Summary
The mechanisms underlying the ability of axons to regrow after injury remain poorly explored at the molecular genetic level. We used a laser injury model in Caenorhabditis elegans mechanosensory neurons to screen 654 conserved genes for regulators of axonal regrowth. We uncover several functional clusters of genes that promote or repress regrowth, including genes classically known to affect membrane excitability, neurotransmission, and synaptic vesicle endocytosis. The conserved Arf Guanine nucleotide Exchange Factor (GEF), EFA-6, acts as an intrinsic inhibitor of regrowth. By combining genetics and in vivo imaging we show that EFA-6 inhibits regrowth via microtubule dynamics, independent of its Arf GEF activity. Among newly identified regrowth inhibitors, only loss of function in EFA-6 partially bypasses the requirement for DLK-1 kinase. Identification of these pathways significantly expands our understanding of the genetic basis of axonal injury responses and repair.
Keywords: C. elegans, genetic screen, axon regeneration, laser axotomy, kinases, EFA6, Slit, Robo, extracellular matrix, DLK kinase, endocytosis, microtubule dynamics
Introduction
Damage to the adult mammalian CNS, in stroke or in spinal cord injury, remains devastatingly untreatable. Despite significant recent advances in our understanding of selected pathways, strategies for treating CNS injury remain limited. Axon injury in mature neurons triggers injury responses and repair pathways (Abe and Cavalli, 2008). These pathways activate regrowth programs whose effectiveness depends on both the intrinsic growth competence of the neuron (Sun and He, 2010) and the local extracellular environment (Busch and Silver, 2007). Much attention has focused on the regrowth-inhibiting properties of CNS myelin components such as Nogo (Schwab, 2010). However, the roles of specific myelin components in vivo remain a matter of debate (Cafferty et al., 2010; Lee et al., 2010).
Compared to the effects of extrinsic cues, less is known about intrinsic mechanisms affecting regrowth competence. Experimental paradigms such as the conditioning lesion show that neuronal sensitivity to extrinsic influences in regeneration is under the control of intrinsic pathways (Enes et al., 2010; Hannila and Filbin, 2008; Ylera et al., 2009). Intrinsic triggers of regrowth include positive injury signaling pathways such as the MAP kinases Erk and JNK, which are activated by injury and retrogradely transported from sites of damage (Perlson et al., 2005). Differences in regenerative ability at different stages also reflect alterations in intrinsic growth capacity (Moore et al., 2009).
Analysis of regeneration-competent neurons in the vertebrate PNS and in model organisms has given insight into pathways that promote axon regrowth after injury (Ambron et al., 1996; Chen et al., 2007). Several studies have used genomic or proteomic approaches to identify regeneration-associated genes (Michaelevski et al., 2010). As yet, a limited number of such genes have been tested for function in vivo. An important goal is to exploit new models for large scale screening and gene discovery that will open up additional therapeutic avenues.
The nematode C. elegans is an emerging model for genetic and chemical screens for factors affecting axon regeneration after injury (Ghosh-Roy and Chisholm, 2010; Samara et al., 2010; Wang and Jin, 2010). Axons labeled with GFP transgenes can be severed precisely with ultrafast laser irradiation (Yanik et al., 2004). Although laser axotomy of single axons differs in the precise mechanism of damage from mechanical severing or crush injuries of vertebrate nerves, at least some regrowth mechanisms are conserved. In C. elegans, as in vertebrate neurons, the second messengers Ca2+ and cAMP are rate limiting for axonal regrowth (Ghosh-Roy et al., 2010; Neumann et al., 2002; Qiu et al., 2002). Pharmacological screening in C. elegans revealed a conserved role for protein kinase C in regenerative growth (Samara et al., 2010). Finally, the Dual Leucine Zipper Kinase/DLK-1 cascade was first demonstrated in C. elegans as essential for axonal regrowth (Hammarlund et al., 2009; Yan et al., 2009) and is required for axon regeneration in Drosophila (Xiong et al., 2010) and likely in mouse (Itoh et al., 2009). These results suggest that axon regrowth after laser surgery involves pathways required in other models of regeneration.
Here we exploit the rich genetic resources of C. elegans to perform a large scale mutation-based screen for genes with roles in adult axon regrowth. We identify many genes required for axonal regrowth, most of which are not required for developmental axon outgrowth and have not previously been implicated in axon regeneration. By analyzing regeneration at multiple time points and in double mutants we order the activity of newly characterized genes relative to each other and to the DLK-1 cascade. Manipulation of the conserved pathways identified here could significantly expand current strategies to augment the regenerative abilities of damaged neurons.
Results
Functional screen for axon regrowth genes
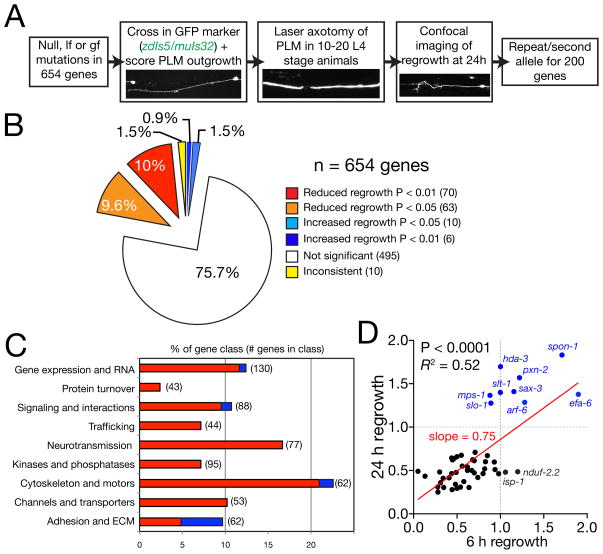
To identify conserved genetic pathways affecting axon regrowth we selected >650 C. elegans genes based on their orthology to human genes and potential neuronal function or known biochemical role (Figure 1A; see Experimental Procedures). We focused on genes not essential for overall health or growth rate; for >90% of the genes, we examined genetic null mutants (Table S1). To assay axon regrowth in vivo we used mechanosensory PLM neurons, which consistently regrow after laser axotomy (Wu et al., 2007). Over 95% of mutants displayed normal PLM axon development; mutants with aberrant development are summarized in Table 3. In the primary screen we severed the PLM axon using femtosecond laser surgery in 10–20 animals per genotype. Under our conditions >95% of PLM neurons survive surgery (Wu et al., 2007). After 2–4 h the proximal axon stump swells and forms a growth cone-like structure that extends over the next 24–48 h. Wild type PLM axons regrow in an error-prone manner and can re-establish synaptic connections in certain genetic backgrounds (Ghosh-Roy et al., 2010). Mutants showing altered regrowth at 24 h (Figure 1B,C; Tables 1,2) were re-tested in a secondary screen (~200 genes). As we sever axons in the mid-L4 stage when animals are growing, reduced regrowth could also reflect developmental delay or arrest in response to our axotomy procedure. We measured the growth of intact neurons in selected strains and found no significant effects on organismal growth rate (Figure S1A).
Figure 1. Overview and results of axon regrowth screen.
(A) Flow chart of screen strategy. (B) Pie chart showing fraction of genes screened displaying significantly reduced or increased regrowth at 24 h. (C) Distribution of increased/decreased regrowth (P < 0.01 and P < 0.05) mutants among nine functional or structural gene classes, shown as % of genes in each class. Color code as (B) except that genes with P < 0.05 (orange, light blue) are omitted. See Table S1 for lists of genes in each class. (D) Total regrowth at 6 h and at 24 h are significantly correlated among 50 genes tested (Pearson r = 0.7, P < 0.0001). Each dot represents a single gene/mutant. Red line, linear regression; slope = 0.72, R2 = 0.52, P < 0.001. Two mitochondrial mutants (isp-1, nduf-2.2) display normal regrowth at 6h and reduced regrowth at 24 h suggesting mitochondrial function becomes important during later regrowth; see also Figure S1.
Table 3.
Roles of axon outgrowth and branching genes in PLM regrowth.
| Gene name | Mutations | Normalized regrowth | P value | PLM Development | Molecular function | Closest human gene |
|---|---|---|---|---|---|---|
| a. Known axon outgrowth mutants
| ||||||
| unc-53 | e404 | 0.12 ± 0.05 | *** | 100% US | NAV | NAV2 |
| unc-115 | e2225 | 0.28 ± 0.04 | ** | 5% OS, 14% branchless | Actin-binding LIM | ABLIM1 |
| unc-73 | e936(lf), rh40 | 0.31 ± 0.1 | *** | 100% US | GEF | TRIO |
| unc-69 | e587, ju69 | 0.43 ± 0.09 | * | 60% US (e587); Normal (ju69) | Kinesin interactor | FEX2 |
| unc-51 | e369, ky347(lf) | 0.47 ± 0.10 | * | 30% US (ky347) | Atg1 kinase | ULK2 |
| unc-76 | e911 | 0.67 ± 0.12 | ns | 95% US, 80% branchless | Kinesin interactor | SCOC |
| unc-16 | e109 | 0.68 ± 0.09 | ns | 7% OS, 10% branchless | JIP1 scaffold | SPAG9 |
| unc-44 | e362 | 0.77 ± 0.22 | * | 5% OS, 5% US | Ankyrin | ANK1 |
| unc-33 | e204 | 0.79 ± 0.12 | ns | 2% OS, 10% branchless | CRMP/TOAD | DPYSL3 |
|
| ||||||
| b. Other mutants with reduced PLM axon outgrowth or branching defects
| ||||||
| deg-3 | u662 | 0.42 ± 0.07 | *** | 50% short axons | nAChR subunit | CHRNA7 |
| pig-1 | gm144 | 0.55 ± 0.08 | * | 40% US | leucine zipper kinase | MELK |
| wsp-1 | gm324 | 0.83 ± 0.08 | ns | 90% branchless | WASP | WASL |
| ddr-1 | ok874 | 0.74 ± 0.12 | ns | 25% branchless | Discoidin domain receptor | DDR2 |
| hst-2 | ok595 | 0.56 ± 0.10 | * | 11% branchless | Heparan sulfotransferase | HS2ST1 |
| klc-1 | ok2609 | 0.82 ± 0.07 | ns | 10% extra branch | Kinesin light chain | hKLC1B |
| klp-10 | ok704 | 0.85 ± 0.06 | ns | 15% extra branch | Kinesin-like | KIF15 |
| nfi-1 | qa524 | 0.79 ± 0.13 | ns | 50% wavy process | Nuclear factor I | NF1A |
| oig-1 | ok1687 | 0.70 ± 0.10 | ns | 26% extra branch | One Ig Domain | PTPRF? |
| rack-1 | tm2262 | 0.74 ± 0.10 | ns | 10% extra branch | Receptor of C- kinase | GNB2L1 |
| ver-3 | gk227 | 1.28 ± 0.17 | ns | 10% extra branch | VEGFR | FLT1 |
| F09A5.1 | ok2286 | 0.86 ± 0.08 | * | 18% extra branch | Spinster | SPINL |
| vang-1 | ok1142 | 0.78 ± 0.10 | ns | 40% proximal branch | Van Gogh/Strabismus | VANGL1 |
| Y38H8A.4 | ok2793 | 0.73 ± 0.07 | * | 20% branchless | Serine/Threonine kinase | TSSK2 |
Table 1.
Selected mutants displaying reduced PLM regrowth
| Gene name | Mutations | Normalized regrowth (24 h) | P value | PLM Development | Molecular function | Closest human gene |
|---|---|---|---|---|---|---|
| 1. Cell Adhesion and ECM
| ||||||
| sdn-1 | zh20 | 0.25 ± 0.06 | <0.001 | Normal | Syndecan | SDC2 |
| sax- 7/lad-1 | ok1244 | 0.30 ± 0.05 | 0.001 | Normal | L1 IgCAM | NRCAM |
| rig-3 | ok2156 | 0.43 ± 0.04 | 0.001 | Normal | GPI-linked IgCAM | NCAM1 |
| rig-4 | ok1160 | 0.52 ± 0.06 | 0.001 | Normal | Sidekick IgCAM | SDK1 |
|
| ||||||
| 2. Channels and transporters
| ||||||
| unc-1 | e719 | 0.67 ± 0.08 | 0.037 | Normal | Stomatin | STOM |
| unc-32 | e189(lf) | 0.41 ± 0.05 | 0.020 | Normal | V0 ATPase | ATP6V0A1 |
| nkb-1 | ok1089 | 0.45 ± 0.05 | 0.039 | 12% US | Na+/K+ ATPase β subunit | ATP1B1 |
|
| ||||||
| 3. Cytoskeleton and transport
| ||||||
| ebp-1 | tm1357 | 0.29 ± 0.02 | 0.001 | Normal | MT End-binding protein | MAPRE1 |
| unc-14 | ju56 | 0.13 ± 0.02 | *** | Normal | RUN domain | RUSC2 |
| unc-104 | e1265 | 0.37 ± 0.12 | 0.009 | Normal | KIF1A kinesin | KIF1B |
| kap-1 | ok676 | 0.46 ± 0.06 | 0.002 | Normal | Kinesin- associated | KIFAP3 |
| jip-1 | km18 | 0.71 ± 0.08 | 0.018 | Normal | JNK interacting protein | MAPK8IP2 |
|
| ||||||
| 4. Protein kinases and phosphatases
| ||||||
| max-2 | ok1904 | 0.35 ± 0.09 | 0.054 | Normal | p21 activated kinase | PAK3 |
| ptp-3 | ok244 | 0.63 ± 0.1 | *** | Normal | LAR receptor tyrosine phosphatase | PTPRD |
| pptr-1 | tm2954 | 0.51 ± 0.09 | 0.031 | Normal | PP2A regulatory subunit | PPP2R5E |
|
| ||||||
| 5. Neurotransmission, Lipid signaling, Metabolism
| ||||||
| dgk-1 | nu62, ok1482 | 0.49 ± 0.07 | 0.001 | Normal | Diacylglycerol kinase q | DGKQ |
| sphk-1 | ok1097 | 0.65 ± 0.05 | 0.039 | Normal | Sphingosine kinase | SPHK1 |
| nduf-2.2 | ok437 | 0.52 ± 0.08 | ** | Normal | NADH Ubiquinone reductase | NDUFS2 |
| isp-1 | qm150 | 0.46 ± 0.09 | 0.017 | Normal | Fe-S protein | UQCRFS1 |
| F45E4.5 | ok560, ok2285 | 0.48 ± 0.09 | 0.008 | Normal | Piccolo/Aczonin | PCLO |
| ric-3 | md158 | 0.34 ± 0.05 | ** | Normal | AChR maturation | RIC3 |
| unc-25 | e156 | 0.61 ± 0.08 | 0.047 | Normal | Glutamic acid decarboxylase | GAD1 |
| cha-1 | p1152 | 0.61 ± 0.08 | <0.001 | Normal | Choline acetyltransferase | CHAT |
| unc-17 | e245, e113 | 0.41 ± 0.09 | <0.001 | 40% Branchless | ACh vesicular transporter | SLC18A3 |
| tph-1 | mg280 | 0.50 ± 0.07 | 0.024 | Normal | Tryptophan hydroxylase | TPH2 |
|
| ||||||
| 6. Cell signaling and protein interaction domains
| ||||||
| cwn-2 | ok895 | 0.48 ± 0.05 | 0.001 | Normal | Wnt ligand | WNT5B |
| tag-60 | ok2292 | 0.30 ± 0.06 | *** | Normal | PDZ domain | SLC9A3R2 |
| tag-68 | gk185 | 0.48 ± 0.07 | *** | Normal | I-Smad | SMAD6 |
| dab-1 | gk291 | 0.51 ± 0.08 | 0.001 | Normal | Disabled | DAB2 |
|
| ||||||
| 7. Membrane trafficking
| ||||||
| unc-26 | e1196 | 0.31 ± 0.05 | 0.002 | Normal | Synaptojanin | SYNJ1 |
| unc-57 | e406 | 0.21 ± 0.04 | 0.001 | Normal | Endophilin | SH3GL3 |
| sec-22 | ok3053 | 0.63 ± 0.04 | 0.017 | 28% OS | TI-VAMP | SEC22B |
| unc-41 | e268 | 0.42 ± 0.08 | <0.001 | Normal | Stonin | STON2 |
|
| ||||||
| 8. Protein degradation, proteases, cell death
| ||||||
| uev-3 | ju639 | 0.51 ± 0.14 | <0.001 | Normal | Ubiquitin E2 ligase variant | UBE2N |
|
| ||||||
| 9. RNA metabolism, chromatin, transcription factors, translation
| ||||||
| alg-1 | gk214 | 0.72 ± 0.15 | 0.012 | Normal | Argonaute-like | EIF2C4 |
| unc-75 | e950 | 0.52 ± 0.06 | 0.001 | Normal | RRM RNA- binding | TNRC4 |
| ngn-1 | ok2200 | 0.41 ± 0.05 | ** | 10% OS | Neurogenin bHLH | NEUROD1 |
| npp-12 | ok335 | 0.64 ± 0.13 | 0.013 | Normal | Gp210 nuclear pore complex | NUP210 |
| spr-1 | ok2144 | 0.55 ± 0.08 | 0.015 | ND | CoREST | RCOR3 |
| xpo-3 | ok1271 | 0.50 ± 0.08 | 0.014 | 60% OS | Exportin-t | XPOT |
| egl-27 | ok151 | 0.44 ± 0.08 | 0.025 | Normal | Nucleosome remodeling | MTA1 |
Genes are classified in nine functional or structural classes. Mutations are genetic or predicted molecular nulls, unless partial loss of function (lf) indicated. Normalized regrowth is relative to matched same-day controls or to pooled controls. Significance levels (*, P < 0.05; **, P < 0.01; ***; P < 0.001) based on Mann-Whitney test in primary screen; Numerical P values are from repeats, after correction for multiple comparisons. ‘PLM development’ is scored in at least 20 worms per genotype; defects in other touch neurons are not indicated here; OS, overshooting axon. ‘Closest human gene’ based on BLASTP score in Wormbase WS213; Ensembl/HGNC gene symbol.
Table 2.
Selected mutants displaying increased PLM regrowth
| Gene name | Mutations | Normalized regrowth | P value | PLM Development | Molecular function | Closest human gene |
|---|---|---|---|---|---|---|
| pxn-2 | ju358 | 1.97 ± 0.19 | 0.009 | Normal | Peroxidasin ECM enzyme | PXDN |
| spon-1 | ju430ts | 1.77 ± 0.41 | (0.082) | Normal | F-spondin ECM | SPON1 |
| vab-19 | e1036cs | 1.63 ± 0.19 | (0.0003) | Normal | Ankyrin repeat protein | KANK1 |
| sax-3 | ky123 | 1.43 ± 0.10 | 0.009 | (mixed defects) | Robo receptor for SLT-1 | ROBO2 |
| efa-6 | ok3533, tm3124 | 1.42 ± 0.11 | 0.014 | 40% OS | Arf6 GEF | PSD4 |
| mps-1 | ok1376 | 1.41 ± 0.10 | 0.043 | Normal | K+ channel accessory subunit | KCNE2 |
| slt-1 | eh15 | 1.39 ± 0.10 | 0.013 | Normal | Slit ligand | SLIT1 |
| hda-3 | ok1991 | 1.58 ± 0.14 | (0.1051) | Normal | Histone deacetylase | HDAC1 |
Conventions as for Table 1. The increased regrowth of pxn-2 mutants (Gotenstein et al., 2010) is included for comparison.
Altered regrowth 24 h post axotomy could reflect defects in growth cone formation or in later processes of axon extension. We analyzed 60 mutants with altered regrowth at 24 h for their effects at 6 h post axotomy, when wild-type axons have just begun to extend (Figure S1B). Most mutants with reduced regrowth at 24 h displayed proportional effects at 6 h (Figure 1D), suggesting these genes act throughout regrowth. However, some mutants displaying increased regrowth at 24 h (e.g. slt-1, sax-3; see Figure 3E,F) did not significantly affect regrowth at 6 h, suggesting these genes affect later axon extension. Conversely, a few mutants (efa-6; see Figure 4C) displayed larger increases in regrowth at 6 h than at 24 h, suggesting a preferential effect on early stages of regrowth.
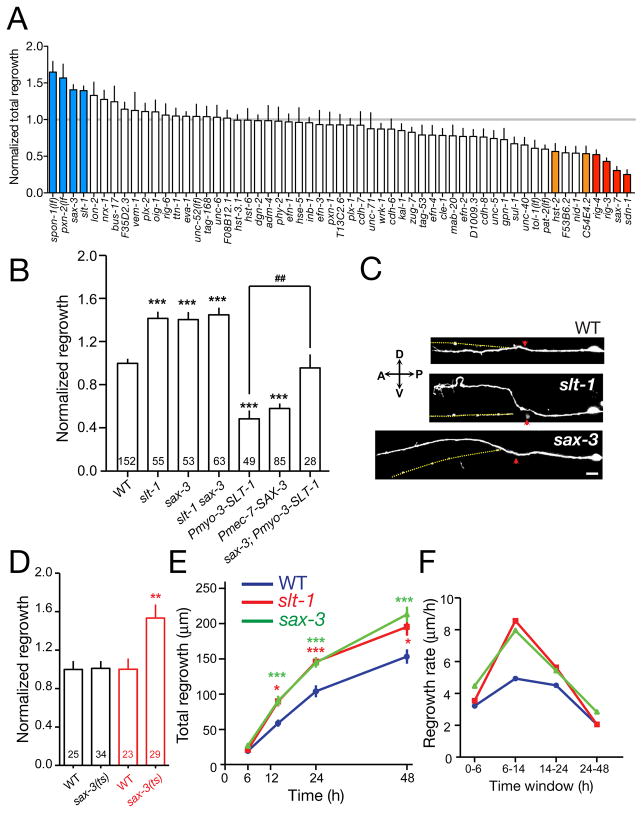
Figure 3. Slit/Robo signals inhibit PLM axon extension.
(A) Normalized 24 h PLM regrowth in mutants affecting axon guidance, cell adhesion, and extracellular matrix. (B) slt-1 and sax-3 mutants display increased PLM regrowth at 24 h; overexpression of SLT-1 in body wall muscles (kyEx441) or of SAX-3 in touch neurons (juEx2219) caused reduced regrowth. The inhibitory effect of SLT-1 overexpression is dependent on sax-3. Regrowth normalized to WT (zdIs5) = 1 ± 0.04 (mean ± SEM). (C) Representative images of PLM axon regrowth in slt-1 and sax-3 mutants at 24 h; red arrows indicate lesion sites, yellow dotted lines indicate original path of PLM. Scale, 10 μm. (D) Reduced SAX-3 activity after axotomy enhances regrowth. When shifted from 20°C to 25°C immediately after axotomy (red), sax-3(ky200ts) mutants displayed increased regrowth compared to unshifted ky200 animals (black). (E, F) slt-1 and sax-3 mutants display faster axon extension in the 6–24 h time period. Statistics, t test; n values in columns; ***, P < 0.001; ##, P < 0.01; *, P < 0.05; ns, not significant.
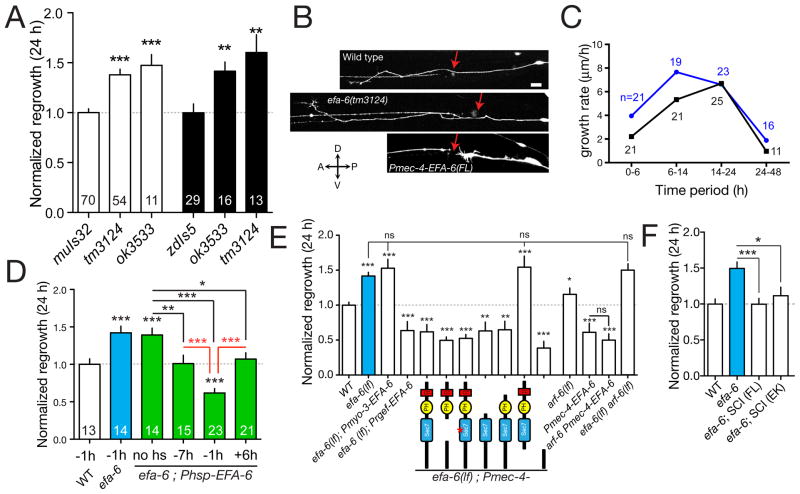
Figure 4. EFA-6 inhibits the early phase of axon regrowth.
(A) PLM axon regrowth at 24 h is increased in efa-6 mutants, normalized to controls (n values in bars). (B) Images of wild type and efa-6 axons at 24 h. Red arrows, site of axotomy. Scale, 10 μm. (C) Axon growth is increased in efa-6 from 0–14 h post axotomy but not later. (D) Inducible overexpression of EFA-6 can inhibit regrowth only at the time of axotomy (time of heat shock relative to axotomy in h). (E) The effect of efa-6(tm3124) on axon regrowth can be reversed by overexpression of EFA-6 using the mec-4 (touch neuron) or rgef-1 (pan-neural) promoters, but not the myo-3 (muscle) promoter. The EFA-6 N-terminus, but not the Sec7 GEF domain, is necessary and sufficient to inhibit regrowth. Domain deletions or point mutations indicated below; n ≥ 9 for each condition. (F) Rescue of efa-6(lf) by single-copy insertions (SCI) of full-length EFA-6 (juSi51) or EFA-6(E447K) (juSi53). n ≥ 11 for each condition. All charts show mean ± SEM; statistics, t test; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. See also Figure S4.
We wished to determine the extent to which genes required for initial regrowth were involved in growth cone formation. By 6 h post axotomy, between 40–60% of wild type PLM axon stumps form growth cones; on average, axons with growth cones at 6 h extend further than those without growth cones (Figure S1C), suggesting growth cones reflect growth rather than stalling. Among 50 mutants tested at 6 h the fraction of growth cones positively correlated with regrowth (R2 = 0.11, P = 0.01, Figure S1D), suggesting many genes required for regrowth affect growth cone formation. At 24 h the fraction of growth cones did not correlate with regrowth (not shown), possibly reflecting a more stochastic presence of growth cones in axon extension. However, mutants displaying increased regrowth at 6 h (e.g. efa-6) did not display a higher fraction of growth cones than wild type (Figure S1D), suggesting the wild type level of growth cone formation is a phenotypic ceiling. We conclude that growth cones correlate with early regrowth but not with overall regrowth at later time points.
Clusters of genes required for axonal regrowth but not for development
We found genes affecting PLM regrowth among all structural and functional classes tested (Figure 1B,C; Tables 1–4). When analyzed as nine gene classes (Figure 1C), genes promoting regrowth (i.e. those displaying reduced growth in loss of function mutants) were more frequent in the ‘cytoskeleton and motors’ and ‘neurotransmission’ classes. Genes inhibiting regrowth (i.e. increased regrowth in loss of function mutants) were concentrated in the ‘cell adhesion/extracellular matrix’ class (Figure 1C). Here for reasons of space we describe our findings on selected genes among channels and transporters, neurotransmitters, and gene expression.
Table 4.
Genes required for normal regrowth of PLM.
| (A) | |
|---|---|
| Seq name | Gene name |
| F48F7.1 | alg-1 |
| F56H1.5 | ccpp-1 |
| ZC416.8b | cha-1 |
| ZK632.6 | cnx-1 |
| W01B6.1 | cwn-2 |
| M110.5a | dab-1 |
| C09E10.2 | dgk-1 |
| T14E8.3 | dop-3 |
| F47A4.2 | dpy-22 |
| Y59A8B.17 | ebp-1 |
| C04A2.3 | egl-27 |
| F42A6.7 | hrp-1 |
| F46C3.3 | hum-4 |
| F42G8.12 | isp-1 |
| F56D12.4 | jip-1 |
| T24H10.7 | jun-1 |
| F08F8.3 | kap-1 |
| F20C5.2 | klp-11 |
| C07H6.7 | lin-39 |
| Y38F1A.10 | max-2 |
| T26A5.3 | nduf-2.2 |
| ZK1290.4 | nfi-1 |
| Y69A2AR.29 | ngn-1 |
| C17E4.9 | nkb-1 |
| T23H12.1 | npp-12 |
| W03G1.6 | pig-1 |
| W08G11.4 | pptr-1 |
| C09D8.1 | ptp-3 |
| W07A12.7 | rhy-1 |
| T14A8.1 | ric-3 |
| C53B7.1 | rig-3 |
| F45E4.7 | rip-1 |
| C18F3.2 | sax-7 |
| F57C7.3 | sdn-1 |
| R31.1 | sma-1 |
| R12B2.1 | sma-4 |
| C34C6.5 | sphk-1 |
| Y52D3.1 | strd-1 |
| F59F5.6 | syd-2 |
| F31E8.3 | tab-1 |
| F37D6.6 | tag-68 |
| ZK1290.2 | tph-1 |
| F39B2.2 | uev-1 |
| F26H9.7 | uev-3 |
| K03E6.2 | unc-1 |
| C52E12.2 | unc-104 |
| F09B9.2a | unc-115 |
| K10D3.2 | unc-14 |
| ZC416.8 | unc-17 |
| Y37D8A.23 | unc-25 |
| JC8.10 | unc-26 |
| ZK637.8a | unc-32 |
| Y50D4C.1 | unc-34 |
| C27H6.1 | unc-41 |
| B0350.2 | unc-44 |
| Y60A3A.1 | unc-51 |
| F45E10.1 | unc-53 |
| T04D1.3 | unc-57 |
| T07A5.6 | unc-69 |
| F55C7.7 | unc-73 |
| C17D12.2 | unc-75 |
| B0041.7 | xnp-1 |
| C49H3.10 | xpo-3 |
| C34D1.5 | zip-5 |
| T23F11.1 | |
| (B) | |
|---|---|
| Seq name | Gene name |
| T07D3.7 | alg-2 |
| K04C2.4 | brd-1 |
| C01G6.8 | cam-1 |
| C34G6.5 | cdc7 |
| F18F11.3 | cdh-8 |
| ZK1236.2 | cec-1 |
| F56D1.6 | cex-1 |
| C02F5.4 | cids-1 |
| Y73F8A.19 | cpna-4 |
| ZK856.1 | cul-5 |
| F25E2.5 | daf-3 |
| C25F6.4 | ddr-1 |
| F11D5.3 | ddr-2 |
| F22B7.10 | dpy-19 |
| W08D2.1 | egl-20 |
| M01D7.7 | egl-30 |
| C08C3.1 | egl-5 |
| F56E3.4 | fax-1 |
| H09G03.2 | frm-8 |
| F47D12.1 | gar-2 |
| Y81G3A.3 | gcn-2 |
| F38A6.3 | hif-1 |
| C18E3.8 | hop-1 |
| K01G5.2 | hpl-2 |
| C34F6.4 | hst-2 |
| F46F2.2 | kin-20 |
| C09H6.2 | lin-10 |
| Y71G12B.20 | mab-20 |
| R10E9.1 | msi-1 |
| F44G3.9 | nhr-111 |
| F26H11.2 | nurf-1 |
| F49E8.4 | pam-1 |
| K06B9.5 | pax-2 |
| T07E3.6 | pdf-1 |
| F57F5.5 | pkc-1 |
| R09E10.7 | pqn-55 |
| Y113G7B.23 | psa-1 |
| Y42H9AR.3 | rabs-5 |
| Y42H9B.2 | rig-4 |
| C18H9.7 | rpy-1 |
| T03D8.3 | sbt-1 |
| F55A4.1 | sec-22 |
| F31E8.2 | snt-1 |
| F20D6.4 | srp-7 |
| K09C8.4 | sul-1 |
| C33D12.6 | tag-312 |
| F39B2.2 | uev-1 |
| F57H12.2 | unc-24 |
| F56A8.7 | unc-64 |
| K11C4.3 | unc-70 |
| C01G10.11 | unc-76 |
| R13A1.4 | unc-8 |
| F25C8.3 | unc-80 |
| C30A5.7 | unc-86 |
| R10E11.3a | usp-46 |
| Y44E3B.1 | zip-4 |
| K10G9.1 | |
| F09G2.1 | |
| Y106G6H.14 | |
| ZK418.9 | |
(A) 70 genes with significantly reduced regrowth in primary screen (P < 0.01), listed by sequence (seq) name and CGC name. (B) 61 genes displaying reduced regrowth with P < 0.05.
Neuronal excitability can promote regrowth (Brushart et al., 2002), but can also act as an intrinsic negative signal via L-type voltage gated calcium channels (Enes et al., 2010). In C. elegans neuronal excitability is generally influenced by the opposing action of voltage-gated calcium and potassium channels (Goodman et al., 1998); the voltage-gated Ca2+ channel EGL-19 is required for regrowth of PLM neurons (Ghosh-Roy et al., 2010). We tested 53 additional channels and associated proteins (Figure S2A), and found a cluster of genes affecting both Ca2+ and Na+ ionic balance to be critical for regrowth, including the Ca2+ channel regulator UNC-80 (Jospin et al., 2007), the Na+ pump NKB-1 (Doi and Iwasaki, 2008), the stomatins UNC-1 and UNC-24 (Sedensky et al., 2004) and the Deg/ENaC Na+ channel UNC-8 (Tavernarakis et al., 1997). Among these genes, UNC-24 and UNC-1 interact with UNC-8 and with the mechanosensory channel complex, suggesting electrical activity regulated by mechanosensory channels could promote regrowth (Bounoutas and Chalfie, 2007). Conversely, loss of function in the BK type K+ channel SLO-1 (Wang et al., 2001) or in the conserved K+ channel regulatory protein MPS-1 (Cai et al., 2005) resulted in enhanced regrowth. As loss of function in K+ channels should tend to increase membrane excitability, these findings suggest excitability promotes PLM regrowth.
PLM regrowth was strongly reduced in mutants affecting chemical neurotransmitters, including acetylcholine (cha-1/ChAT and unc-17/vesicular ACh transporter), GABA (unc-25/GAD), and biogenic amines (tph-1/Tryptophan hydroxylase) (Figure S2B). Mutants affecting ACh synthesis or packaging (cha-1, unc-17), or AChR biosynthesis (ric-3) displayed reduced regrowth, suggesting a neurotransmitter role of ACh is important. PLM expresses AChRs containing the DEG-3 subunit (Treinin and Chalfie, 1995), and we find that deg-3 mutants display strongly reduced regrowth (Table 3). Although deg-3(u662) mutants also display aberrant PLM development, PLM morphology was normal in other cholinergic mutants tested (cha-1, etc), suggesting the requirement for ACh in regrowth is separable from any role in development.
Mechanosensory neurons are neither GABAergic nor receive GABAergic input, suggesting an indirect role of GABA in regrowth. Notably, regrowth did not require genes involved in GABA vesicular packaging (unc-46, unc-47) or the postsynaptic muscle GABA receptor (unc-49). GABA has nonsynaptic growth-promoting roles in vertebrate neuronal development (Akerman and Cline, 2007) and a trophic role in regenerating vertebrate neurons (Shim and Ming, 2010; Toyoda et al., 2003). Speculatively, regenerating neurons may become more dependent on trophic factors whose roles in development are masked by genetic redundancy.
The DLK-1 MAPK cascade is essential for axon regrowth after injury (Hammarlund et al., 2009; Yan et al., 2009). We screened over 80 additional protein kinases, representing ~1/4 of all conserved C. elegans kinases (Manning, 2005), as well as selected protein phosphatases (Figure S3). In addition to the members of the DLK-1 MAPK cascade, several cytosolic kinases were important for regrowth, including the stress-activated KGB/MEK-1 pathway, the p21-activated kinase MAX-2 and the Atg1 kinase UNC-51 kinase. Of these, only MAX-2 and UNC-51 have been previously linked to axonogenesis in C. elegans (Lucanic et al., 2006; Ogura et al., 1994); UNC-51, but not MAX-2 is required for PLM developmental outgrowth (Table 3). We also find that PKC-1/protein kinase C can promote PLM regrowth, consistent with a recent report (Samara et al., 2010). Additionally, among 12 protein phosphatases tested, we identified the LAR-like receptor tyrosine phosphatase PTP-3 (Ackley et al., 2005) and the PP2A regulatory subunit PPTR-1 as critical for regrowth (Table 1; Figure S3C). LAR has been implicated in axon regrowth in vertebrates (Xie et al., 2001). To our knowledge PP2A has not been linked to axon regrowth. In C. elegans PPTR-1 negatively regulates Akt signaling (Padmanabhan et al., 2009) and ribonucleoprotein (RNP) particle stability (Gallo et al., 2010). Loss of function in akt-1 or akt-2 did not significantly affect regrowth (Figure S3A). AKT-1 and AKT-2 could play redundant roles; alternatively PPTR-1 may promote regrowth via RNP stabilization.
Axonal injury induces pervasive changes in gene expression (Yang et al., 2006) and our previous studies implicated bZip proteins in regrowth (Ghosh-Roy et al., 2010; Yan et al., 2009). We tested 130 additional genes implicated in RNA metabolism, transcription and translation, as well as specific transcription factors. The Argonaute-like protein ALG-1 (Grishok et al., 2001) was critical for regrowth, implying a regrowth-promoting role for microRNAs. Several proteins affecting chromatin remodeling were required, including the SWI/SNF complex component XNP-1/ATR-X. Conversely, loss of function in the histone deacetylase HDA-3/HDAC3 improved regrowth (Table 2); as loss of HDA-3 function is neuroprotective in a C. elegans model of polyglutamine toxicity (Bates et al., 2006), HDA-3 may act generally to repress neuroprotective genes. Of 63 transcription factors tested, the neurogenin bHLH family member NGN-1 (Nakano et al., 2010) showed a strong requirement (Table 1). As PLM neuron differentiation was normal in ngn-1 mutants, NGN-1/neurogenin may specifically promote regrowth. The range of gene expression regulators identified here underscores the complexity of the changes in gene expression following axonal injury.
Axon regrowth requires genes functioning in synaptic vesicle endocytosis
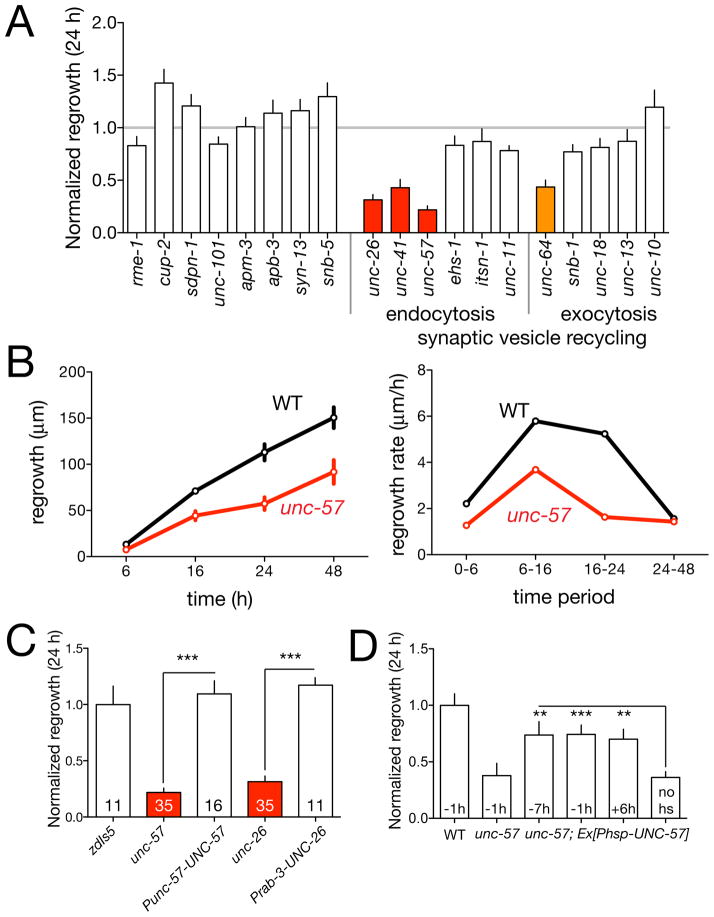
Axon regrowth was strongly reduced in a cluster of mutants previously thought to be dedicated to synaptic vesicle (SV) recycling (Figure 2A), including unc-26/Synaptojanin, unc-57/Endophilin, and unc-41/Stonin. These are ‘core module’ proteins or ‘secondary effectors’ in SV endocytosis (Dittman and Ryan, 2009). In contrast, genes involved in SV exocytosis, such as unc-13/mUnc13, unc-18/mUnc18, or unc-10/Rim, were not required for regrowth (Figure 2A). Both unc-26 and unc-57 mutants displayed significantly reduced regrowth at 6 h; unc-57 mutants displayed reduced regrowth from 6 h to 24 h, but not from 24–48 h (Figure 2B). Expression of UNC-57 driven by its own promoter, or pan-neural expression of UNC-26 rescued axon regrowth defects, supporting the view that the SV endocytosis genes are required cell-autonomously for axon regrowth (Figure 2C). To address whether UNC-57 acts continuously in regrowth we expressed it under the control of a heat shock promoter and induced UNC-57 expression by heat shock at times before and after axotomy. Heat shock-induced expression of UNC-57 either 7 h before or 6 h after axotomy could rescue the defects of unc-57 mutants (Figure 2D), suggesting a continuous requirement in regenerative growth. As we sever the PLM axon at sites distant from synapses, and SV exocytosis genes appear to be dispensable for axon regrowth, the requirement for SV endocytosis genes in regrowth may be independent of known roles in SV recycling.
Figure 2. Regrowth requires a subset of synaptic vesicle recycling genes.
(A) Normalized regrowth in mutants lacking selected synaptic vesicle and trafficking genes (mean ± SEM). (B) Timecourse of regrowth in unc-57/Endophilin mutants (mean ± SEM); growth rates plotted for each time period. (C) Transgenic rescue of the unc-26 and unc-57 regrowth phenotypes. (D) Heat shock induced expression of UNC-57 can rescue the unc-57 regrowth phenotypes when animals are heat shocked before or after axotomy. Statistics, t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Permissive and inhibitory roles of extracellular factors
Axon regeneration is influenced in many ways by the extracellular environment We tested ~60 genes encoding extracellular matrix components, putative cell adhesion proteins, and cell surface receptors (Figure 3A). Several such genes were required for regrowth (Table 1), including the cell surface proteoglycan SDN-1/Syndecan (Rhiner et al., 2005), the L1CAM ortholog SAX-7/LAD-1 (Chen et al., 2001), the novel GPIlinked IgCAM RIG-3 and the IgCAM RIG-4/Sidekick (Schwarz et al., 2009). In vertebrate axons L1 is upregulated after injury and required for regrowth (Becker et al., 2004); however syndecans or sidekick family members have not previously been implicated in axon regeneration. Conversely, loss of function in several putative basement membrane components, such as spon-1/F-spondin (Woo et al., 2008) or pxn-2/Peroxidasin (Gotenstein et al., 2010) resulted in enhanced regrowth (Table 2). In vertebrates the ‘glial scar’ is an ECM barrier to CNS regeneration (Busch and Silver, 2007); although C. elegans does not encode orthologs of glial scar components such as chondroitin sulfate proteoglycans, these observations raise the possibility that the basement membrane forms an analogous barrier to PLM regrowth.
Wnt signals regulate the polarity of PLM neurite outgrowth in development (Hilliard and Bargmann, 2006). We find PLM regrowth involves distinct Wnt signals. For example the Wnt CWN-2 is not required for PLM development yet is required for regrowth (Table 1). CWN-2 is expressed anterior to PLM, suggesting it could be permissive or attractive in PLM regrowth, similar to its roles in other neurons (Kennerdell et al., 2009; Song et al., 2010).
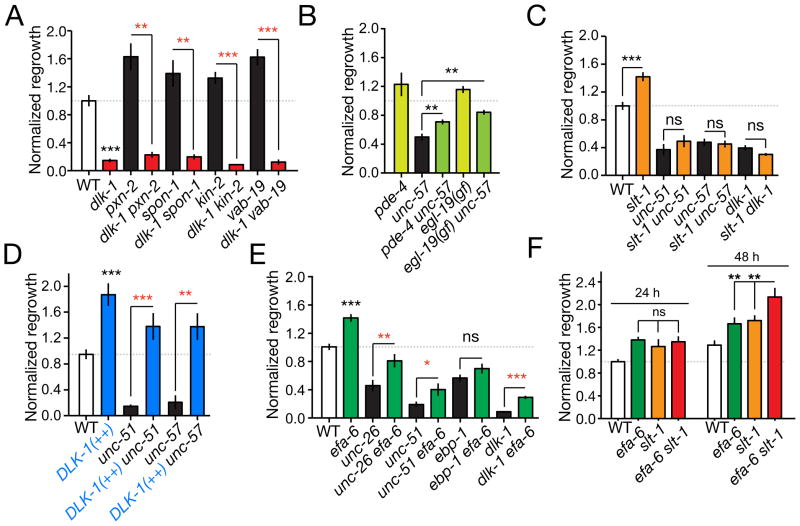
Among tested axon guidance pathways, Slit-Robo signaling had an inhibitory effect on regeneration. Both slt-1/Slit and sax-3/Robo null mutants displayed increased PLM regrowth, and slt-1 sax-3 double mutants showed no further enhancement in axon regeneration than either single mutant (Figure 3B,C). Further, overexpression of SAX-3 in touch neurons inhibited PLM regrowth, indicating SAX-3/Robo can act cell autonomously to restrain regrowth (Figure 3B). Constitutive expression of SLT-1 from body wall muscles also reduced PLM regrowth in a SAX-3-dependent manner (Figure 3B). In development, SAX-3 activity has a minor role in promoting PLM outgrowth (Li et al., 2008). To address whether SAX-3 acts at the time of regrowth or earlier we performed temperature shift experiments on sax-3(ky200ts) (Zallen et al., 1998), and found that animals shifted to the restrictive temperature immediately post-axotomy exhibited increased regrowth equivalent to sax-3 null mutants (Figure 3D), indicating that SAX-3 acts at the time of regrowth. Last, we addressed when in regrowth SLT-1 and SAX-3 signals acted. slt-1 and sax-3 mutants displayed normal regrowth from 0–6 h then increased regrowth during the 6–24 h period (Figure 3E,F), suggesting SLT-1 and SAX-3 signals inhibit extension of the regrowing axon. In ventrally guided AVM axons, SLT-1 signals play repulsive roles in development and regrowth (Gabel et al., 2008; Hao et al., 2001). In contrast, in PLM neurons SAX-3/Robo appears to switch from a growth-promoting role during development to an inhibitory role in regrowth.
The conserved signaling protein EFA-6 is an intrinsic inhibitor of regeneration
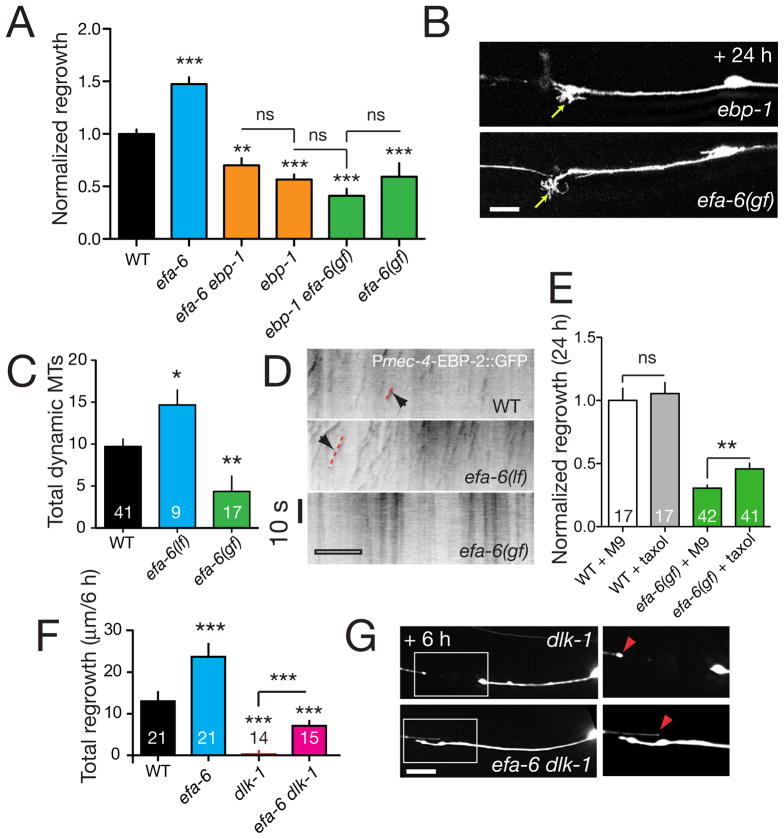
Among the few genes with inhibitory effects on regrowth we focused on EFA-6, the C. elegans member of the EFA6 (Exchange Factor for Arf6) family. EFA6 proteins contain a variable N-terminal region, a Sec7 homology domain with GEF activity specific to ARF6 GTPases (Franco et al., 1999), a pleckstrin homology (PH) domain, and a coiled-coil domain (Figure S4A). C. elegans efa-6 mutants displayed mild PLM axon overshooting in development (Figure S4B,C) and enhanced regrowth of PLM (Figure 4A,B). Cell-type specific transgenic expression of EFA-6 from pan-neural or touch neuron-specific promoters, but not from a muscle-specific promoter, rescued efa-6 developmental defects (Figure S4C) and inhibited PLM regrowth after axotomy both in efa-6(lf) (Figure 4E) and efa-6(+) backgrounds (not shown; see also Figure 5), indicating EFA-6 acts cell autonomously and that PLM regrowth is sensitive to EFA-6 levels. In contrast to slt-1 or sax-3, efa-6 mutants displayed enhanced regrowth during the 0–6 h period (Figure 1D, 4C), implying EFA-6 acts early in regrowth. Furthermore, heat shock induced EFA-6 overexpression 1 h before axotomy inhibited PLM regrowth, whereas induction earlier or later had little effect (Figure 4D).
Figure 5. EFA-6 negatively regulates axonal microtubule dynamics downstream or in parallel to DLK-1.
(A) Normalized regrowth of efa-6, ebp-1, and double mutants. efa-6(lf) does not bypass the requirement for ebp-1, nor does EFA-6 overexpression enhance the regrowth reduction of ebp-1 mutants. (B) Expanded but immotile growth cone like structures formed in severed axon stumps in ebp-1(lf) mutants and EFA-6 overexpressors at 24 h post axotomy; cf. the lack of growth cones in axon stumps of dlk-1 mutants (panel F). (C,D) Analysis of MT dynamics in regrowing axons; number of dynamic MTs (EBP-2::GFP nucleation events) detected in kymographs is indicated in bars (C). efa-6(tm3124) mutants display increased numbers of dynamic MTs. Overexpression of the EFA-6 N-terminus (efa-6(gf), juEx3533) decreases the number of dynamic MTs. (D) Kymographs of MT dynamics in PLM axons 3 h post axotomy in the 40 μm region proximal to the site of axotomy, visualized with Pmec-4-EBP-2::GFP (juEx2843); scale, 10 μm. (E) Microinjection of taxol increases regrowth in efa-6(gf) animals compared to buffer-injected controls. The effect of taxol on wild type (muIs32) is not significant. (F) Regrowth 6 h post axotomy is increased in dlk-1 efa-6 double mutants compared to dlk-1 single mutants. (G) Images of dlk-1 single and double mutant axons at 6 h post axotomy. The region in the boxed area is enlarged at right. Red arrowheads indicate end of distal fragment closest to axotomy site. Scales, 10 μm. All charts show mean ± SEM; statistics, t test or Mann-Whitney test; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
To investigate the mechanism underlying EFA-6 function, we next examined arf-6(lf) mutants. arf-6(lf) mutants displayed modestly increased regrowth and did not further enhance efa-6(lf) in regrowth (Figure 4E). However, EFA-6-overexpressing transgenes potently inhibited regrowth in arf-6(lf) backgrounds (Figure 4E), suggesting EFA-6 acts on regrowth independent of ARF-6. To dissect which functional domains of EFA-6 were important in axon regrowth we expressed mutant EFA-6 lacking either the Sec7 domain, the PH domain, or the C-terminal coiled coil domain (Table S2). Each of these ‘gain of function’ transgenes rescued efa-6 developmental overgrowth (Figure S4B) and inhibited regrowth, as did constructs in which the conserved catalytic residue of the Sec7 domain was mutated (E447K). In contrast, expression of an EFA-6 variant lacking the N-terminus did not block PLM regrowth (Figure 4E). As overexpression of EFA-6 might affect non-physiological pathways, we made single-copy insertion transgenes expressing full length EFA-6 or the E447K mutant and found that both rescued efa-6 developmental and regrowth phenotypes (Figure 4F), suggesting a GEF-independent role for EFA-6 in inhibiting regrowth.
Our recent studies on C. elegans embryos indicate that EFA-6 regulates microtubule (MT) growth by promoting MT catastrophe (O’Rourke et al., 2010). Axon regeneration also involves precise regulation of axonal MT dynamics (Erturk et al., 2007). In our screen we found that the MT plus-end binding protein EBP-1 (Srayko et al., 2005) was required for regrowth (Figure 5A). The reduced regrowth of ebp-1 mutants could not be bypassed by efa-6(lf) and was not further decreased by EFA-6 overexpression (Figure 5A). Notably, the morphology of the axon stumps in ebp-1 mutants resembled those in EFA-6 overexpressors (Figure 5B), suggesting the increased regrowth in efa-6 mutants might reflect decreased axonal MT dynamics.
To test whether EFA-6 affected axonal MT dynamics we expressed the MT plus-end binding protein EBP-2 fused to GFP (see Experimental Procedures). End binding protein GFP fusions are established markers of growing ends of MTs in vertebrate neurons (Stepanova et al., 2003) and in C. elegans embryos (Srayko et al., 2005). In wild type axons within 3 h of axotomy, before overt regrowth, axonal MTs (defined as motile EBP-2::GFP puncta) became highly dynamic close to the severed end of the axon (arrows, Figure 5D). In contrast, in efa-6(lf) mutants axonal MTs were more abundant and regrew for longer times and distances than in the wild type (Figure 5C,D). Conversely, in EFA-6 overexpressing axons the number of dynamic axonal MTs was significantly reduced (Figure 5D,E). Axonal MT dynamics were normal in arf-6 mutants (not shown), suggesting enhanced regrowth in efa-6 mutants is mainly due to the microtubule destabilizing role of EFA-6.
To directly address whether the reduced regrowth in EFA-6 overexpressors is due to destabilization of the MT cytoskeleton we tested whether the MT stabilizing drug taxol could overcome regrowth inhibition. Delivery of taxol by microinjection into the body cavity did not affect regrowth in the wild type, yet significantly rescued regrowth of EFA-6-overexpressing axons (Figure 5E). Conversely, incubation in colchicine blocked axonal regrowth (not shown). These findings show that MT polymerization is critical for C. elegans axon regrowth and support a specific role for EFA-6 promoting catastrophe of axonal MTs.
Interactions among regrowth pathways
Our screen identified many genes with positive and negative influences on PLM axonal regrowth. To address how these pathways interact we analyzed regrowth in double and triple mutants. Genetic backgrounds that elevate cAMP signaling (kin-2) display enhanced PLM axon regeneration but do not overcome the block in regrowth in dlk-1 mutants (Ghosh-Roy et al., 2010) (Figure 6A). Examination of double mutants between dlk-1 and other enhanced-regrowth mutants revealed similar dependence on dlk-1 (Figure 6A). In contrast, elevated Ca2+ or cAMP signaling in egl-19(gf)/VGCC or pde-4(lf)/Phosphodiesterase mutants enhanced axon regrowth in unc-57/Endophilin mutants (Figure 6B), suggesting Ca2+ and cAMP act in parallel to UNC-57 and upstream of DLK-1. However, lack of SLT-1 did not promote regrowth in unc-51/Atg1 or unc-57/Endophilin mutants (Figure 6C). These findings suggest Slit/Robo signals play a modulatory role, dependent on intrinsic pathways such as DLK-1, UNC-51, and UNC-57.
Figure 6. Interactions among growth-promoting and growth-inhibiting pathways.
(A) dlk-1(lf) is epistatic to mutants displaying enhanced regrowth. In all panels regrowth is normalized to wild type at 24 h; mean ± SEM. (B) Loss of function in pde-4 or gain of function in egl-19 partly suppress unc-57. (C) unc-51, unc-57, and dlk-1 are epistatic to slt-1 in regrowth. (D) DLK-1 overexpression (Prgef-1-DLK-1, juEx2789) can fully suppress the reduced regrowth of unc-51 and unc-57 mutants. (E) Loss of function in efa-6 partly suppresses the reduced regrowth of unc-26, unc-51, and dlk-1, but does not suppress ebp-1 (ebp-1 efa-6 data from Fig. 5 are included for comparison). (F) efa-6 slt-1 double mutants display enhanced regrowth compared to single mutants at 48 h post axotomy. All statistics, t test; n ≥ 10 for each condition except panel D (n ≥ 5); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As loss of function in dlk-1 and other regrowth-promoting genes results in similar phenotypes, we used a gain of function effect caused by overexpression of DLK-1 [dlk-1(++)] to address their order of activity. Overexpression of DLK-1 is sufficient to enhance PLM axon regeneration (Yan et al., 2009). DLK-1 overexpression completely suppressed the regrowth defects of unc-51/Atg1 and unc-57/Endophilin mutants (Figure 6E), consistent with DLK-1 acting downstream or in parallel to UNC-51/ATG1 and the SV endocytosis genes. Loss of function in RPM-1, a negative regulator of DLK-1, did not suppress unc-57/Endophilin regrowth defects (not shown), consistent with previous findings that PLM regrows normally in rpm-1 mutants (Yan et al., 2009).
Among all double mutants tested, only efa-6(lf) suppressed regeneration defects of dlk-1 mutants (Figure 5F, 6E). In efa-6 dlk-1 double mutants the proximal stumps of severed axons extended significantly further than in dlk-1(lf) although they did not form growth cones (Figure 5G). efa-6 mutations also partially suppressed the regrowth defects of unc-26/Synaptojanin and unc-51/Atg1 mutants (Figure 6E), consistent with EFA-6 acting downstream or in parallel to DLK-1, UNC-26 and UNC-51 in axon regrowth.
Genes with inhibitory roles, such as slt-1 and efa-6, affect different stages of regrowth and therefore likely act in distinct pathways. To test whether elimination of multiple inhibitory pathways could further enhance regrowth relative to single mutants, we analyzed slt-1 efa-6 double mutants. We found that regrowth at the 24 h time point was not further enhanced in slt-1 efa-6 double mutants compared to the highest single mutant (Figure 6F). However regrowth at 48 h post axotomy was significantly enhanced in efa-6 slt-1 double mutants compared to single mutants. Thus, the combined loss of two inhibitory pathways can result in further increases in regrowth at later time points.
Discussion
Our results establish the feasibility of systematic genetic screening for axon regeneration phenotypes using genetically amenable model organisms. Our findings underscore the molecular complexity of axon regeneration, and provide a genetic framework for a more comprehensive understanding of axonal repair and regrowth mechanisms.
Genetic complexity of axon regrowth
As a forward genetic phenotype-based screen in axon regeneration remains technically challenging, we have focused on systematic large-scale testing of conserved candidate genes. Our selection of candidates is by necessity biased, and we plan to expand the screen to reduce this bias. Nonetheless, our analysis supports the view that regenerative axon regrowth requires many genetic pathways in addition to those defined in developmental axon outgrowth, polarity, or guidance. In addition to core factors presumably required for growth cone formation or navigation in many contexts, regenerative regrowth involves pathways that sense damage and trigger the resumption of developmental programs that may be repressed in mature neurons. By focusing on genes nonessential for development or PLM outgrowth we have identified candidates with relatively specific effects on regrowth.
Overall, ~10% of genes tested in our screen displayed significant effects on axonal regrowth (<60% of normal regrowth). In addition to such genes with ‘strong’ requirements we found a similar number of genes with smaller yet significant effects, displaying regrowth 80–60% of the wild type (Table 4B). At least some of these genes may define pathways with modulatory or partly redundant roles. Most such genes have only been examined at the 24 h time point; future studies could address whether such genes have greater effects at different time points or in different genetic backgrounds.
A function for the synaptic vesicle endocytic pathway in regrowth
Among genes required for regenerative regrowth we identified several unexpected functional clusters, including genes implicated in synaptic vesicle (SV) endocytosis and in neurotransmission. The requirement for SV recycling genes seems independent of their role in synaptic function as other genes critical for synaptic transmission (unc-13, unc-18) did not affect regrowth. Endocytic trafficking could play several roles in axon regrowth: repair of damage to the plasma membrane, vesicular transport of retrograde injury signals, and membrane addition in axon extension (Tuck and Cavalli, 2010). Endocytosis can inhibit axon growth by internalization of Nogo (Joset et al., 2010). Although SV endocytosis genes are required at multiple times in regrowth, the requirement for UNC-57/Endophilin could be bypassed by elevated DLK-1 activity. We therefore favor the interpretation that the SV endocytosis genes may be required for vesicles that function in injury signaling. For example, the Drosophila DLK family member Wallenda associates with retrogradely transported vesicles, and transport is important for the response to injury (Xiong et al., 2010). The finding that SV endocytosis is critical for regrowth can be placed in a broader context of evidence that synaptic growth is neuroprotective (Massaro et al., 2009).
EFA-6 is an intrinsic inhibitor of regrowth
Precise regulation of microtubule (MT) dynamics is emerging as a critical factor in axonal regenerative growth (Erturk et al., 2007; Hellal et al., 2011; Sengottuvel et al., 2011; Stone et al., 2010), yet few intrinsic MT regulators in regrowth have been identified. Our analysis reveals EFA-6 as a negative regulator of axon regrowth that affects axonal MT dynamics. Although named for its presumed GEF activity for Arf6 small GTPases, the Sec7 GEF domain of EFA-6 is not essential for its effects on regrowth. Instead, growth-inhibitory effects of EFA-6 are mediated by its N-terminus, a region that lacks well-defined domains (Cox et al., 2004) but which shares motifs with other EFA6 family members (O’Rourke et al., 2010). Although EFA-6 can inhibit regrowth independently of its Arf GEF activity, arf-6 mutants themselves displayed modestly increased regrowth. EFA-6 could inhibit regrowth by two mechanisms, one involving ARF-6 activation and the other involving its N-terminus.
By characterizing EBP-2::GFP dynamics we find that efa-6 mutant axons display increased MT dynamics after axotomy, consistent with studies in the early embryo (O’Rourke et al., 2010). The suppression of the EFA-6 regrowth inhibition effect by taxol supports the model that the reduced regrowth of EFA-6 overexpressing axons is a consequence of destabilized MTs. EFA-6 could directly or indirectly destabilize growing MTs, and a key question is how EFA-6 affects MT dynamics. Members of the mammalian EFA6 family are expressed in neurons but their functions have yet to be studied in detail (Sakagami, 2008). It will be important to determine whether mammalian EFA6 family members also affect axonal regrowth.
Genetically defined stages and pathways in axon regrowth
A key outcome of our screen has been the identification of pathways with inhibitory influences on axon regrowth, indicating that PLM axon regrowth in the wild type is restrained by intrinsic and extrinsic inhibitory influences. Several mutants display similarly increased regrowth suggesting PLM axons cannot extend faster than 6–8 μm/h. Nevertheless total regrowth can be further increased, as in slt-1 efa-6 double mutants, by prolonging the period over which axons extend. As in vertebrate spinal cord regeneration, where combinatorial therapies can enhance regrowth (Kadoya et al., 2009), reduction in multiple inhibitory pathways may be needed to optimise regrowth in C. elegans. A remaining question is whether these inhibitory pathways account for the inability of certain C. elegans neurons to regrow in the wild type (Gabel et al., 2008; Wu et al., 2007).
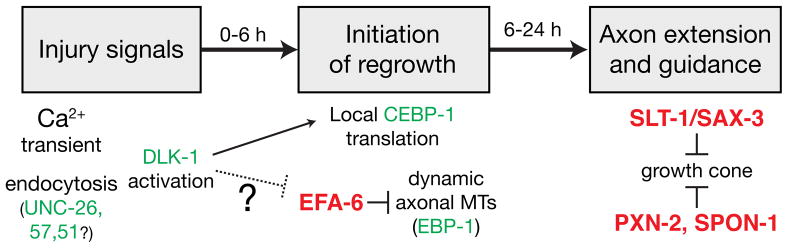
Overall our analysis suggests the following model for PLM axon regrowth (Figure 7). Axonal injury triggers a calcium transient that activates cAMP and PKA signaling upstream of DLK-1 (Ghosh-Roy et al., 2010). In parallel, SV endocytosis may be activated to form signaling vesicles. Such vesicles could transport DLK-1 itself, or other injury signals. DLK-1 kinase is activated and triggers local translation (Yan et al., 2009). Each of these pathways is critical either for competence of injured axons to regrow or for the initial stages of regeneration in which the proximal stump re-establishes a growth cone. Axonal MTs become highly dynamic after axotomy but their growth is restrained by factors such as EFA-6. As the newly reformed growth cone extends, it navigates a microenvironment composed of permissive and inhibitory environmental signals. Inhibitory signals include basement membrane components and Slit and Robo signals. As our studies have focused on axons capable of regeneration it will be important to test whether pathways defined here are limiting in axons that do not spontaneously regrow.
Figure 7. A genetic pathway for PLM axon regrowth.
Summary of relationships between regrowth-promoting (green) and regrowth-inhibiting (red) genes in PLM regrowth, incorporating data reported here and previous results (Ghosh-Roy et al., 2010; Yan et al., 2009).
Experimental Procedures
C. elegans genetics, transgenes, and candidate gene set
We maintained C. elegans on NGM agar plates as described (Brenner, 1974). Animals were grown at 20°C unless stated otherwise. For deletion mutations obtained from the C. elegans gene knockout consortium (ok, gk mutations) or the Japan National Bioresource Project (tm mutations), we backcrossed the mutant at least two times to N2 wild type. For selected ‘hit’ genes we retested the mutant after a second round of outcrossing, and found consistent effects on regrowth. Deletions were genotyped by PCR; primer sequences are available on request. Transgenes were generated by standard procedures (see Online Supplementary Methods).
We chose a set of 654 genes based on the following criteria (1) recognizable C. elegans – human similarity, as assessed by ‘best BLAST score’ in Wormbase; (2) viable mutant strain; (3) known structural or functional category (e.g. kinase, channel); (4) expression in neurons (Wormbase). Some genes were prioritized based on RNAi screens for synaptic function (Sieburth et al., 2005) or axonal guidance (Schmitz et al., 2007). A few genes were selected based on expression in touch neurons (Zhang et al., 2002).
Femtosecond laser axotomy and imaging
We performed laser axotomy essentially as described (Wu et al., 2007). To immobilize worms for EBP-2::GFP imaging without anesthetics we used 12.5% agarose pads and a suspension of 0.1 μm diameter polystyrene beads (Polysciences) under the cover slip (C. Fang-Yen, personal communication). For live imaging of EBP-2::GFP we collected 200 frames of 114 msec exposure each every 230 msec using the spinning disk confocal and generated kymographs using Metamorph™ (Molecular Devices) from a 40 μm ROI on the PLM axon proximal to the cut site.
To apply taxol to regrowing axons in vivo we grew animals on NGM agar plates containing 5 μM taxol (Sigma) for 24 h prior to axotomy. 1 h before axotomy we injected 2–5 nl of 50 μM taxol in M9 buffer into the body cavity using standard injection protocols, and then recovered the animals on taxol-containing plates for 30 min. We axotomized PLM using our standard protocol except with 50 μM taxol in solutions. Control animals were injected with M9 buffer and cultured without taxol. Animals injected with buffer or taxol were healthy and grew at normal rates.
Statistical analysis
The distribution of total regrowth length of axons in wild type and controls passed standard tests of normality. In preliminary analysis we used the Student t-test or the Mann-Whitney test. Among 650 such two-way comparisons, 33 are expected to be significant at the 0.05 level by chance. Most genes discussed here displayed effects significant at the 0.01 level (red bars in bar charts of regrowth); we also discuss some genes that gave repeatable results at the 0.05 level (orange bars). To compare regrowth between experiments with different control means we normalized each experimental data point by dividing it by its control mean. To correct for multiple comparisons we used two approaches. First, most genes displaying significant differences in the primary screen were either repeated, or re-tested with a second allele, in many cases by a different experimenter. We then calculated adjusted P values for the set of repeat experiments using the Benjamini-Hochberg correction for False Discovery Rate (Benjamini and Hochberg, 1995). All other statistical analyses used Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank Alison Hughes, Niousha Saghafi, Amanda Rajapaksa, Sunny Sun, Peg Scott, Caroline Yu, Laura Toy, and other members of our labs for strain construction. We thank Johann Gagnon-Bartsch and Terry Speed for advice on statistical analysis; Cori Bargmann, Gian Garriga, and Erik Jorgensen for reagents; Chris Fang-Yen for the bead immobilization protocol; and Emily Troemel for comments on the manuscript. We thank the C. elegans Gene Knockout Consortium and the Japanese National Bioresource Project for deletion mutations, and the Caenorhabditis Genetics Center for strains. L.C., A.D.C., and Y.J. designed the screen. Z. Wu performed axotomy, imaging, and technical development. L.C. constructed strains and analyzed efa-6; Z. Wang analyzed slt-1/sax-3 signaling. A.G.-R. designed and performed MT imaging and analysis. L.C., Z. Wang, T.H., A.G.-R. and D.Y. contributed to the screen and analyzed the results. S.O’R. and B.B. provided reagents and unpublished data for efa-6. L.C., Z. Wang, Y.J. and A.D.C. wrote the manuscript. Z. Wang is a Fellow of the Jane Coffin Childs Memorial Fund. Z. Wu is an Associate and Y.J. is an Investigator of the Howard Hughes Medical Institute. Supported by grants from the NIH to B.B. (R01 GM049859 and GM058017), Y.J. (R01 NS035546), and A.D.C. (R01 NS057317).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe N, Cavalli V. Nerve injury signaling. Curr Opin Neurobiol. 2008;18:276–283. doi: 10.1016/j.conb.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackley BD, Harrington RJ, Hudson ML, Williams L, Kenyon CJ, Chisholm AD, Jin Y. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman CJ, Cline HT. Refining the roles of GABAergic signaling during neural circuit formation. Trends Neurosci. 2007;30:382–389. doi: 10.1016/j.tins.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Ambron RT, Zhang XP, Gunstream JD, Povelones M, Walters ET. Intrinsic injury signals enhance growth, survival, and excitability of Aplysia neurons. J Neurosci. 1996;16:7469–7477. doi: 10.1523/JNEUROSCI.16-23-07469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Lieberoth BC, Morellini F, Feldner J, Becker T, Schachner M. L1.1 is involved in spinal cord regeneration in adult zebrafish. J Neurosci. 2004;24:7837–7842. doi: 10.1523/JNEUROSCI.2420-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bounoutas A, Chalfie M. Touch sensitivity in Caenorhabditis elegans. Pflugers Arch. 2007;454:691–702. doi: 10.1007/s00424-006-0187-x. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushart TM, Hoffman PN, Royall RM, Murinson BB, Witzel C, Gordon T. Electrical stimulation promotes motoneuron regeneration without increasing its speed or conditioning the neuron. J Neurosci. 2002;22:6631–6638. doi: 10.1523/JNEUROSCI.22-15-06631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch SA, Silver J. The role of extracellular matrix in CNS regeneration. Curr Opin Neurobiol. 2007;17:120–127. doi: 10.1016/j.conb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Cafferty WB, Duffy P, Huebner E, Strittmatter SM. MAG and OMgp synergize with Nogo-A to restrict axonal growth and neurological recovery after spinal cord trauma. J Neurosci. 2010;30:6825–6837. doi: 10.1523/JNEUROSCI.6239-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SQ, Hernandez L, Wang Y, Park KH, Sesti F. MPS-1 is a K+ channel beta-subunit and a serine/threonine kinase. Nat Neurosci. 2005;8:1503–1509. doi: 10.1038/nn1557. [DOI] [PubMed] [Google Scholar]
- Chen L, Ong B, Bennett V. LAD-1, the Caenorhabditis elegans L1CAM homologue, participates in embryonic and gonadal morphogenesis and is a substrate for fibroblast growth factor receptor pathway-dependent phosphotyrosine-based signaling. J Cell Biol. 2001;154:841–855. doi: 10.1083/jcb.200009004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Yu WM, Strickland S. Peripheral regeneration. Annu Rev Neurosci. 2007;30:209–233. doi: 10.1146/annurev.neuro.30.051606.094337. [DOI] [PubMed] [Google Scholar]
- Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittman J, Ryan TA. Molecular circuitry of endocytosis at nerve terminals. Annu Rev Cell Dev Biol. 2009;25:133–160. doi: 10.1146/annurev.cellbio.042308.113302. [DOI] [PubMed] [Google Scholar]
- Doi M, Iwasaki K. Na+/K+ ATPase regulates the expression and localization of acetylcholine receptors in a pump activity-independent manner. Mol Cell Neurosci. 2008;38:548–558. doi: 10.1016/j.mcn.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enes J, Langwieser N, Ruschel J, Carballosa-Gonzalez MM, Klug A, Traut MH, Ylera B, Tahirovic S, Hofmann F, Stein V, et al. Electrical activity suppresses axon growth through Ca(v)1.2 channels in adult primary sensory neurons. Curr Biol. 2010;20:1154–1164. doi: 10.1016/j.cub.2010.05.055. [DOI] [PubMed] [Google Scholar]
- Erturk A, Hellal F, Enes J, Bradke F. Disorganized microtubules underlie the formation of retraction bulbs and the failure of axonal regeneration. J Neurosci. 2007;27:9169–9180. doi: 10.1523/JNEUROSCI.0612-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel CV, Antoine F, Chuang CF, Samuel AD, Chang C. Distinct cellular and molecular mechanisms mediate initial axon development and adult-stage axon regeneration in C. elegans. Development. 2008;135:1129–1136. doi: 10.1242/dev.013995. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Chisholm AD. Caenorhabditis elegans: a new model organism for studies of axon regeneration. Dev Dyn. 2010;239:1460–1464. doi: 10.1002/dvdy.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Wu Z, Goncharov A, Jin Y, Chisholm AD. Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase. J Neurosci. 2010;30:3175–3183. doi: 10.1523/JNEUROSCI.5464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Hall DH, Avery L, Lockery SR. Active currents regulate sensitivity and dynamic range in C. elegans neurons. Neuron. 1998;20:763–772. doi: 10.1016/s0896-6273(00)81014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, Chisholm AD. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010;137:3603–3613. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannila SS, Filbin MT. The role of cyclic AMP signaling in promoting axonal regeneration after spinal cord injury. Exp Neurol. 2008;209:321–332. doi: 10.1016/j.expneurol.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao JC, Yu TW, Fujisawa K, Culotti JG, Gengyo-Ando K, Mitani S, Moulder G, Barstead R, Tessier-Lavigne M, Bargmann CI. C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron. 2001;32:25–38. doi: 10.1016/s0896-6273(01)00448-2. [DOI] [PubMed] [Google Scholar]
- Hellal F, Hurtado A, Ruschel J, Flynn KC, Laskowski CJ, Umlauf M, Kapitein LC, Strikis D, Lemmon V, Bixby J, et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 2011;331:928–931. doi: 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI. Wnt signals and frizzled activity orient anterior-posterior axon outgrowth in C. elegans. Dev Cell. 2006;10:379–390. doi: 10.1016/j.devcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Itoh A, Horiuchi M, Bannerman P, Pleasure D, Itoh T. Impaired regenerative response of primary sensory neurons in ZPK/DLK gene-trap mice. Biochem Biophys Res Commun. 2009;383:258–262. doi: 10.1016/j.bbrc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Joset A, Dodd DA, Halegoua S, Schwab ME. Pincher-generated Nogo-A endosomes mediate growth cone collapse and retrograde signaling. J Cell Biol. 2010;188:271–285. doi: 10.1083/jcb.200906089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jospin M, Watanabe S, Joshi D, Young S, Hamming K, Thacker C, Snutch TP, Jorgensen EM, Schuske K. UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr Biol. 2007;17:1595–1600. doi: 10.1016/j.cub.2007.08.036. [DOI] [PubMed] [Google Scholar]
- Kadoya K, Tsukada S, Lu P, Coppola G, Geschwind D, Filbin MT, Blesch A, Tuszynski MH. Combined intrinsic and extrinsic neuronal mechanisms facilitate bridging axonal regeneration one year after spinal cord injury. Neuron. 2009;64:165–172. doi: 10.1016/j.neuron.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell JR, Fetter RD, Bargmann CI. Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development. 2009;136:3801–3810. doi: 10.1242/dev.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Geoffroy CG, Chan AF, Tolentino KE, Crawford MJ, Leal MA, Kang B, Zheng B. Assessing spinal axon regeneration and sprouting in Nogo-, MAG-, and OMgp-deficient mice. Neuron. 2010;66:663–670. doi: 10.1016/j.neuron.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kulkarni G, Wadsworth WG. RPM-1, a Caenorhabditis elegans protein that functions in presynaptic differentiation, negatively regulates axon outgrowth by controlling SAX-3/robo and UNC-5/UNC5 activity. J Neurosci. 2008;28:3595–3603. doi: 10.1523/JNEUROSCI.5536-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M, Kiley M, Ashcroft N, L’Etoile N, Cheng HJ. The Caenorhabditis elegans P21-activated kinases are differentially required for UNC-6/netrin-mediated commissural motor axon guidance. Development. 2006;133:4549–4559. doi: 10.1242/dev.02648. [DOI] [PubMed] [Google Scholar]
- Manning G. WormBook. 2005. Genomic overview of protein kinases; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro CM, Pielage J, Davis GW. Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. J Cell Biol. 2009;187:101–117. doi: 10.1083/jcb.200903166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelevski I, Segal-Ruder Y, Rozenbaum M, Medzihradszky KF, Shalem O, Coppola G, Horn-Saban S, Ben-Yaakov K, Dagan SY, Rishal I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci Signal. 2010;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S, Ellis RE, Horvitz HR. Otx-dependent expression of proneural bHLH genes establishes a neuronal bilateral asymmetry in C. elegans. Development. 2010;137:4017–4027. doi: 10.1242/dev.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Bradke F, Tessier-Lavigne M, Basbaum AI. Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron. 2002;34:885–893. doi: 10.1016/s0896-6273(02)00702-x. [DOI] [PubMed] [Google Scholar]
- O’Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat Cell Biol. 2010;12:1235–1241. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994;8:2389–2400. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, Tissenbaum HA. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Rhiner C, Gysi S, Frohli E, Hengartner MO, Hajnal A. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 2005;132:4621–4633. doi: 10.1242/dev.02042. [DOI] [PubMed] [Google Scholar]
- Sakagami H. The EFA6 family: guanine nucleotide exchange factors for ADP ribosylation factor 6 at neuronal synapses. Tohoku J Exp Med. 2008;214:191–198. doi: 10.1620/tjem.214.191. [DOI] [PubMed] [Google Scholar]
- Samara C, Rohde CB, Gilleland CL, Norton S, Haggarty SJ, Yanik MF. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci U S A. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci U S A. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab ME. Functions of Nogo proteins and their receptors in the nervous system. Nat Rev Neurosci. 2010;11:799–811. doi: 10.1038/nrn2936. [DOI] [PubMed] [Google Scholar]
- Schwarz V, Pan J, Voltmer-Irsch S, Hutter H. IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Dev. 2009;4:13. doi: 10.1186/1749-8104-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedensky MM, Siefker JM, Koh JY, Miller DM, 3rd, Morgan PG. A stomatin and a degenerin interact in lipid rafts of the nervous system of Caenorhabditis elegans. Am J Physiol Cell Physiol. 2004;287:C468–474. doi: 10.1152/ajpcell.00182.2003. [DOI] [PubMed] [Google Scholar]
- Sengottuvel V, Leibinger M, Pfreimer M, Andreadaki A, Fischer D. Taxol facilitates axon regeneration in the mature CNS. J Neurosci. 2011;31:2688–2699. doi: 10.1523/JNEUROSCI.4885-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Ming GL. Roles of channels and receptors in the growth cone during PNS axonal regeneration. Exp Neurol. 2010;223:38–44. doi: 10.1016/j.expneurol.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Ch’ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Song S, Zhang B, Sun H, Li X, Xiang Y, Liu Z, Huang X, Ding M. A Wnt-Frz/Ror-Dsh pathway regulates neurite outgrowth in Caenorhabditis elegans. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol Biol Cell. 2010;21:767–777. doi: 10.1091/mbc.E09-11-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Shreffler W, Wang S, Driscoll M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron. 1997;18:107–119. doi: 10.1016/s0896-6273(01)80050-7. [DOI] [PubMed] [Google Scholar]
- Toyoda H, Ohno K, Yamada J, Ikeda M, Okabe A, Sato K, Hashimoto K, Fukuda A. Induction of NMDA and GABAA receptor-mediated Ca2+ oscillations with KCC2 mRNA downregulation in injured facial motoneurons. J Neurophysiol. 2003;89:1353–1362. doi: 10.1152/jn.00721.2002. [DOI] [PubMed] [Google Scholar]
- Treinin M, Chalfie M. A mutated acetylcholine receptor subunit causes neuronal degeneration in C. elegans. Neuron. 1995;14:871–877. doi: 10.1016/0896-6273(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Tuck E, Cavalli V. Roles of membrane trafficking in nerve repair and regeneration. Commun Integr Biol. 2010;3:209–214. doi: 10.4161/cib.3.3.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jin Y. Genetic dissection of axon regeneration. Curr Opin Neurobiol. 2010 doi: 10.1016/j.conb.2010.08.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Saifee O, Nonet ML, Salkoff L. SLO-1 potassium channels control quantal content of neurotransmitter release at the C. elegans neuromuscular junction. Neuron. 2001;32:867–881. doi: 10.1016/s0896-6273(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Woo WM, Berry EC, Hudson ML, Swale RE, Goncharov A, Chisholm AD. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development. 2008;135:2747–2756. doi: 10.1242/dev.015289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Ghosh-Roy A, Yanik MF, Zhang JZ, Jin Y, Chisholm AD. Caenorhabditis elegans neuronal regeneration is influenced by life stage, ephrin signaling, and synaptic branching. Proc Natl Acad Sci U S A. 2007;104:15132–15137. doi: 10.1073/pnas.0707001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yeo TT, Zhang C, Yang T, Tisi MA, Massa SM, Longo FM. The leukocyte common antigen-related protein tyrosine phosphatase receptor regulates regenerative neurite outgrowth in vivo. J Neurosci. 2001;21:5130–5138. doi: 10.1523/JNEUROSCI.21-14-05130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wang X, Ewanek R, Bhat P, Diantonio A, Collins CA. Protein turnover of the Wallenda/DLK kinase regulates a retrograde response to axonal injury. J Cell Biol. 2010;191:211–223. doi: 10.1083/jcb.201006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xie Y, Chai H, Fan M, Liu S, Liu H, Bruce I, Wu W. Microarray analysis of gene expression patterns in adult spinal motoneurons after different types of axonal injuries. Brain Res. 2006;1075:1–12. doi: 10.1016/j.brainres.2005.12.060. [DOI] [PubMed] [Google Scholar]
- Yanik MF, Cinar H, Cinar HN, Chisholm AD, Jin Y, Ben-Yakar A. Neurosurgery: functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- Ylera B, Erturk A, Hellal F, Nadrigny F, Hurtado A, Tahirovic S, Oudega M, Kirchhoff F, Bradke F. Chronically CNS-injured adult sensory neurons gain regenerative competence upon a lesion of their peripheral axon. Curr Biol. 2009;19:930–936. doi: 10.1016/j.cub.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Yi BA, Bargmann CI. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell. 1998;92:217–227. doi: 10.1016/s0092-8674(00)80916-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.