Abstract
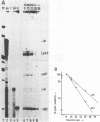
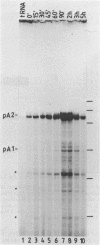
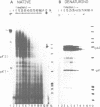
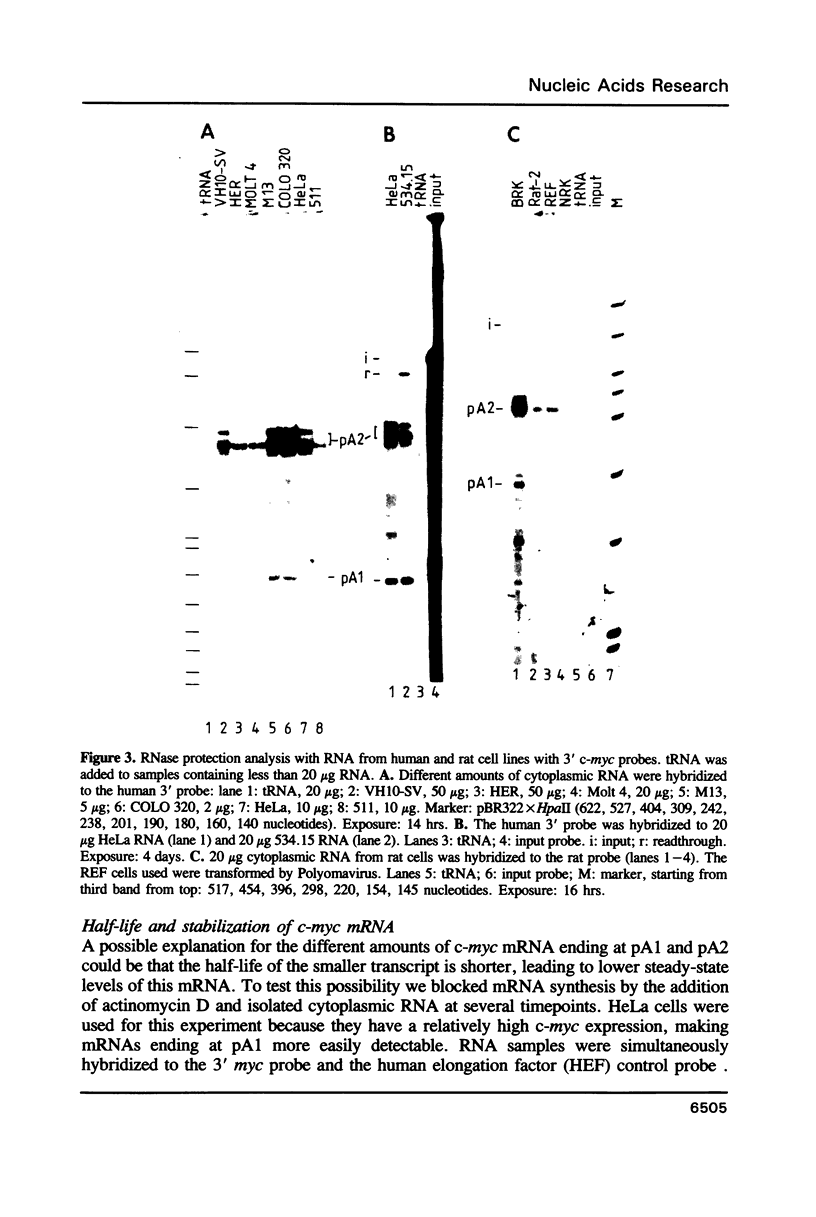
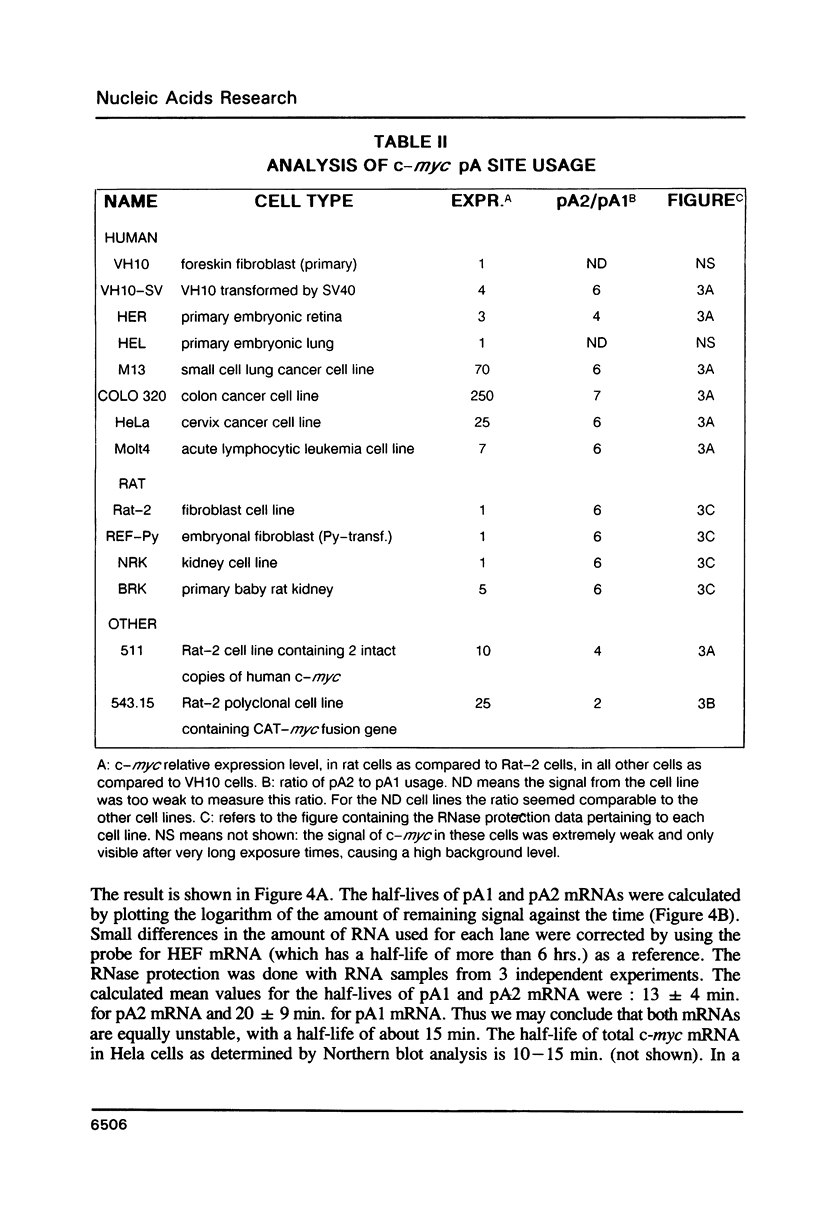
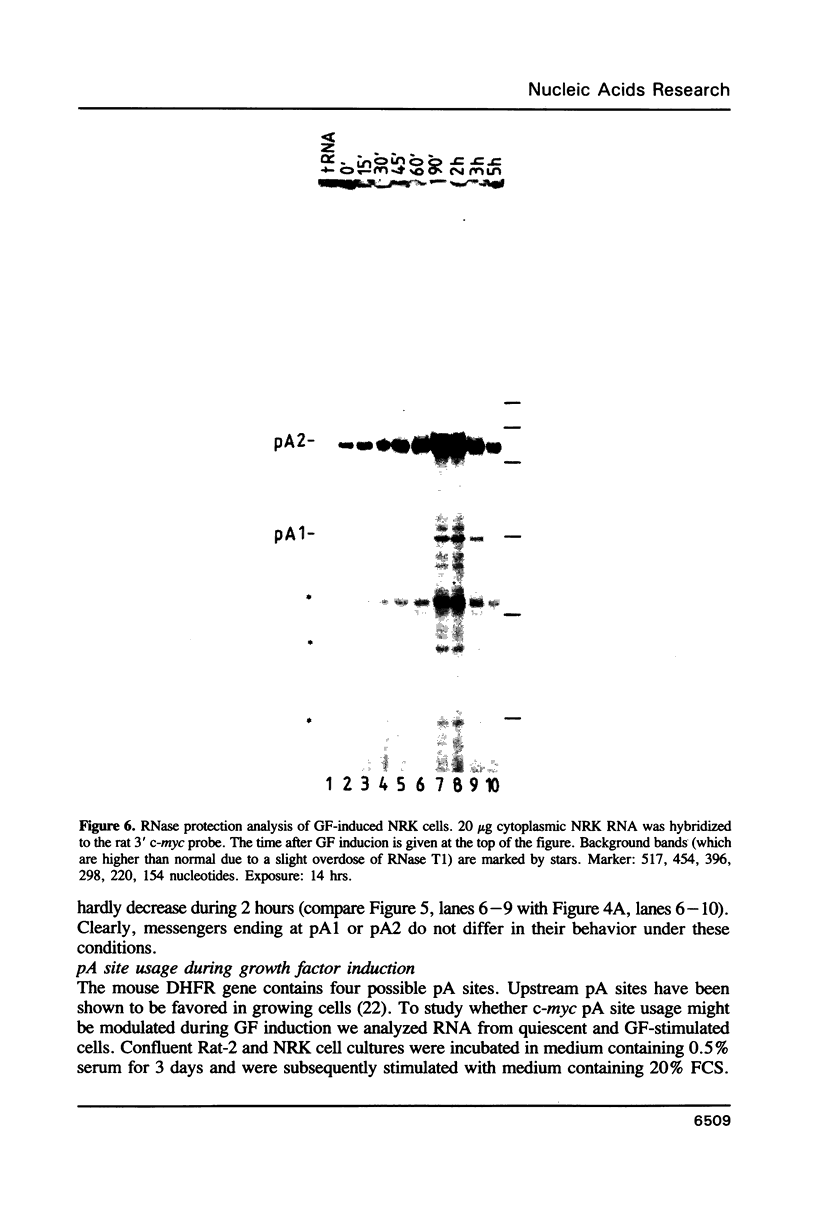
The c-myc gene contains 2 well conserved polyadenylation (pA) sites. In all human and rat cell lines from various differentiation stages and tissue types the amount of mRNA terminating at the second pA site is 6-fold higher than the amount ending at the upstream site. This is not due to a difference in stability of the two mRNA types and therefore must be due to preferential usage of the downstream site. The usage of the pA sites is not altered during growth factor induction of quiescent cells. We have not been able to detect differences in behavior between mRNAs ending at either pA site. Both types of mRNA are induced upon treatment of cells with cycloheximide. Furthermore, we have shown that the poly(A) tail of c-myc mRNA is lost during degradation of the messenger, as was described previously for c-myc mRNA in an in vitro system. The time required for the loss of the poly(A) tail is similar to the half-life of c-myc mRNA.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams J. M., Gerondakis S., Webb E., Corcoran L. M., Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alitalo K., Koskinen P., Mäkelä T. P., Saksela K., Sistonen L., Winqvist R. myc oncogenes: activation and amplification. Biochim Biophys Acta. 1987 Apr 20;907(1):1–32. doi: 10.1016/0304-419x(87)90016-3. [DOI] [PubMed] [Google Scholar]
- Alitalo K., Schwab M., Lin C. C., Varmus H. E., Bishop J. M. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1707–1711. doi: 10.1073/pnas.80.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Brands J. H., Maassen J. A., van Hemert F. J., Amons R., Möller W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur J Biochem. 1986 Feb 17;155(1):167–171. doi: 10.1111/j.1432-1033.1986.tb09472.x. [DOI] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome R. M., Cole C. N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol. 1988 Nov;8(11):4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Marty L., Piechaczyk M., el Sabrouty S., Dani C., Jeanteur P., Blanchard J. M. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 1985 Mar 11;13(5):1431–1442. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Makino R., Kawamura H., Arisawa A., Yoneda K. Characterization of rat c-myc and adjacent regions. Nucleic Acids Res. 1987 Aug 25;15(16):6419–6436. doi: 10.1093/nar/15.16.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R. J., Sharp P. A. Growth-dependent expression of dihydrofolate reductase mRNA from modular cDNA genes. Mol Cell Biol. 1983 Sep;3(9):1598–1608. doi: 10.1128/mcb.3.9.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Cooper J. A., Hunter T., Verma I. M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984 Dec 20;312(5996):711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Linial M., Gunderson N., Groudine M. Enhanced transcription of c-myc in bursal lymphoma cells requires continuous protein synthesis. Science. 1985 Dec 6;230(4730):1126–1132. doi: 10.1126/science.2999973. [DOI] [PubMed] [Google Scholar]
- Müller R., Bravo R., Burckhardt J., Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984 Dec 20;312(5996):716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Goldflam S., Vuocolo G. A. Accurate and efficient transcription of human c-myc genes injected into Xenopus laevis oocytes. Mol Cell Biol. 1985 Jun;5(6):1434–1441. doi: 10.1128/mcb.5.6.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottenburg C., Varmus H. E. Features of the chicken c-myc gene that influence the structure of c-myc RNA in normal cells and bursal lymphomas. Mol Cell Biol. 1986 Aug;6(8):2800–2806. doi: 10.1128/mcb.6.8.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmsdorf H. J., Schönthal A., Angel P., Litfin M., Rüther U., Herrlich P. Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res. 1987 Feb 25;15(4):1643–1659. doi: 10.1093/nar/15.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck R. P., Hirai S. I., Yaniv M., Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988 Aug 11;334(6182):535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Swartwout S. G., Preisler H., Guan W. D., Kinniburgh A. J. Relatively stable population of c-myc RNA that lacks long poly(A). Mol Cell Biol. 1987 Jun;7(6):2052–2058. doi: 10.1128/mcb.7.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp W. C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981 Aug;113(1):408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Yang J. Q., Remmers E. F., Marcu K. B. The first exon of the c-myc proto-oncogene contains a novel positive control element. EMBO J. 1986 Dec 20;5(13):3553–3562. doi: 10.1002/j.1460-2075.1986.tb04682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehner Z. E., Paterson B. M. Characterization of the chicken vimentin gene: single copy gene producing multiple mRNAs. Proc Natl Acad Sci U S A. 1983 Feb;80(4):911–915. doi: 10.1073/pnas.80.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Larco J. E., Todaro G. J. Epithelioid and fibroblastic rat kidney cell clones: epidermal growth factor (EGF) receptors and the effect of mouse sarcoma virus transformation. J Cell Physiol. 1978 Mar;94(3):335–342. doi: 10.1002/jcp.1040940311. [DOI] [PubMed] [Google Scholar]