disease is caused directly by mono-allelic deletions due to haploinsufficiency for one or more genes. In the case of del(5q) MDS, genetic lesions on the non-deleted allele have not been identified, despite extensive searches for microdeletions or mutations that lead to homozygous inactivation.
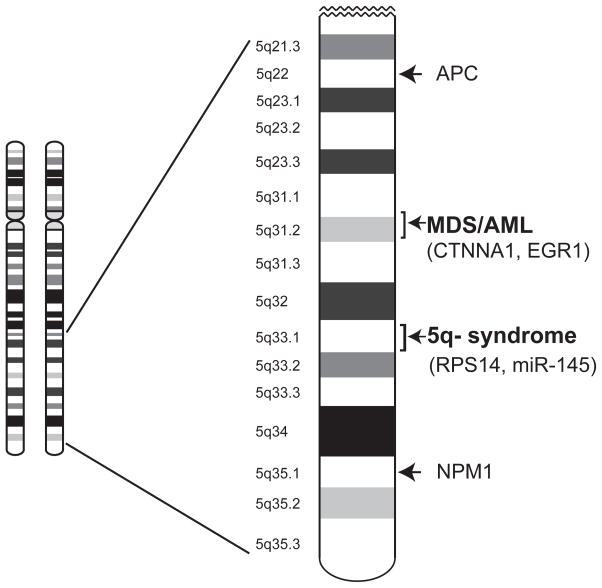
While the vast majority of patients with del(5q) MDS have large deletions, rare patients have smaller chromosomal deletions that have enable geneticists to localize common deleted regions (CDR) that are minimally necessary for a clinical phenotype. Two CDRs have been reported (Figure 1). The CDR located more distally on Chromosome 5q is minimally sufficient for the 5q- syndrome and is located at 5q52-33.8,9 The CDR located more proximal to the centromere is located at 5q31.10–12 Most patients have deletions that encompass both loci, but the CDRs provide a starting place for the search for critical genes.7
Figure 1.
Schema of the common deleted regions on Chromosome 5q for MDS and key genes.
Pathogenesis of anemia in del(5q) MDS
Anemia is the most prominent cytopenia in patients with the 5q- syndrome. The anemia is macrocytic, and patients are generally transfusion-dependent. Given the stability of the disease, with low rates of progression to acute myeloid leukemia, iron overload from chronic transfusions can be a significant cause of morbidity and mortality.7,13–15
The distal CDR, associated with the 5q- syndrome, contains 40 genes.9 Conditional deletion of this entire region recapitulates the severe macrocytic anemia in a murine model.16 Sequencing of genes and microarray-based analysis of copy number have failed to identify mutations or microdeletions that would lead to homozygous inactivation, fulfilling Knudson’s two hit hypothesis for a tumor suppressor.17 An alternative explanation is that heterozygous deletion of a critical gene is pathogenic. Since deletions of one allele are large, and no abnormalities have been reported on the allele, genetic studies have not been able to identify a crucial gene. Functional studies are an alternative approach to the molecular dissection of the 5q deletion and the identification of pathologic roles for individual genes.
The RPS14 gene was identified as a critical gene for the erythroid phenotype of the 5q- syndrome using an RNA interference screen.18 Each of the 40 genes in the 5q32-33 CDR were targeted by short hairpin RNAs, enabling a systematic evaluation of the effects of decreased expression of each gene. The shRNAs were introduced into primary human hematopoietic stem and progenitor cells using lentiviral vectors in order to examine the effects on hematopoietic differentiation. Only shRNAs targeting the RPS14 gene caused a severe block in erythroid differentiation. Moreover, forced over-expression of RPS14 in cells with del(5q) MDS rescued erythroid differentiation. Expression of RPS14 in del(5q) MDS is approximately 50% of expression non-del(5q) MDS, and the non-deleted allele is not deleted, demonstrating that RPS14 is a haploinsufficiency disease gene.18–21
Heterozygous inactivating mutations have been described in a congenital syndrome, Diamond Blackfan anemia (DBA).22,23 DBA patients have a severe macrocytic anemia, analogous to the erythroid defect in the 5q- syndrome. Approximately 25% of cases of DBA have mutations in the RPS19 gene, and mutations have now been reported in more than 8 ribosomal genes.22,24–27 The mutations are universally heterozygous, and functional studies indicate that homozygous inactivation of most if not all ribosomal genes would not be tolerated in a mammalian cell. In aggregate, the genetic data strongly implicate haploinsufficiency for ribosomal genes in the pathogenesis of macrocytic anemias of both DBA and the 5q- syndrome.23
Studies of both del(5q) MDS and DBA have demonstrated that induction of p53 is essential for the erythroid failure in the setting of ribosomal gene haploinsufficiency.16,28,29 Decreased expression of RPS14 and RPS19 causes a dramatic increase in total p53 levels, expression of p53 target genes including p21, and cell cycle arrest.30 In murine models, the macrocytic anemia associated with heterozygous loss of RPS14 (in combination with 8 adjacent genes) or heterozygous mutation of RPS19 in a p53 null background.16 These studies indicate that haploinsufficiency for ribosomal protein genes causes a p53-mediated cell cycle arrest in erythroid progenitor cells and a consequent macrocytic anemia.
Pathogenesis of dysmegakaryocyopoiesis
While thrombocytopenia is common in MDS in general, some patients with del(5q) MDS have an elevated platelet count, and patients commonly have hypolobated micromegakaryocytes.13 Patients with DBA do not have thrombocytosis, indicating that haploinsufficiency for a ribosomal gene does not generally cause an elevated platelet count or the distinctive megakaryocyte morphology. 23
A microRNA cluster is located within the CDR associated with the 5q- syndrome, containing miR-143 and miR-145. In addition, miR146a is located just telomeric to the CDR. Expression of miR-145 and miR-146a is lower in patients with del(5q) MDS compared with MDS patients with an intact Chromosome 5. In both human and murine cells, decreased expression of miR-145, or miR145 and miR-146a together, cause an increased expression of miR-145.31,32
While miR-145 and miR-146a likely regulate the expression of many genes, several critical targets have been identified. Two key regulators of the innate immune response, Toll-interluekin-1 receptor domain-containing adaptor protein (TIRAP) and tumor necrosis factor receptor-associated factor-6 (TRAF6) are targeted by miR-145 and miR-146a, respectively. In mice, decreased expression of both miR-145 and miR146, or forced over-expression of TRAF6, causes thrombocytosis and the hypolobated micromegaryocyte characteristic of the 5q- syndrome.32
The FLI-1 gene, encoding a critical transcriptional regulator of megakaryocyte differentiation, is also a target of miR-145.31 Decreased expression of miR-145 causes increased FLI-1 levels, with consequent increase in megkaryocyte production. In aggregate, these data indicate that the effects of haploinsufficiency for RPS14 and heterozygous deletion of miR145 and miR146a are integrated in the phenotype of the 5q- syndrome, and that the 5q- syndrome is a contiguous gene syndrome.
Clonal selection, disease progression, and other aspects of disease phenotype
The del(5q) lesion is present in the hematopoietic stem cell compartment and can be found in all lineages.33,34 Cells harboring del(5q) gain clonal advantage in the bone marrow, and the deletion remains present in leukemias that result from progression of del(5q) MDS. A systematic examination of all genes on 5q for genes that might contribute to clonal advantage of the del(5q) clone has not been reported. However, experiments in murine models have provided functional evidence for the role of individual genes in hematopoietic stem cell function and leukemic progression.7
The EGR1 gene is located in the 5q31 common deleted region. In a murine model with heterozygous inactivation of EGR1, hematopoietic stem cells have increased proliferation and mobilization from the bone marrow.35 In addition, mice with heterozygous loss of EGR1 develop leukemia with increased frequency and decreased latency in mice treated with a DNA alkylating agent, N-ethyl-nitrosourea (ENU), to induce secondary mutations.36 These findings indicate that heterozygous deletion of EGR1 may play a functional role in the pathogenesis of MDS and AML in patients with del(5q).
The APC gene is located on 5q23 is deleted in more than 95% of cases with del(5q) MDS. APC is a negative regulator of beta catenin function, and inactivating mutations in APC are pathogenic in colon cancer. Heterozygous deletion of APC in a mouse model causes expansion of the long-term hematopoietic stem cell population, but decreased repopulation of secondary recipients in bone marrow transplantation assays.37 Similarly, mice bearing the APC(min) allele, resulting in heterozygous loss of function for APC, have increased repopulating potential in primary bone marrow transplants, but decreased repopulation potential of secondary recipients due to loss of quiescence in the hematopoietic stem cell compartment.38 In both models, heterozygous inactivation of APC results in MDS or MDS/MPN phenotype. Given the role of beta catenin in stem cell self-renewal and hematologic malignancy, APC haploinsufficiency may contribute to the pathogenesis of del(5q) MDS through increased beta catenin activity.37,38
Multiple other genes have been implicated in the pathogenesis of del(5q) MDS, but the precise role of these genes in the phenotype of patients with del(5q) has not been established. The SPARC gene is located in the common deleted region for the 5q- syndrome, and homozygous inactivation of the SPARC gene in mice causes thrombocytopenia and decreased erythroid colony formation.39 SPARC has haploinsufficient expression in del(5q) MDS, and treatment with lenalidomide increases expression of the gene.19,40 The DIAPH1 gene encodes a protein involved in actin dynamics.41 In one proposed model, this gene may integrate the effects of haploinsufficiency for multiple genes on 5q, including RPS14 and EGR1.41
Phenotypic heterogeneity in patients with del(5q) MDS
The vast majority of patients (>95%) with del(5q) MDS have large deletions that encompass both defined CDRs and additional segments of the chromosome, but not all patients with isolated deletions of Chromosome 5q have the full collection of features ascribed to the 5q- syndrome. Additional molecular abnormalities, independent of Chromosome 5q, also contribute to the phenotype of patients with del(5q), including other cytogenetic abnormalities, somatic mutations in individual genes, and aberrant epigenetic alterations.42,43 Each of these abnormalities has the potential to alter hematopoietic differentiation and blood counts, the percentage of blast cells in the bone marrow, progression to leukemia, and overall survival. Thus the phenotypic heterogeneity of patients with del(5q) MDS is not surprising, but further research is required to define the precise contributions of these additional molecular abnormalities.
The size of deletions on Chromosome 5q is likely to alter disease phenotype, but this has yet to be demonstrated conclusively. Rare patients with small deletions have been extremely informative for the identification of common deleted regions, but the number of patients with these deletions is small, making strong genotype-phenotype associations difficult. Patients with monosomy for Chromosome 5 nearly always associated with multiple cytogenetic abnormalities, while del(5q) occurs commonly in isolation. It is also possible that deletions that encompass NPM1 on distal 5q may have a more aggressive phenotype given the genetic instability caused by NPM1 haploinsufficiency.44 Future studies with precise mapping of deletion boundaries in large numbers of patients will be necessary to determine whether the extent of deletions correlate with clinical phenotype.
The state of the bone marrow microenvironment in MDS pathogenesis is poorly understood, but is a possible contributor to disease phenotype. For example, osteoblasts are thought to be a critical component of the hematopoietic stem cell niche.45,46 Mice with dysfunctional osteoblasts due to selective inactivation of micro-RNA processing in osteoblast progenitor cells develop dysplasia of myeloid lineage cells with ineffective hematopoiesis.47 The role of the bone marrow microenvironment in patients with MDS remains to be determined.
Phenotypic heterogeneity in MDS patients with del(5q) is the inevitable consequence of the numerous influences on the del(5q) clone. These influences include additional clonal genetic abnormalities, epigenetic alterations, and perhaps an aberrant microenvironment and the status of normal (non-MDS) hematopoietic stem and progenitor cells.
Conclusions: validating haploinsufficiency disease genes
Increasing evidence supports the hypothesis that heterozygous deletions of Chromosome 5q in MDS causes haploinsufficiency for multiple genes that alter hematopoiesis. The phenotype encompassed by the 5q- syndrome is likely generated by the integration of effects from decreased expression of multiple genes. Specific aspects of the clinical phenotype have now been ascribed to distinct genes.
Validation of the functional importance of candidate genes within heterozygous deletions presents particular challenges. In general, genetic evidence of recurrent somatic mutations, translocations, or copy number alterations for a particular gene has provided incontrovertable evidence of the importance for of this gene in a type of cancer. This type of evidence has not been obtained for genes within the 5q deletion in MDS, despite a great deal of effort, and may never be found. Large heterozygous deletions may be the only recurrent genetic event. In this case, only functional studies can demonstrate the importance of a particular gene, but these studies are challenging and can lead to false conclusions.
A particularly appealing approach to heterozygous deletions is to evaluate all genes within a common deleted region systematically. This has been accomplished in the 5q32-33 common deleted region using two approaches, RNA interference and a series of conditionally floxed mice.18,48 The examination of all genes within a common deleted region removes the bias inherent in the examination of single candidate gene in isolation.
In theory, haploinsufficiency can be rescued by forced overexpression of the gene. In the case of del(5q) MDS, rescue of some aspect of the MDS phenotype by overexpression of a candidate gene in hematopoietic stem and progenitor cells that bear the 5q deletion provides powerful evidence in support of a pathogenic role for that gene. This experiment is associated with a number of technical challenges: viable progenitor cells from MDS patient bone marrow aspirates must be obtained, purified, and cultured; these cells must be efficiently transduced with a virus expressing the relevant cDNA or control; and expression of the cDNA must approximate endogenous expression, rescuing haploinsufficiency. For example, overexpression of the RPS14 gene in del(5q) MDS cells rescues erythroid differentiation.18 Such experiments are important for the validation of key candidate genes.
Murine models of heterozygous inactivation of candidate genes can provide a critical demonstration of the effects of haploinsufficiency for a gene in vivo. While the phenotype of murine models of hematologic malignancy does not always fully recapitulate the human disease, mice with conditional deletion of the 5q- syndrome common deleted region causes the expected macrocytic anemia.48 Murine models have also provided support for the effects of heterozygous loss of NPM1, EGR1, and APC.35–38,44
Functional studies, in vitro and in vivo, have provided critical insights into the biology of del(5q) MDS. Activation of p53 in response to heterozygous loss of RPS14, or other ribosomal genes that are mutated in Diamond Blackfan anemia, leads to a macrocytic anemia in multiple models. Anemia, thrombocytosis, and clonal advantage are likely due to the combined effects of haploinsufficiency for multiple genes. These studies provide an example for the elucidation of critical genes within other recurrent heterozygous deletions in human malignancies.
Acknowledgments
Rachel Murphy assisted with figure generation. This work was supported by grants This work was funded by the NIH (grants R01 HL082945 and P01 CA108631) and the Burroughs-Wellcome Fund (CAMS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Giagounidis AA, Germing U, Aul C. Biological and prognostic significance of chromosome 5q deletions in myeloid malignancies. Clin Cancer Res. 2006;12:5–10. doi: 10.1158/1078-0432.CCR-05-1437. [DOI] [PubMed] [Google Scholar]
- 2.Giagounidis AA, Germing U, Wainscoat JS, Boultwood J, Aul C. The 5q- syndrome. Hematology. 2004;9:271–277. doi: 10.1080/10245330410001723824. [DOI] [PubMed] [Google Scholar]
- 3.Teerenhovi L. Specificity of haematological indicators for ‘5q- syndrome’ in patients with myelodysplastic syndromes. Eur J Haematol. 1987;39:326–330. doi: 10.1111/j.1600-0609.1987.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berghe H, Michaux L. 5q-, twenty-five years later: a synopsis. Cancer Genet Cytogenet. 1997;94:1–7. doi: 10.1016/s0165-4608(96)00350-0. [DOI] [PubMed] [Google Scholar]
- 5.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, Sokal G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251:437–438. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 6.Van den Berghe H, Vermaelen K, Mecucci C, Barbieri D, Tricot G. The 5q-anomaly. Cancer Genet Cytogenet. 1985;17:189–255. doi: 10.1016/0165-4608(85)90016-0. [DOI] [PubMed] [Google Scholar]
- 7.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23:1252–1256. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 8.Boultwood J, Fidler C, Lewis S, Kelly S, Sheridan H, Littlewood TJ, Buckle VJ, Wainscoat JS. Molecular mapping of uncharacteristically small 5q deletions in two patients with the 5q- syndrome: delineation of the critical region on 5q and identification of a 5q- breakpoint. Genomics. 1994;19:425–432. doi: 10.1006/geno.1994.1090. [DOI] [PubMed] [Google Scholar]
- 9.Boultwood J, Fidler C, Strickson AJ, Watkins F, Gama S, Kearney L, Tosi S, Kasprzyk A, Cheng JF, Jaju RJ, Wainscoat JS. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 10.Fairman J, Chumakov I, Chinault AC, Nowell PC, Nagarajan L. Physical mapping of the minimal region of loss in 5q- chromosome. Proc Natl Acad Sci U S A. 1995;92:7406–7410. doi: 10.1073/pnas.92.16.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai F, Godley LA, Joslin J, Fernald AA, Liu J, Espinosa R, 3rd, Zhao N, Pamintuan L, Till BG, Larson RA, Qian Z, Le Beau MM. Transcript map and comparative analysis of the 1.5-Mb commonly deleted segment of human 5q31 in malignant myeloid diseases with a del(5q) Genomics. 2001;71:235–245. doi: 10.1006/geno.2000.6414. [DOI] [PubMed] [Google Scholar]
- 12.Zhao N, Stoffel A, Wang PW, Eisenbart JD, Espinosa R, 3rd, Larson RA, Le Beau MM. Molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases to 1–1.5 Mb and preparation of a PAC-based physical map. Proc Natl Acad Sci U S A. 1997;94:6948–6953. doi: 10.1073/pnas.94.13.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giagounidis AA, Aul C. The 5q- syndrome. Cancer Treat Res. 2008;142:133–148. [PubMed] [Google Scholar]
- 14.Mathew P, Tefferi A, Dewald GW, Goldberg SL, Su J, Hoagland HC, Noel P. The 5q- syndrome: a single-institution study of 43 consecutive patients. Blood. 1993;81:1040–1045. [PubMed] [Google Scholar]
- 15.Nimer SD. Clinical management of myelodysplastic syndromes with interstitial deletion of chromosome 5q. J Clin Oncol. 2006;24:2576–2582. doi: 10.1200/JCO.2005.03.6715. [DOI] [PubMed] [Google Scholar]
- 16.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2009;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boultwood J, Pellagatti A, Cattan H, Lawrie CH, Giagounidis A, Malcovati L, Della Porta MG, Jadersten M, Killick S, Fidler C, Cazzola M, Hellstrom-Lindberg E, Wainscoat JS. Gene expression profiling of CD34+ cells in patients with the 5q- syndrome. Br J Haematol. 2007;139:578–589. doi: 10.1111/j.1365-2141.2007.06833.x. [DOI] [PubMed] [Google Scholar]
- 20.Oliva EN, Cuzzola M, Nobile F, Ronco F, D’Errigo MG, Lagana C, Morabito F, Galimberti S, Cortelezzi A, Aloe Spiriti MA, Specchia G, Poloni A, Breccia M, Ghio R, Finelli C, Iacopino P, Alimena G, Latagliata R. Changes in RPS14 expression levels during lenalidomide treatment in Low- and Intermediate-1-risk myelodysplastic syndromes with chromosome 5q deletion. Eur J Haematol. 2010 doi: 10.1111/j.1600-0609.2010.01473.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Valencia A, Cervera J, Such E, Sanz MA, Sanz GF. Lack of RPS14 promoter aberrant methylation supports the haploinsufficiency model for the 5q- syndrome. Blood. 2008;112:918. doi: 10.1182/blood-2008-05-159707. [DOI] [PubMed] [Google Scholar]
- 22.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 23.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 25.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 26.Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr, Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellagatti A, Marafioti T, Paterson JC, Barlow JL, Drynan LF, Giagounidis A, Pileri SA, Cazzola M, McKenzie AN, Wainscoat JS, Boultwood J. Induction of p53 and up-regulation of the p53 pathway in the human 5q- syndrome. Blood. 2010;115:2721–2723. doi: 10.1182/blood-2009-12-259705. [DOI] [PubMed] [Google Scholar]
- 29.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2010 doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar MS, Narla A, Nonami A, Dimitrova N, Ball B, McAuley JR, Chin C, Chen CY, Kutok JL, Galili N, Raza A, Attar E, Gilliland DG, Jacks T, Ebert BL. Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. In submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R, Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson L, Astrand-Grundstrom I, Arvidsson I, Jacobsson B, Hellstrom-Lindberg E, Hast R, Jacobsen SE. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood. 2000;96:2012–2021. [PubMed] [Google Scholar]
- 34.Nilsson L, Eden P, Olsson E, Mansson R, Astrand-Grundstrom I, Strombeck B, Theilgaard-Monch K, Anderson K, Hast R, Hellstrom-Lindberg E, Samuelsson J, Bergh G, Nerlov C, Johansson B, Sigvardsson M, Borg A, Jacobsen SE. The molecular signature of MDS stem cells supports a stem-cell origin of 5q myelodysplastic syndromes. Blood. 2007;110:3005–3014. doi: 10.1182/blood-2007-03-079368. [DOI] [PubMed] [Google Scholar]
- 35.Min IM, Pietramaggiori G, Kim FS, Passegue E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Fernald AA, Anastasi J, Le Beau MM, Qian Z. Haploinsufficiency of Apc leads to ineffective hematopoiesis. Blood. 2010;115:3481–3488. doi: 10.1182/blood-2009-11-251835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane SW, Sykes SM, Al-Shahrour F, Shterental S, Paktinat M, Lo Celso C, Jesneck JL, Ebert BL, Williams DA, Gilliland DG. The Apc(min) mouse has altered hematopoietic stem cell function and provides a model for MPD/MDS. Blood. 2010;115:3489–3497. doi: 10.1182/blood-2009-11-251728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann S, O’Kelly J, Raynaud S, Funk SE, Sage EH, Koeffler HP. Common deleted genes in the 5q- syndrome: thrombocytopenia and reduced erythroid colony formation in SPARC null mice. Leukemia. 2007;21:1931–1936. doi: 10.1038/sj.leu.2404852. [DOI] [PubMed] [Google Scholar]
- 40.Pellagatti A, Jadersten M, Forsblom AM, Cattan H, Christensson B, Emanuelsson EK, Merup M, Nilsson L, Samuelsson J, Sander B, Wainscoat JS, Boultwood J, Hellstrom-Lindberg E. Lenalidomide inhibits the malignant clone and up-regulates the SPARC gene mapping to the commonly deleted region in 5q- syndrome patients. Proc Natl Acad Sci U S A. 2007;104:11406–11411. doi: 10.1073/pnas.0610477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisenmann KM, Dykema KJ, Matheson SF, Kent NF, DeWard AD, West RA, Tibes R, Furge KA, Alberts AS. 5q- myelodysplastic syndromes: chromosome 5q genes direct a tumor-suppression network sensing actin dynamics. Oncogene. 2009;28:3429–3441. doi: 10.1038/onc.2009.207. [DOI] [PubMed] [Google Scholar]
- 42.Patnaik MM, Lasho TL, Finke CM, Gangat N, Caramazza D, Holtan SG, Pardanani A, Knudson RA, Ketterling RP, Chen D, Hoyer JD, Hanson CA, Tefferi A. WHO-defined ‘myelodysplastic syndrome with isolated del(5q)’ in 88 consecutive patients: survival data, leukemic transformation rates and prevalence of JAK2, MPL and IDH mutations. Leukemia. 2010;24:1283–1289. doi: 10.1038/leu.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haase D, Germing U, Schanz J, Pfeilstocker M, Nosslinger T, Hildebrandt B, Kundgen A, Lubbert M, Kunzmann R, Giagounidis AA, Aul C, Trumper L, Krieger O, Stauder R, Muller TH, Wimazal F, Valent P, Fonatsch C, Steidl C. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 44.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 45.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 46.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 47.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, Aung Z, Matza M, Merkenschlager M, Lin C, Rommens JM, Scadden DT. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]