Summary
Muscle regeneration declines with aging and myopathies, and reprogramming of differentiated muscle cells to their progenitors can serve as a robust source of therapeutic cells. Here, we used the Cre-Lox method to specifically label post-mitotic primary multinucleated myotubes and then utilized small molecule inhibitors of tyrosine phosphatases and apoptosis to de-differentiate these myotubes into proliferating myogenic cells, without gene over expression. The reprogrammed, fusion competent, muscle precursor cells contributed to muscle regeneration in vitro and in vivo and were unequivocally distinguished from reactivated reserve cells due to the lineage marking method. The small molecule inhibitors down-regulated cell cycle inhibitors and chromatin remodeling factors known to promote and maintain the cell fate of myotubes, facilitating cell fate reversal. Our findings enhance understanding of cell-fate determination and create novel therapeutic approaches for improved muscle repair.
Introduction
Skeletal muscle represents a classic example of terminal differentiation wherein myogenic proliferating cells expressing Pax7 and MyoD permanently withdraw from the cell cycle upon serum deprivation and physiologically fuse into multinucleated myotubes expressing muscle differentiation markers myogenin and eMyHC (Okazaki and Holtzer, 1966; Olson, 1992; Rudnicki and Jaenisch, 1995). The regenerative capacity of muscle stem cells declines upon aging and in certain pathologies exemplified by Duchenne muscular dystrophy. Hence studying reprogramming of terminally differentiated muscle cells to their proliferating progenitors holds not only theoretical value but is also therapeutically relevant. The reprogramming from myotubes to myogenic precursor cells is particularly challenging since myogenic proliferating cells not only undergo post-mitotic arrest, but also physically fuse with each other to form multinucleated myotubes during their terminal differentiation. Once these cells terminally differentiate, they are incapable of re-entering into mitosis even when switched to serum rich medium (Endo and Nadal-Ginard, 1986, 1998; Stockdale and Holtzer, 1961). In contrast, reserve cells (myoblasts which remain mono-nucleated upon serum withdrawal) can re-enter cell cycle when switched back to the mitogen-high serum rich growth medium (Carnac et al., 2000; Friday and Pavlath, 2001; Yoshida et al., 1998). Several advances have been previously made in the field of muscle de-differentiation. Over-expression of cyclin D1 and cdk4/6 or knocking down cell cycle inhibitors alone or in combination is insufficient for myotubes to enter mitosis (Latella et al., 2001; Tiainen et al., 1996). Studies in C2C12 cells have shown that a fraction of myotubes derived from this cell line can de-differentiate in the presence of newt extract, myoseverin, or when msx1 or twist are over-expressed (Duckmanton et al., 2005; Hjiantoniou et al., 2008; McGann et al., 2001; Odelberg et al., 2000; Rosania et al., 2000). However, the rare dedifferentiated cells were not tested for their ability to contribute to muscle regeneration in vivo. Earlier work has also reported that C2C12 myotubes responsive to thrombin activated serum response factor triggers expression of immediate early genes but is not sufficient for S phase entry (Loof et al., 2007). Interestingly, the same group also demonstrated that H3K9 di-methylation remains unperturbed in C2C12 myotubes in the presence of serum as opposed to salamander myotubes which readily enter cell proliferation. A recent study has shown deletion in Ink4a locus in C2C12 immortalized cell lines which provides an advantage to C2C12 cells to enter cell cycle upon knockdown of Rb. Knockdown of pRb in conjunction with Arf can induce cell cycle entry in primary myocytes but not in primary myotubes where nuclei get arrested at the onset of mitosis (Pajcini et al., 2010). Nevertheless, the process of de-differentiation of primary multi-nucleated myotubes is still not well understood and most of the previous studies relied on the over-expression of exogenous genes. Some of the previous studies have employed single myocyte and myotube isolation and that can lead to preferential selection of those myotubes that survive such process and does not clear ambiguity of reserve cells which can come along with myotubes. Sparse plating of myoblasts was also tried, but that prevents formation of multinucleated myotubes and limits the study to myocytes. To address these challenges, we performed muscle reprogramming studies in differentiated lineage marked primary myotubes generated by the physiological fusion of Rosa26-Lox-YFP myoblasts with Cre-expressing myoblasts; where the multinucleated myotube cell fate results in the recombination of YFP locus and expression of YFP. Our work critically examined and identified small molecule inhibitors that are necessary and sufficient for the de-differentiation of myotubes to their progenitor cells without forced expression of specific genes. Briefly, these studies demonstrate that in the presence of tyrosine phosphatase (BpV) and apoptosis (Q-VD) inhibitors, Cre-Lox lineage marked myotubes exhibited altered morphology, down regulated terminal differentiation markers, upregulated markers of myogenic progenitor cells, attenuated the cell cycle inhibitors p21, p15, and p16. Based on BrdU incorporation the de-differentiation efficiency was ~12%. To further validate labeling technique and de-differentiation of labeled myotubes, the lineage marked myotubes followed by its de-differentiation into mononucleated cells were captured by time lapse microscopy. The de-differentiated proliferating cells maintained their myogenic identity and were capable of expansion in culture, as well as re-differentiation into myotubes in vitro and in vivo. Furthermore, at the level of molecular mechanism, this work established that phosphatase and apoptosis inhibitors caused down-regulation of a number of chromatin remodeling factors and components that are necessary for the maintenance of terminal myogenic differentiation, thus predisposing myotubes for muscle precursor cell fate. The controlled reprogramming from post mitotic multinucleated terminally differentiated cell fate into a progenitor cell fate of the same lineage provides new insights into adult tissue formation and enables unique clinical strategies for enhancing muscle regeneration.
Results
Fusion-dependent lineage marking of primary myotubes
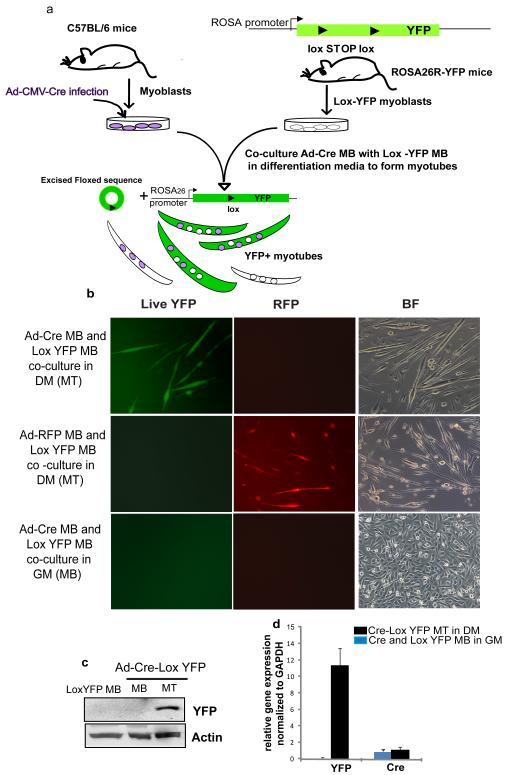
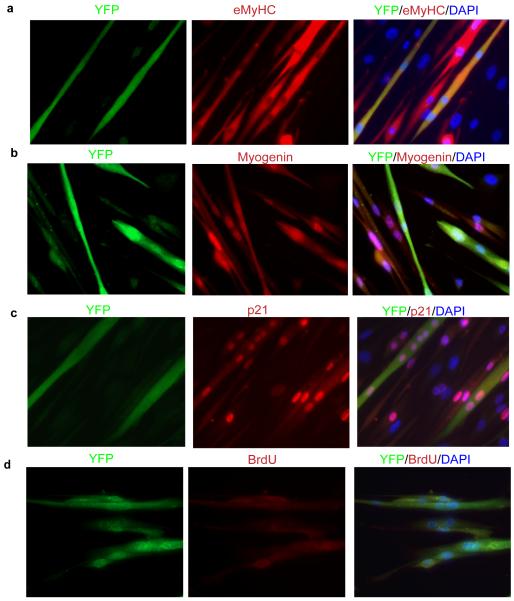
To overcome instances of mistaken cell identity during reprogramming studies, we first established a method to genetically and irreversibly label terminally differentiated myotubes using the Cre-Lox technique (Figure 1a). The Cre-Lox system has been widely used for tissue specific disruption of genes, utilizing the P1 bacteriophage Cre recombinase which specifically recognizes Lox sites and excises any DNA sequence flanked by these sites (Nagy, 2000). The schematic of the system is represented in Figure 1a. Cre recombinase was expressed exogenously by adenoviral mediated infection (Ad-Cre) in wild type primary myoblasts (MB). Ad-Cre infected myoblasts (Cre-MB) were co-cultured with Lox-YFP myoblasts (Lox-YFP MB) derived from Rosa26 YFP reporter mice (Srinivas et al., 2001) in the ratio of 1:2 so that more number of Lox YFP myonuclei coexist with Ad-Cre myonuclei in single myotube and yield high expression of YFP. In standard low mitogen differentiation-promoting medium (DMEM, 2% horse serum), these co-cultured MB within 72-96 hours physiologically fused into YFP expressing myotubes (hence forth, YFP+ myotubes) where both Cre and Lox-YFP myonuclei co-existed (Figure 1b). No mononucleated cells expressing YFP were observed, thus confirming the validity of this lineage marking strategy. Adenoviral infection control was also performed by co-culturing Ad-RFP infected MB with Lox-YFP MB in differentiation medium (DM) and this did not yield YFP+ myotubes (Figure 1b).The YFP+ myotubes accounted for around 70% of total myotubes formed within 96 hours. Of these, around 60% of YFP+ myotubes, had 2-4 myonuclei, while 30% had 5-7 myonuclei with an average number of 4.5 myonuclei per YFP+ myotubes. Non-YFP myotubes that arose from syngeneic fusion events of Cre-MB or Lox-YFP MB were also detected. This labeling strategy was captured by time lapse microscopy encompassing total number of 4 days from the co-culture of Cre and Lox YFP myoblasts to their fusion into multinucleated myotubes (Supplementary movie S1). Representative images of time lapse microscopy are shown in Supplementary Figure S1. To further rule out any possibility of YFP expression without physiological myoblast fusion specific control experiments were conducted. The Cre–MB were co-cultured with Lox-YFP MB in mitogenic growth medium (GM; Ham’s F10, 20% BGS, 9ng/ml bFGF-2) where cells remained mononucleated and did not fuse into myotubes (Figure 1b). After 96 hours of co-culture in GM, cells were processed for western blotting (Figure 1c), qRT-PCR analysis (Figure 1d) and immunostaining (Supplementary Figure S1) for YFP expression. No YFP expression was observed by any of these techniques. A more stringent control experiment was performed to check the possibility of horizontal transfer of Cre recombinase, without complete fusion process. For this, Cre myoblasts were cultured in differentiation medium to form myotubes. Later, Lox YFP MB (2× 105 cells) were added to 96 hour old Cre expressing myotubes and cultures switched to mitogenic growth medium for 72–96 hours. As seen in Supplementary Figure S1e, both epifluorescent imaging and anti-YFP staining confirmed the absence of YFP expressing mononucleated cells without physiological fusion of Cre-MB and Lox-YFP MB into myotubes. These results clearly show that indeed YFP+ myotubes arose only from the fusion of Cre and Lox-YFP MB in DM. The results obtained and quantified with YFP live-direct fluorescence were completely consistent with the data produced using anti-YFP immuno-fluorescence and both assays were routinely employed throughout these studies. The YFP+ myotubes were positive for muscle differentiation markers myogenin and eMyHC, and for CDK inhibitor, p21 (which indicates the post-mitotic state) (Figure 2a-c). Furthermore, these YFP+ myotubes did not incorporate BrdU, confirming that all YFP+ marked cells produced by fusion of Cre and Lox-YFP MB have exited cell cycle and are in post-mitotic arrest by 96 hours in DM (Figure 2d). These findings establish an unambiguous lineage marking of terminally differentiated myotubes that is dependent on physiological myoblast fusion.
Figure 1. Lineage marking of primary myotubes by Cre-Lox method A Schematic of the system.
a. Wild type myoblasts (MB) derived from C57BL/6 mice were infected with Ad-Cre and subsequently co-cultured with Lox-YFP MB obtained from Rosa 26-YFP reporter mice in differentiation medium (DM) to form myotubes. The fusion of these two populations of MB led to the excision of stuffer sequence (green circle) by Cre recombinase activity to give rise to lineage marked YFP expressing myotubes (green). Self fusion among the two populations of MB will give rise to YFP negative myotubes (colorless). These lineage marked myotubes were then used in de-differentiation studies. Fusion-dependent, Cre-Lox mediated labeling of myotubes upon co-culture of Ad-Cre MB with Lox-YFP MB in DM. b. As described in Figure a, wild type MB were co-cultured with Lox-YFP MB (1:2 ratio) in DM to induce formation of myotubes. Endogenous YFP fluorescence in myotubes was observed by 72-96 hours as shown by epifluorescent images. In control infection with control Ad-RFP virus, no YFP fluorescence was observed. No YFP expression was observed upon co-culture of Ad-Cre MB and Lox YFP MB in GM where myoblasts did not undergo physiological fusion to form myotubes. c. Western blotting to determine YFP expression using lysates from Lox-YFP MB, Ad-Cre MB co-cultured with Lox-YFP MB in GM and parallel in DM. YFP protein was observed in the myotubes which arose from fusion of Ad-Cre and Lox-YFP MB in DM. d. qRT-PCR analysis for YFP gene expression. RNA was extracted from Ad-Cre and Lox-YFP MB co-cultured in GM and in DM for 96 hours to detect the levels of YFP and Cre recombinase by qRT-PCR. Data was normalized to internal control GAPDH. Error bars indicate mean and standard deviation, n=3. YFP mRNA levels was only observed in Ad-Cre-Lox-YFP+ myotubes while Cre recombinase expressed in both the co-cultures of Ad-Cre and Lox-YFP MB in GM and in DM. The fusion-dependent marking of myotubes was clearly and robustly mediated by this adaptation of the Cre-Lox method, and no mononucleated cells expressed YFP..
See also Figure S1
Figure 2. Immunodetection of YFP and muscle specific markers.
a-d. YFP+ myotubes obtained after Cre-Lox fusion express muscle differentiation marker and does not incorporate BrdU. Cre-Lox YFP+ myotubes cultures were co-immunostained with muscle differentiation markers eMyHC (a) myogenin (b) p21 (c) and DNA synthesis label BrdU (d) along with anti-YFP antibody. Representative images are shown.
Combination of a tyrosine phosphatase inhibitor (BpV) and apoptosis inhibitor (Q-VD) reprograms lineage marked myotubes into proliferating YFP+ mononucleated cells
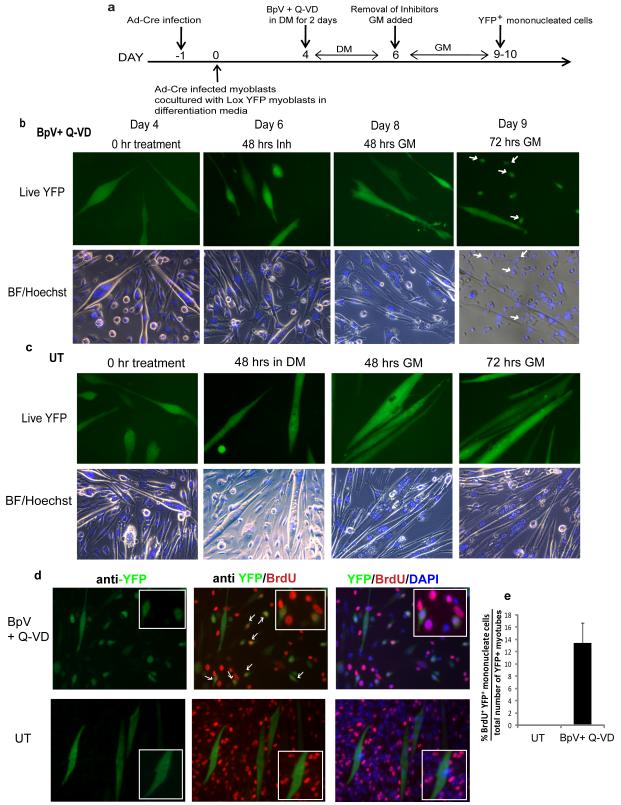
Myotube formation involves many events such as changes in cytoskeletal assembly, sequential expression of differentiation specific genes, modulation of signaling pathways and up-regulation of tyrosine phosphatases (Bennett and Tonks, 1997; Delgado et al., 2003; Lassar et al., 1994; Weintraub, 1993). Hence we reasoned that global transient inactivation of tyrosine phosphatases would reset signaling in myotubes, making them receptive to mitogens present in growth medium conditions and propelling them into cell cycle as well as towards less differentiated state. Earlier BpV (a tyrosine phosphatase inhibitor) was reported to delay differentiation of dividing C2C12 into myotubes (Castaldi et al., 2007) and other studies also indicated that a small percentage of myotubes that enter S-phase upon over-expression of genes fail to proliferate and succumb to apoptosis (Endo and Nadal-Ginard, 1998; Latella et al., 2000). Therefore, we reasoned that if BpV was able to trigger the process of myotube de-differentiation and that the addition of an apoptosis inhibitor (Q-VD) in our studies may help in survival of those myotubes that might undergo massive restructuring of cell cytoskeleton simultaneously with the breakage of post-mitotic arrest. Using our lineage marking technique for myotubes, we then explored whether BpV+ Q-VD (hence forth inhibitor mix) was capable to reprogram already differentiated primary myotubes to their muscle progenitor fate. For de-differentiation assays, inhibitor mix (10uM each) was added to YFP+ myotube cultures daily for two days after which cultures were switched to GM (Figure 3a). Remarkably, in the presence of inhibitor mix, considerable number of YFP+ myotubes showed altered morphology and cleaved into small cells which were followed by the appearance of YFP+ mononucleated progeny of the de-differentiated myotubes (Figure 3b). When myotubes produced by the fusion of primary myoblasts were treated with BpV alone, cell death occurred as described in previous studies (Rumora et al., 2003). Interestingly, in the presence of BpV alone, myotubes did show apoptosis and few of them gave rise to YFP+ mononucleated cells albeit at very low frequency (~1.18%) in comparison to inhibitor mix treatment which augmented the de-differentiation frequency to around ~12-13% (Supplementary Figure S2 a, c and d) These results demonstrate that the inhibition of apoptosis is a critical requirement for the de-differentiation of multinucleated primary myotubes. In control experiments, no YFP+ mononucleated cells were observed in untreated YFP+ myotubes that were switched to GM for 72 hours (Figure 3c), or in the presence of Q-VD alone (Supplementary Figure S2 b). Furthermore, the YFP+ cells derived from the post-mitotic multinucleated myotubes engaged in proliferation, where around 12-13% of total lineage marked population (was found to incorporate BrdU during a 24 hour BrdU labeling interval (Figure 3d and e). The former myotube identity of these YFP+ cells clearly discriminated them from the reactivation of reserve YFP− myoblasts, which also proliferated and expanded when myotube cultures were switched to the highly mitogenic GM (Figure 3d). To rule out any spurious YFP expression in the absence of Cre expressing cells and in the presence of inhibitor mix, 4 day old Lox YFP myotube cultures were treated with the inhibitor mix and then switched to growth medium. No YFP expression was ever observed in these cultures. This clearly establishing that Lox YFP cells do not spontaneously express YFP upon addition of inhibitor mix in the absence of Cre recombinase (Supplementary Figure S4 a). To capture reprogramming of lineage marked myotubes, we also performed time lapse microscopy where Ad-Cre-Lox YFP myotubes were labeled for 96 hours followed by the addition of inhibitor mix for another 48 hours and the cultures were then switched to GM. Live cell imaging was performed for the total period of 4 days where inhibitor mix treated YFP+ multinucleated myotubes gave rise to YFP+ mononucleated cells (Supplementary movie S2). Representative images of the time lapse imaging can be seen in Supplementary Figure S3. A combination of a small molecule tyrosine phosphatase inhibitor, BpV and an apoptosis inhibitor, Q-VD was necessary and sufficient for such reprogramming of terminally differentiated muscle cells. Since, inhibitor of tyrosine phosphatases induces apoptosis, the possibility that some aspect of apoptosis may mediate the reprogramming of multinucleated myotubes to undergo de-differentiation was also examined. 0.2uM doxorubicin, a classic inducer of apoptosis which has been studied in muscle (Latella et al., 2004) was added to Cre-Lox myotube cultures for 48 hrs either alone or in combination with the apoptosis inhibitor. This was followed by removal of drugs and switching cultures to growth media conditions for 72-96 hours. We observed reduced apoptosis by doxorubicin in the presence of apoptosis inhibitor but no YFP+ mononucleated cells were observed in these cultures in spite of altered morphology of myotubes (Supplementary Figure S4 b and c). These suggests that apoptosis does not mediate the de-differentiation of myotubes.
Figure 3. BpV with Q-VD de-differentiates the irreversibly-labeled YFP+ myotubes to YFP+ proliferating mononucleated cells.
a. Myotube de-differentiation strategy. MB infected with Ad-Cre were co-cultured with Lox-YFP MB in DM for 4 days to give rise to YFP+ myotubes. These were treated with 10uM BpV + 10uM Q-VD in parallel with other experimental conditions for two days in DM. The treated myotubes were then switched to myoblast GM which was replaced fresh every day. YFP+ mononucleated cells were observed around day 10. De-differentiation of YFP+ myotubes to YFP+ proliferative cells. b. YFP+ myotubes cultures were treated with the BpV+ Q-VD and photographed every day. The addition of BpV+ Q-VD led to morphological changes and when switched to GM these cells expanded as YFP+ mononucleated cells in 72 hours (white arrow shows YFP+ mononucleated cells). Representative high magnification images of de-differentiation experiment over the course of 10 days with live Hoechst is shown by epifluorescent microscopy. c. No-treatment (UT): Untreated YFP+ myotubes were grown in similar conditions and did not show any de-differentiation events. These data demonstrate that inhibitor mix is necessary and sufficient for de-differentiation of genetically labeled myotubes into expanding mononucleated cells. d. Reprogrammed YFP+ mononucleated cells rapidly divide. Cre-Lox-YFP+ myotubes reprogrammed as depicted in Figure 3b and c, were pulsed with BrdU for 24 hours and co-stained with anti YFP and BrdU antibodies. Arrows indicate representative BrdU+YFP+ cells in treated conditions. Untreated cultures of YFP+ myotubes do not show any YFP+ mononucleated cells though BrdU incorporation is seen in non YFP cycling mononucleated cells. Inset shows magnified images. e. Quantification of percent of BrdU+ /YFP+ mononucleated cells out of total number of YFP+ myotubes (shown are the mean and standard deviations ,n=3 p< 0.05). Note that many reserve myoblasts re-entered cell cycle and incorporated BrdU in GM (both in the presence of BpV+ Q-VD and in control untreated cultures); these cells, however, were reliably distinguished in our experiments by the absence of YFP.
In parallel to these experiments, we also addressed a possible role of Oct4 in myotube reprogramming considering the pivotal reprogramming activity of Oct4 and the ability of this transcriptional factor alone to reprogram neural stem cells (Kim et al., 2009). We used our published method (Conboy and Conboy, 2010) to activate endogenous muscle stem (satellite) cells by injury into hind limb muscle of Tet-Oct4 mice (Hochedlinger et al., 2005) and derived primary myoblasts which were kept in DM to form myotubes and then treated or untreated with doxycycline (dox) for 24 and 48 hours, to induce Oct4 protein and mRNA expression (Supplementary Figure S5). These Tet-Oct4 myoblasts were infected with Ad-Cre and co-cultured with Lox-YFP MB in DM for 96 hours, where myotubes were readily formed (Supplementary Figure S5). No significant changes in morphology of myotubes were observed up to 48 hrs of Oct4 induction followed by growth media incubation for additional 4 days (Supplementary Figure S5d). In concert with earlier studies, where it has been reported that induction of Oct4 in the differentiated cells of the intestine and hair follicle has no effect on their cellular phenotype (Hochedlinger et al., 2005), our results showed that Oct4 induction up to 2 days was not sufficient to induce any morphological changes in myotubes or to promote their de-differentiation in GM (Supplementary Figure S5).
YFP+ mononucleated cells from de-differentiated myotubes re-express markers of myogenic progenitors, down-regulate markers of myotubes, attenuate p21 and retain their myogenic potential to fuse into de-novo myotubes
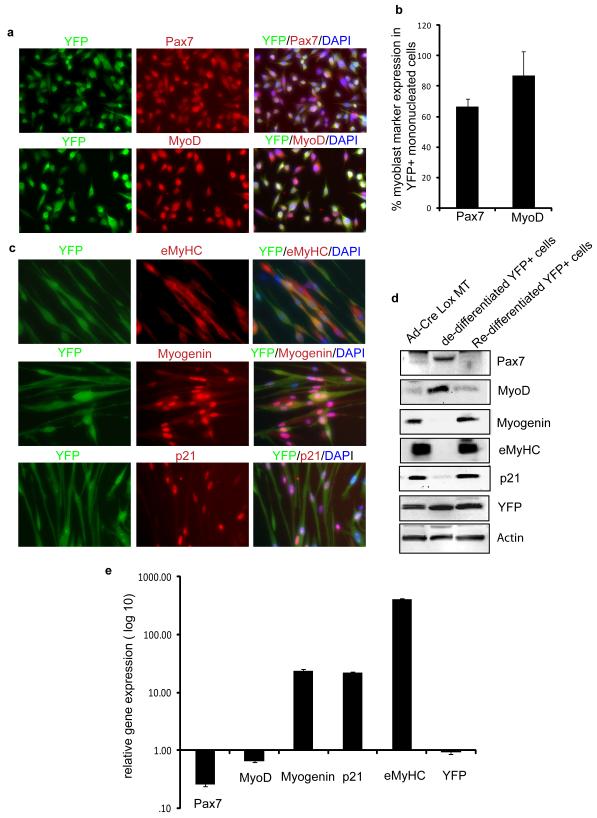
To further assess the properties of reprogrammed cells, mono-nucleated YFP+ progeny of de-differentiated myotubes was, FACS sorted ,and expanded in culture (Supplementary Figure S6 a). These YFP+ mononucleated cells were immunostained with antibody against Ki67 (proliferation marker) and BrdU(S phase marker) along with YFP. As shown in supplementary Figure S6 c, these FACS sorted expanded YFP+ cells positively immunostained for Ki67 and incorporated BrdU. Further, cell cycle analysis by propidium iodide DNA staining confirmed that these cells can proliferate and exist in different phases of cell cycle (Supplementary Figure S6 e). To confirm that the actively dividing reprogrammed YFP+ cells are indeed myogenic, they were analyzed for the myogenic markers Pax7, MyoD1 and differentiation markers myogenin, eMyHC and Cdk inhibitor, p21 (Figure 4a). Based on the quantification of the immunofluorescence, around 70% of YFP+ mononucleated cells expressed high levels of Pax7 and ≥ 90% expressed MyoD1 (Figure 4b). The differentiation capability of the YFP+ mononucleated cells was tested by switching the cultures to DM where normal primary myoblasts exit cell cycle and fuse into multinucleated myotubes. The YFP+ precursor cells were found to retain their myogenic potential as they underwent rapid physiological fusion de-novo into myotubes that expressed typical muscle differentiation markers, eMyHC and myogenin and p21 (Figure 4c) Thus, the markers of terminal differentiation that were down-regulated upon myotube reprogramming with inhibitor mix treatment, were up-regulated again when YFP+ myogenic progenitor cells differentiated into de-novo myotubes in the mitogen-low differentiation medium (Figure 4d). The changes in marker gene expression was also validated at transcriptional level by qRT-PCR which clearly showed the up-regulation of eMyHC, p21 and myogenin and down regulation of Pax7 and MyoD mRNA levels upon differentiation of YFP+ mono-nucleated cells (Figure 4e). Thus, based on the profile of myogenic markers and the functional properties, dedifferentiated genetically labeled progeny of primary myotubes acquired the fate of muscle precursor cells or myoblasts.
Figure 4. Genetically-labeled progeny of de-differentiated myotubes have functional and genetic attributes of muscle progenitor cells.
a. Co-immunostaining of FACS-sorted, proliferating YFP+ mononucleated cells for (i) Pax7 and (ii) MyoD along with anti-YFP antibody was performed and representative images are shown. b. Histogram quantifies Pax7 and MyoD expressing YFP+ mononucleated cells which represents mean and standard deviation of three independent experiments. c. De-differentiated, FACS sorted, YFP+ cells were expanded in GM and cultured in DM for 96 hours where myoblasts typically form myotubes; cultures were co-immunostained with antibodies specific to YFP and to myotube specific marker (i) eMyHC ii) myogenin, as well as (ii) the CDK inhibitor p21. d. Western blotting with antibodies specific for Pax7, MyoD, eMyHC, p21, myogenin and YFP was performed using protein extracts from Cre-Lox-YFP myotubes, de-differentiated YFP+ mononucleated cells and re-differentiated YFP+ cells as indicated. Actin served as loading control. e. Gene expression analysis of muscle differentiation markers. qRT-PCR data in log scale for Pax7, MyoD, myogenin, p21 and eMyHC depicts the relative gene expression of re-differentiated myotubes to de-differentiated YFP+ cells. The data represents the mean and standard error for three independent experiments.
See also Figure S6
Reprogrammed YFP+ cells can contribute to in vivo muscle regeneration
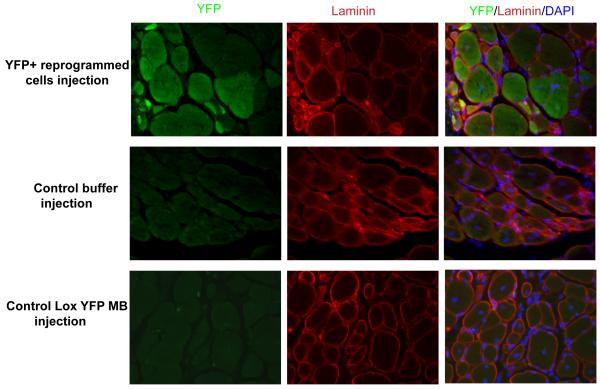
The ultimate test was to make sure that these reprogrammed cells could contribute to in vivo muscle regeneration under physiological conditions. The dividing de-differentiated YFP+ cells were expanded in GM for ~1.5-2 weeks and injected into cardiotoxin injured Tibialis Anterior (TA) of immuno-deficient NOD-SCID mice. Injections of un-recombined Lox-YFP myoblasts and buffer medium served as negative controls. 2 weeks of post injection, muscles were dissected out, sectioned and immunostained for YFP and laminin. YFP+ reprogrammed cells readily fused with regenerating myofibres and contributed to muscle repair in vivo (Figure 5). These results establish that post-mitotic myotubes can de-differentiate into functional, proliferating myogenic precursor cells that regenerate muscle tissue after an injury. Importantly, cultured muscle precursor cells that eagerly regenerate muscle in vivo were produced from terminally differentiated primary myotubes without an exogenous gene expression, making this method therapeutically feasible.
Figure 5. Reprogrammed YFP+ proliferative cells contribute to in vivo muscle regeneration.
FACS sorted YFP+ proliferating mononucleated cells were expanded in GM and injected in cardiotoxin injured Tibialis Anterior (TA) immuno-compromised NOD-SCID mice. 2-3 weeks later, TA muscles were dissected out, sectioned at 10um and co stained with YFP and laminin to visualize YFP+ myofibres. Control buffer and Lox YFP myoblast injected TA muscle did not show any YFP+ myofibres.
Inhibitor mix treatment modulates gene expression in myotubes
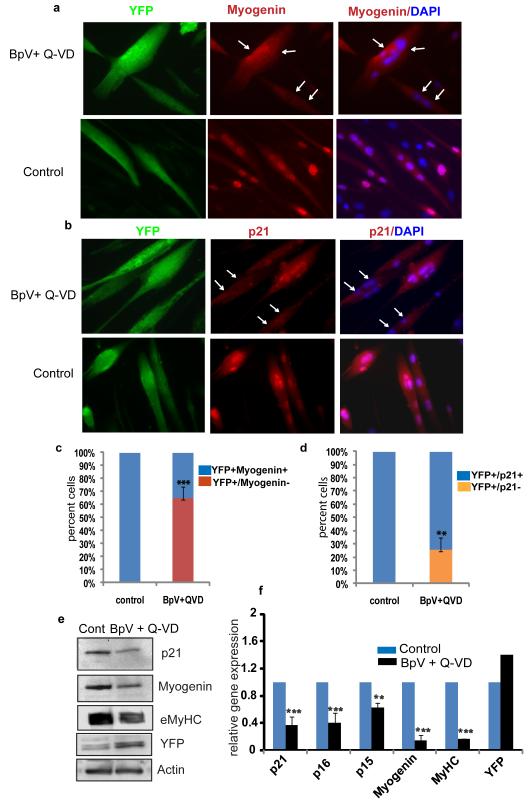
To address the mechanism by which inhibitor mix facilitated de-differentiation of YFP+ myotubes, we analyzed early changes in expression of eMyHC (a terminal muscle differentiation marker), myogenin (a muscle marker expressed on onset of differentiation), p21, p15 and p16 (cdk inhibitors) in Ad-Cre-Lox YFP+ myotubes treated with inhibitor mix for 48 hours (Figure 6a, b). By immunofluorescence, over 60% of the myonuclei in YFP+ myotubes down regulated myogenin as compared to untreated myotubes whereas the levels of p21, a negative regulator of mitosis that plays an important role in cell-cycle arrest (Bunz et al., 1998; Cayrol et al., 1998) were found to be attenuated in approximately 25% of the myonuclei present in YFP+ myotubes (Figure 6c, d). The downregulation of eMyHC, p21, p15, p16 and myogenin in Ad-Cre-Lox-YFP+ myotubes treated with inhibitor mix was also confirmed by western blotting experiments and qRT-PCR analysis (Figure 6e and f). These results demonstrate that the myogenic cell fate is reversed at the genetic level in multi-nucleated myotubes, before they split into single dividing cells. We also observed that inhibitor treatment increased the expression of YFP in Cre-Lox myotubes both at protein and RNA level. As shown earlier in supplementary Figure S4 a, Lox YFP myotubes do not express YFP spontaneously upon inhibitor mix treatment and in the absence of Cre. However when YFP locus is recombined, the broad range phosphatase inhibitor mix which is known to modulate numerous signaling pathways by acting at the transcriptional as well as post transcriptional levels might activate ROSA locus at the transcriptional level thereby increasing YFP expression.
Figure 6. Molecular analysis of reprogramming in genetically labeled myotubes.
Inhibitor mix treatment down regulates muscle differentiation marker in Cre Lox-YFP myotubes. a and b. 4 day old Ad-Cre-Lox-YFP myotubes were untreated/ treated with inhibitor mix for 48 hours, followed by immuno-detection of myogenin (a), p21 (b) and YFP (green), using antibodies specific for these proteins. Myogenin and p21 were down-regulated in a subset of YFP+ myotubes (shown by white arrows). Control myotubes did not change expression of muscle differentiation markers. c and d. The histogram quantifies the percent of YFP+/myogenin +, YFP+ myogenin− cells and YFP+/p21+ and YFP+/p21− in the experiment shown in Figure 6a and b (n=3 ± S.D.; p***<0.001, p**<0.05). e and f. Ad-Cre-Lox-YFP myotubes untreated/ treated with BpV+ Q-VD for 48 hours were analysed for protein and mRNA levels. Protein lysates were subjected to western blotting for antibodies against p21, myogenin and eMyHC. Actin served as a loading control. q-RT-PCR was performed on RNA lysates for gene expression of p21, p15, p16, myogenin and eMyHC. Data was normalized to GAPDH and represents mean and standard deviation of three independent experiments each done in triplicates (n=3 ± S.D.; p***<0.001, p**<0.05). Untreated sample was taken as 1.
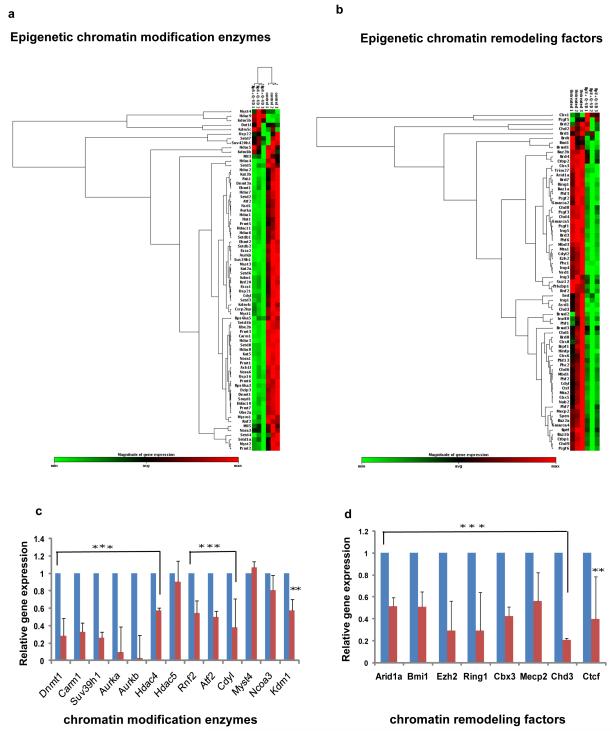
Studies have also shown that upon myotube differentiation, widespread chromatin remodeling occurs and genes necessary for differentiation are activated while those for proliferation are repressed (Forcales and Puri, 2005; Guasconi and Puri, 2009; McKinsey et al., 2002; Palacios and Puri, 2006; Sartorelli and Caretti, 2005). Since a subset of labeled myotubes enter cell cycle and proliferate in GM, we reasoned that inhibitor mix have a global effect and perturbation of signaling pathways would in turn affect chromatin remodeling thereby facilitating reprogramming of myotubes to their progenitor cells. To analyze these changes during inhibitor mix treatment, PCR arrays for chromatin enzymes and chromatin remodeling factors was performed on untreated and inhibitor mix treated Cre-Lox YFP myotube cultures (Figure 7). As summarized in Figure 7c and d, Carm1, Suv39h1, SWI/SNF complex components which have been earlier shown to promote myogenic differentiation (Ait-Si-Ali et al., 2004; Chen et al., 2002; de la Serna et al., 2001) were downregulated upon inhibitor mix treatment along with other histone methyltransferases. Supplementary table S1 and S2 provides a complete list of chromatin factor and enzyme genes modulated by inhibitor mix treatment. These findings suggest that inhibitor mix down-regulates the chromatin factors and enzymes dedicated to the maintenance of differentiated state in primary myotubes, enabling them to respond to the growth factors present in serum and de-differentiate to YFP+ proliferating progenitor cells.
Figure 7. Inhibitor mix treatment modulates chromatin remodeling factors and enzymes.
a and b. Clustergram analysis of chromatin remodeling factors and enzymes for Ad-Cre-Lox YFP myotubes treated and untreated with BpV+Q-VD (inhibitor mix) for 48 hours using SA Biosciences/Qiagen PCR arrays. 0.5ug RNA isolated from three independent set of experiments of Ad-Cre-Lox YFP myotubes were reverse transcribed and gene expression profile monitored. c and d. Histogram representation for few set of genes normalized by Hprt gene levels. Control untreated was taken as 1. (n=3 ± S.D.; p***<0.001, p**<0.05).
Discussion
Our studies explored the small molecule pharmacological approach to de-differentiation and reprogramming that does not involve over-expression of exogenous genes, which is the currently searched for method in the field of cell reprogramming .The use of a broad pharmacological inhibitor of tyrosine phosphatases simultaneously with the inhibition of apoptosis was, in our hands, sufficient to induce actual reprogramming of terminally differentiated post-mitotic multinucleated skeletal muscle cells into their progenitors. The use of small molecule inhibitors for reprogramming studies has high translational significance. The apoptosis inhibitors used in our studies is reversible treatment, has short half life and is not present in cells when expanded in vitro for transplantation experiments. Hence the reprogrammed myogenic progenitor cells transplanted with the aim to alleviate myopathic conditions will not be resistant to apoptosis and consequentially will not pose risk of cancers. In this work the irreversible cell-fate lineage marking of myotubes was based on the fact that terminally differentiated skeletal muscle cells are normally produced by the fusion of myoblasts. Thus, Lox-YFP Rosa 26 nuclei and Cre containing nuclei at some point coexisted in a multinucleated cell in order to produce YFP+ myotubes. The Cre-Lox myotubes labeling method efficiently distinguishes reserve cells from multinucleated myotubes. Our data shows that 4 day old cultures of YFP+ primary myotubes express typical muscle differentiation markers such as myogenin, eMyHC, express high levels of cdk inhibitor p21 and do not incorporate BrdU which strongly suggests that these YFP marked cells are indeed terminally differentiated (Figure 2a-d). The observation that dividing YFP+ mononucleated myogenic progeny were obtained from YFP+ myotubes unambiguously establishes the reprogramming step toward a less differentiated precursor cell. Importantly, such genetic labeling for the first time demonstrates de-differentiation of mature 4-day old multinucleated primary myotubes into proliferating fusion-competent myoblasts that expand in vitro and repair muscle in vivo.
The calculation of reprogramming efficiency from terminally differentiated myotubes to the muscle progenitor cells is complicated by the fact that heterogenous Cre and Lox YFP myoblasts form YFP+ myotubes where varying number of both myonuclei co-exist in same multinucleated myotube. Further, only Lox YFP myonuclei and not Ad-Cre MB nuclei co-existing in YFP+ myotubes will give rise to YFP+ mono-nucleated cells when labeled myotube de-differentiates. Moreover, the de-differentiation of myotubes that are produced by syngeneic fusion events were not accounted for in our YFP labeling method. Hence, we estimated efficiency by two different methods. By method 1 total number of YFP+ mononucleated cells were divided by the total number of YFP+ myotubes before inhibitor treatment. Based on this method, efficiency was estimated ~12-13% in the presence of inhibitor mix as compared to BpV alone which was around ~ 1.18% (Figure S2d). By method 2, the total number of YFP+ mononucleated cells was divided by an estimated number of Lox YFP myonuclei present in all labeled YFP+ myotubes before inhibitor treatment (see methods section).This method gave an estimate of ~ 5% in presence of inhibitor mix and ~0.4% in presence of BpV alone (Figure S2d and methods section). For the above reasons, we feel these calculations give very conservative estimates of de-differentiation. No matter, the method of quantification, BpV alone gave poor reprogramming efficiency and apoptotic inhibitor was needed to augment the de-differentiation likely by facilitating the survival of those myotubes which undergo reprogramming in the presence of phosphatase inhibitor.
Recent reports have shown that cells expressing higher levels of anti proliferative genes and those involved in senescence are indeed difficult to reprogram (Li et al., 2009; Utikal et al., 2009). Since myotubes are post-mitotically arrested cells which express high levels of CDK inhibitors, these may have low reprogramming efficiency. Studies have indicated that experimental down regulation of CDK inhibitors in post-mitotic myotubes results in accumulation of DNA damage, hinders cell cycle reentry and cause DNA fragmentation and apoptosis (Pajalunga et al., 2010) It has been shown that dividing cells robustly repair DNA damage (Nouspikel and Hanawalt, 2002) and since our de-differentiated cells are cultured in mitogen-high growth medium (following the BpV and apoptosis inhibitor treatments) and are actively dividing, any DNA damage accumulated in myotubes is likely to be repaired during the process of DNA replication. Notably, our results definitively demonstrate that reprogrammed myotubes give rise to functional muscle progenitor cells which form new muscle in vitro and in vivo, hence no irreversible DNA damage or mutations occurred to compromise the myogenic properties of the de-differentiated cells. Recent studies has also shown that inhibition of Rb and p16/p19 can induce cell cycle entry in post mitotic myocytes (Pajcini et al., 2010). Our work conducted on 4 day old mature myotubes not only downregulated CDK inhibitors and muscle differentiation markers but also decreased gene expression of chromatin remodelers that maintain differentiated state. The role of chromatin organization in establishment and maintenance of cell fates has been well defined. It has also been shown to play a role in commitment of myoblast to terminally differentiated myotubes as different signaling pathways have been shown to modulate chromatin signaling in muscle progenitor cells upon differentiation (Caretti et al., 2004; de la Serna et al., 2001; McKinsey et al., 2002; Palacios and Puri, 2006; Saccone and Puri). Recent work has also demonstrated the stage specific role of Ezh2 in muscle regeneration where Ezh2 occupies the Pax7 regulatory sequences in differentiated state (Palacios et al., 2010). This suggests that in spite of downregulation of Ezh2 upon differentiation as reported earlier (Caretti et al., 2004), a certain level of Ezh2 along with other polycomb members would be required to maintain muscle progenitor genes in repressed state. Our PCR arrays on Cre-Lox myotubes after inhibitor mix treatment demonstrate significant down regulation of Ezh2 and other polycomb members as compared to untreated myotubes suggesting the creation of sensitized background in myotubes that may force them to re-express muscle progenitor genes and enter cell cycle. The conversion of cell fates due to enhanced cell fate plasticity in polycomb compromised backgrounds has been documented in other model systems such as flies (Lee et al., 2005) and worms (Yuzyuk et al., 2009). In this regard, our work highlights the fact that key players in maintaining and establishing terminally differentiated state need to be erased transiently at epigenetic level in order to switch a differentiated state to a less differentiated state within the same cell lineage.
Since we used a generic tyrosine phosphatase and apoptosis inhibitor which is expected to modulate signaling in many biochemical pathways, the down-regulation of specific chromatin remodeling factors predisposed primary myotubes to respond to the mitogens of GM thereby changing their cell fate to that of proliferating myogenic precursor cells. Interestingly, while upon addition of inhibitor mix there were profound changes in the morphology of many myotubes, only a subset of these myotubes de-differentiated into proliferating mono-nucleated precursor cells. Since there is temporal progression towards the degree of terminal differentiation in myotubes there might be a specific time window when myotubes would be more responsive to the treatment and capable of undergoing cell fate reversal. Future work would determine the exact role of chromatin remodeling factors in acquiring muscle progenitor cell fate from multinucleated post-mitotic differentiated state. As there is temporal progression of gene expression upon myotube differentiation, it would be interesting to study in the future whether the inhibitor mix is sufficient to induce de-differentiation in mature myofibres formed in vivo or indeed other factors are required to yield regenerative cells.
Significance
Use of pharmacological inhibitors to modulate different signaling pathways without gene over expression is therapeutically relevant in coaxing differentiated cells to yield regenerative cells. Our data highlights the combinatorial use of tyrosine phosphatase and apoptosis inhibitors for primary myotube de-differentiation to yield myogenic proliferating cells which aided in muscle regeneration both in vitro and in vivo in SCID mice. The novel myotube labeling technique developed for this study served as a powerful tool to clearly show the origin of reprogrammed cells and to sort them away from reactivated reserve myoblasts. Importantly, small molecule inhibitors induced changes in myotubes at epigenetic level and facilitated them to enter proliferative state in mitogen rich medium. All together, the novel labeling technique employed the use of small molecule inhibitors advance the ongoing research in regenerative medicine and would enable unique clinical strategies for enhancing tissue regeneration.
Experimental Procedure
Animal strains
B6;129-Gt(ROSA)26Sortm1(rtTA*M2)Jae Col1a1tm2(tetOPou5f1)Jae/J strain (Stock Number: 006911), B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J strain (Stock Number: 006148), NOD.CB17-Prkdcscid/J (Stock Number: 001303) and C57BL6/J (2-3 months old) mice were obtained from pathogen free breeding colonies at The Jackson Laboratories. Animals were housed at the Northwest Animal Facility, University of California, Berkeley and procedures were performed in accordance to administrative panel on the Office of Laboratory Animal Care, UC Berkeley.
Reagents, antibodies, western blotting and immunofluorescence
BpV(phen) (Alexis Biochemicals), Doxycycline (Sigma) and apoptosis inhibitor (Q-VD-OPh, Non O methylated Cat No. 551476, Calbiochem), BrdU (Sigma), ECM (Sigma), Hoechst 33342 (Sigma), DNase1 (Sigma), propidium iodide (molecular probes), Rnase A (Fermentas) and mouse bFGF (R&D) were purchased. Antibodies to BrdU (ab6326), GFP (ab6556 and ab13970), Ki67 (abcam ab155800) and Oct4 (ab18976) were from Abcam. Antibodies against Actin (rabbit polyclonal) were from Sigma, Pax7 was from DSHB (Develpmental Studies Hybridoma Bank), eMyHC from DSHB and Upstate, mouse monoclonal antibodies against Myogenin, MyoD and p21 were from Santa Cruz Biotechnology. For western blotting, the cells were lysed in RIPA buffer (50mM Tris-Cl pH7.6, 150mM NaCl, 0.1% SDS, 1% NP-40, 0.25% sodium deoxycholate, 1mM sodium orthovanadate, 1mM NaF, 1mM PMSF, 1mM EDTA, 1X Protease inhibitor, Sigma) and protein concentration determined by Bradford Assay. 30 ug protein was run on precast 4-20% Gels (BioRad), then transferred to nitrocellulose membrane for 2 hrs and protein expression detected by BioRad Gel Doc/Chemi Doc imaging system and Quantity One Software. For immunofluorescence, cells were fixed with 4% PFA for 15 minutes at room temperature followed by permeabilization with 0.25% Triton-X 100 for 12 minutes and blocked for one hour in blocking buffer (1%BGS + 0.1% Na-Azide in 1XPBS) followed by primary antibody incubation in blocking buffer for 2 hours or overnight and secondary antibody incubation for 1 hour in blocking buffer with Alexa fluorophore conjugated species specific secondary antibody (Invitrogen). For BrdU labeling, cells were pulsed with 10uM BrdU for either 2 or 24 hours, fixed with 4% PFA for 15 mins, permeabilized with 0.25% Triton-X 100 for 12 minutes followed by DNase1 treatment (0.2 units/uL) for 30 minutes at room temperature. For all immunofluorescence, cells were mounted with mounting media containing DAPI (Prolong Gold Antifade, Invitrogen) to visualize nuclei in all immunostaining experiments.
Primary myoblast isolation
Primary myoblasts were obtained by isolation of satellite cells from these mice as described previously (Conboy and Conboy, 2010). TA and Gastrocnemius muscle of the different transgenic mice were injected with total 5ug cardiotoxin (Sigma) dissolved in 1XPBS and muscle was dissected out after 3 day injury as described (Conboy and Conboy, 2010). In brief, muscle underwent enzymatic digestion at 37°C in DMEM (CellGro) + 1% Penicillin/ Streptomycin + 250 units/ml Collagenase Type II (Sigma) solution for one and half hour on rocker with slight agitation. Bulk myofibers were purified by repeated rounds of trituration, sedimentation and washing to remove interstitial cells, tendons etc. Satellite cells were purified from these bulk myofibres by incubation with 0.5 units/ml dispase at 37°C for half an hour followed by sedimentation, washing, and fine mesh straining. The satellite cells were then cultured on 1:500 ECM (Sigma) coated plates in myoblast growth medium containing Ham’s F-10 nutrient mixture (Gibco)+ 20% Bovine Growth Serum (BGS; Hyclone) + 9 ng/ml bFGF (R&D) +1% Penicillin/Streptomycin. Later, proliferating fusion competent myoblasts were pre-plated to remove fibroblasts from culture.
Adenovirus infection and primary myotube labeling
Wild type MB or Tet-Oct4 MB obtained were infected with Ad-CMV-Cre or Ad-RFP control virus (Vector BioLabs) for 4-6 hours, washed off and cultured in GM for 24 hours. Ad-Cre MB were then lifted off the plates, counted and co-cultured with Lox-YFP myoblasts in differentiation inducing medium DMEM (Cellgro) + 2% horse serum (Sigma) + 1% Penicillin/Streptomycin (BD Falcon) on 12 well ECM coated plates. The 96 hour old myotubes were visualized for YFP expression, treated with different experimental conditions and photographed everyday using Zeiss epifluorescence microscope Axio Observer A.1 fitted with Zeiss EYFP filter (BP 500/20 FT515 BP 535/30) at 10X and 20X objective.
De-differentiation assay
WT myoblasts orTet-Oct4 (3×105) myoblasts upon 4-6 hour infection with 300 MOI of Ad-Cre virus were washed with growth medium twice to get rid of any residual virus and incubated for another 24 hours in growth medium at 37°C in 5% CO2 incubator. These myoblasts were then lifted off from the plates by addition of 1X PBS, counted (3×104 cells/well) and co-cultured with 7×104 Lox-YFP myoblasts (obtained from B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J mice strain) per well in differentiation inducing medium DMEM (Cellgro) + 2% horse serum + 1% Penicillin/Streptomycin on 12 well (BD Falcon) on ECM (1:500) coated 12 well plates. Later, 96 hour old YFP+ myotube cultures were treated with BpV (phen) (10uM) along with apoptosis/caspase inhibitor (Q-VD-OPh) (10uM) daily for 2 days with and without doxycycline (2ug/ml) in differentiation medium. The inhibitors were then withdrawn and cells were switched to growth medium containing bFGF (9ng/ml). The treated cells were fed fresh growth medium and bFGF everyday over a period of time and the morphology of YFP expressing myotubes was routinely visualized by epifluorescence microscope using YFP filter and photographed using 10X and 20X objectives. For live hoechst staining, 4μM of hoechst was added everyday to YFP labeled myotube cultures, incubated for 10 minutes at 37°C in 5% CO2 incubator, washed off to remove unlabeled hoechst and photographed.
Calculation of myotube reprogramming efficiency
The labeled YFP+ myotubes per well in 4 day old Cre-Lox myotube cultures were determined to be ~2600. After inhibitor treatment, ~ 350 YFP+ mononucleated cells were found in culture. Hence reprogramming efficiency was calculated as the percent of YFP+ mononucleated cells out of the total number of labeled YFP+ myotubes which was determined ~13.46%. Another method was based on the determination of Lox YFP myonuclei in labeled myotubes. As an average number of myonuclei present in each YFP+ myotube is ~4 and for myotube labeling, Cre and Lox YFP myoblasts were cultured in ratio of 1:2 hence Lox YFP myonuclei were estimated to be ~ 6940 [ (2600×4)×2/3]. Therefore, reprogramming efficiency calculated was the percent of YFP+ mononucleated cells out of an estimated total number of Lox YFP myonuclei in YFP+ myotubes and was determined ~ 5.04% [(350/6940)×100] (see Figure S2 d).
Cell transplantation
1× 106 reprogrammed YFP+ mononucleated cells expanded in GM in culture were re-suspended in medium containing Ham’s F-10 with 2% BGS and injected in 24 hours cardiotoxin pre injured Tibialis Anterior (TA) muscles of NOD-SCID mice (4 weeks old) obtained from Jackson lab. Control injections were performed with medium alone or Lox YFP myoblasts re-suspended in medium. After 2 -3 weeks of cell injections, TA muscles were dissected out and fixed in 4% paraformaldehyde (PFA) for minimum of 2 hours and subsequently washed three times with 1X PBS for total of 45 minutes. The muscles were then sequentially transferred to 2%, 5% and10% sucrose in PBS with slight agitation at room temperature for 30 minutes-1hour. Finally muscles were left overnight in 20% sucrose at 4°C and frozen in liquid nitrogen cooled iso-pentane in OCT embedding medium. 10um muscle sections were cut and mounted on superfrost slides. 300um serially spaced muscle sections from whole block were post-fixed with 4% PFA for 10 minutes and permeabilized for 10 minutes with 0.25% Triton X-100 and blocked in goat serum. Primary antibodies against YFP (chicken polyclonal GFP, Abcam) and laminin (rat laminin from Sigma) were added overnight followed by 1 hour incubation in the respective fluorochrome conjugated secondary antibodies (Goat anti Chicken 488 and Goat anti rat 546) from Molecular probes.
RNA isolation and qRT-PCR
RNA isolation was performed by RNAeasy Kit (Qiagen) according to manufacture recommendations. For real time-qPCR, transcribed cDNA was diluted 1:5 for every sample and 1ul of this diluted cDNA was used for 25ul PCR reaction containing 80nM forward and reverse primers, 12.5 ul of 2X iQ Syber Green mix (Bio-Rad), ran on iQ5 cycler (Bio-Rad) and data analysed by iQ5 optical system software. The values normalized against internal control GAPDH and plotted using ΔΔCt method. The primers against specific genes for qRT-PCR are: GAPDH-F 5′-GGGAAGCCCATCACCATCT-3′ and GAPDH-R 5′-GCCTCACCCCATTTGATGTT-3′; Myogenin-F 5′-GACCCTACAGACGCCCACAA and Myogenin-R-5′-CCGTGATGCTGTCCACGAT-3′;MyoD-F 5′-CGGCTCTCTCTGCTCCTTTG-3′ and MyoD-R-5′-GAGTCGAAACACGGGTCATCA-3′; Pax7-F-5′-CCCTCAGTGAGTTCGATTAGC-3′ and Pax7-R-5′-CCTTCCTCGTCGTCCTCTTTC-3′; p21-F-5′GAACATCTCAGGGCCGAAAA-3′and p21-R-5′-TGCGCTTGGAGTGATAGAAATC-3′; eMyHC-F-5′-AGAGGACGTGTATGCCATGA-3′ and eMyHC-R-5′-TGGCCATGTCCTCAATCTTGT-3′; Cre-recombinase-F-5′-GCCGGGTCAGAAAAAATGG and Cre recombinase-R- 5′-AGGGCGCGAGTTGATAGCT-3′; eYFP F-5′ -GCACGACTTCTTCAAGTCCGCCATGCC-3′ and eYFP R 5′-GCG GATCTTGAAGTTCACCTTGATGCC-3′. Primers for p15 and p16 have been described elsewhere (Li et al., 2009). For SA biosciences/Qiagen PCR arrays, 0.5 ug RNA was reverse transcribed using SA biosciences RT kit and PCR arrays carried out according to manufacture instructions. Web based PCR array data analysis tool of SA biosciences was utilized for data analysis.
Flow cytometry
De-differentiated YFP+ mononucleated cells were lifted off the plates by addition of PBS and re-suspended in PBS+ 5% BSA, filtered through 40uM filter (BD Falcon) to remove any aggregates and placed on ice. Cell sorting was performed on Cytopeia Influx sorter with gating on YFP+ population set by a comparison with negative control sample of Lox-YFP myoblasts. The sorted cells were replated on ECM coated tissue culture dishes containing myoblast growth medium with 9ng/ml bFGF and assessed for different markers. For cell cycle analysis, de-differentiated cells were expanded in culture, pelleted down and fixed in ice cold 70% ethanol overnight at −20°C. Next day, cells were pelleted down at 1400rpm for 5 minutes at 4°C and ethanol was removed. Cells were washed once with ice cold 1X DPBS and re-suspended in DNA staining solution (RNase 0.1mg/ml, Propidium iodide 2ug/ml, 0.05% Triton X 100 in 1X DPBS) for 30 minutes at room temperature. The cells were pelleted down, washed once with ice cold DPBS and resuspended in DPBS+ 3% BSA for FACS analysis.
Statistical Analysis
p values were determined using student’s t-test (2 samples equal variance, 2 tailed).
Supplementary Material
Highlights.
Irreversible lineage marking of multinucleated myotubes by Cre-Lox method.
Small molecule inhibitors reprogram lineage marked muscle to their progenitor cells.
Reprogrammed myogenic progenitor cells contribute to muscle regeneration in vivo.
Acknowledgements
We would like to thank Hector Nolla for FACS sorting (UC Berkeley flow cytometry facility). We acknowledge the CHPS Imaging core and Paul Herzmark for assistance with time lapse imaging microscopy. Thanks to Mike Conboy, Conboy lab members and Vivek Chopra for critical feedback, help and support. This work was supported by NIH/NIA AG 027252, CIRM RN1-00532 grant to IMC and CIRM postdoctoral fellowship to PP.
Abbreviations
- YFP
Yellow fluorescent protein
- RFP
Red fluorescent protein
- dox
doxycycline
- DM
differentiation medium
- GM
growth medium
- BpV
bis-peroxovanadium
- bFGF
basic fibroblast growth factor
- eMyHC
embryonic myosin heavy chain
- GAPDH
glyceraldehyde phosphate dehydrogenase
- MB
myoblasts
- MT
myotubes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
P.P: conceived project, designed and performed experiments, collected, assembled analyzed and interpreted data and wrote manuscript; I.M.C: conceived project, directed the study, interpreted the data and wrote the manuscript. The authors declare no competing financial interests
References
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–1291. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnac G, Fajas L, L’Honore A, Sardet C, Lamb NJ, Fernandez A. The retinoblastoma-like protein p130 is involved in the determination of reserve cells in differentiating myoblasts. Curr Biol. 2000;10:543–546. doi: 10.1016/s0960-9822(00)00471-1. [DOI] [PubMed] [Google Scholar]
- Castaldi L, Serra C, Moretti F, Messina G, Paoletti R, Sampaolesi M, Torgovnick A, Baiocchi M, Padula F, Pisaniello A, et al. Bisperoxovanadium, a phospho-tyrosine phosphatase inhibitor, reprograms myogenic cells to acquire a pluripotent, circulating phenotype. FASEB J. 2007;21:3573–3583. doi: 10.1096/fj.06-7454com. [DOI] [PubMed] [Google Scholar]
- Cayrol C, Knibiehler M, Ducommun B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene. 1998;16:311–320. doi: 10.1038/sj.onc.1201543. [DOI] [PubMed] [Google Scholar]
- Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Conboy IM. Preparation of adult muscle fiber-associated stem/precursor cells. Methods Mol Biol. 2010;621:149–163. doi: 10.1007/978-1-60761-063-2_10. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- Delgado I, Huang X, Jones S, Zhang L, Hatcher R, Gao B, Zhang P. Dynamic gene expression during the onset of myoblast differentiation in vitro. Genomics. 2003;82:109–121. doi: 10.1016/s0888-7543(03)00104-6. [DOI] [PubMed] [Google Scholar]
- Duckmanton A, Kumar A, Chang YT, Brockes JP. A single-cell analysis of myogenic dedifferentiation induced by small molecules. Chem Biol. 2005;12:1117–1126. doi: 10.1016/j.chembiol.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Endo T, Nadal-Ginard B. Transcriptional and posttranscriptional control of c-myc during myogenesis: its mRNA remains inducible in differentiated cells and does not suppress the differentiated phenotype. Mol Cell Biol. 1986;6:1412–1421. doi: 10.1128/mcb.6.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Nadal-Ginard B. Reversal of myogenic terminal differentiation by SV40 large T antigen results in mitosis and apoptosis. J Cell Sci. 1998;111(Pt 8):1081–1093. doi: 10.1242/jcs.111.8.1081. [DOI] [PubMed] [Google Scholar]
- Forcales SV, Puri PL. Signaling to the chromatin during skeletal myogenesis: novel targets for pharmacological modulation of gene expression. Semin Cell Dev Biol. 2005;16:596–611. doi: 10.1016/j.semcdb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Friday BB, Pavlath GK. A calcineurin- and NFAT-dependent pathway regulates Myf5 gene expression in skeletal muscle reserve cells. J Cell Sci. 2001;114:303–310. doi: 10.1242/jcs.114.2.303. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjiantoniou E, Anayasa M, Nicolaou P, Bantounas I, Saito M, Iseki S, Uney JB, Phylactou LA. Twist induces reversal of myotube formation. Differentiation. 2008;76:182–192. doi: 10.1111/j.1432-0436.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Scholer HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009 doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Skapek SX, Novitch B. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr Opin Cell Biol. 1994;6:788–794. doi: 10.1016/0955-0674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Latella L, Lukas J, Simone C, Puri PL, Bartek J. Differentiation-induced radioresistance in muscle cells. Mol Cell Biol. 2004;24:6350–6361. doi: 10.1128/MCB.24.14.6350-6361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella L, Sacchi A, Crescenzi M. Long-term fate of terminally differentiated skeletal muscle cells following E1A-initiated cell cycle reactivation. Cell Death Differ. 2000;7:145–154. doi: 10.1038/sj.cdd.4400592. [DOI] [PubMed] [Google Scholar]
- Latella L, Sacco A, Pajalunga D, Tiainen M, Macera D, D’Angelo M, Felici A, Sacchi A, Crescenzi M. Reconstitution of cyclin D1-associated kinase activity drives terminally differentiated cells into the cell cycle. Mol Cell Biol. 2001;21:5631–5643. doi: 10.1128/MCB.21.16.5631-5643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loof S, Straube WL, Drechsel D, Tanaka EM, Simon A. Plasticity of mammalian myotubes upon stimulation with a thrombin-activated serum factor. Cell Cycle. 2007;6:1096–1101. doi: 10.4161/cc.6.9.4141. [DOI] [PubMed] [Google Scholar]
- McGann CJ, Odelberg SJ, Keating MT. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc Natl Acad Sci U S A. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair (Amst) 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- Odelberg SJ, Kollhoff A, Keating MT. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Holtzer H. Myogenesis: fusion, myosin synthesis, and the mitotic cycle. Proc Natl Acad Sci U S A. 1966;56:1484–1490. doi: 10.1073/pnas.56.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Interplay between proliferation and differentiation within the myogenic lineage. Dev Biol. 1992;154:261–272. doi: 10.1016/0012-1606(92)90066-p. [DOI] [PubMed] [Google Scholar]
- Pajalunga D, Puggioni EM, Mazzola A, Leva V, Montecucco A, Crescenzi M. DNA replication is intrinsically hindered in terminally differentiated myotubes. PLoS ONE. 2010;5:e11559. doi: 10.1371/journal.pone.0011559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Puri PL. The epigenetic network regulating muscle development and regeneration. J Cell Physiol. 2006;207:1–11. doi: 10.1002/jcp.20489. [DOI] [PubMed] [Google Scholar]
- Rosania GR, Chang YT, Perez O, Sutherlin D, Dong H, Lockhart DJ, Schultz PG. Myoseverin, a microtubule-binding molecule with novel cellular effects. Nat Biotechnol. 2000;18:304–308. doi: 10.1038/73753. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- Rumora L, Barisic K, Maysinger D, Zanic Grubisic T. BpV (phen) induces apoptosis of RINm5F cells by modulation of MAPKs and MKP-1. Biochem Biophys Res Commun. 2003;300:877–883. doi: 10.1016/s0006-291x(02)02952-2. [DOI] [PubMed] [Google Scholar]
- Saccone V, Puri PL. Epigenetic regulation of skeletal myogenesis. Organogenesis. 6:48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale FE, Holtzer H. DNA synthesis and myogenesis. Exp Cell Res. 1961;24:508–520. doi: 10.1016/0014-4827(61)90450-5. [DOI] [PubMed] [Google Scholar]
- Tiainen M, Pajalunga D, Ferrantelli F, Soddu S, Salvatori G, Sacchi A, Crescenzi M. Terminally differentiated skeletal myotubes are not confined to G0 but can enter G1 upon growth factor stimulation. Cell Growth Differ. 1996;7:1039–1050. [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111(Pt 6):769–779. doi: 10.1242/jcs.111.6.769. [DOI] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.