Summary
In this review, I discuss the evidence for differential natural selection acting across enzymes in the glycolytic pathway in Drosophila. Across the genome, genes evolve at very different rates and possess markedly varying levels of molecular polymorphism, codon bias and expression variation. Discovering the underlying causes of this variation has been a challenge in evolutionary biology. It has been proposed that both the intrinsic properties of enzymes and their pathway position have direct effects on their molecular evolution, and with the genomic era the study of adaptation has been taken to the level of pathways and networks of genes and their products. Of special interest have been the energy-producing pathways. Using both population genetic and experimental approaches, our laboratory has been engaged in a study of molecular variation across the glycolytic pathway in Drosophila melanogaster and its close relatives. We have observed a pervasive pattern in which genes at the top of the pathway, especially around the intersection at glucose 6-phosphate, show evidence for both contemporary selection, in the form of latitudinal allele clines, and inter-specific selection, in the form of elevated levels of amino acid substitutions between species. To further explore this question, future work will require corroboration in other species, expansion into tangential pathways, and experimental work to better characterize metabolic control through the pathway and to examine the pleiotropic effects of these genes on other traits and fitness components.
Key words: Drosophila, metabolism, metabolic control, molecular evolution, systems
Introduction
Understanding the causes of the different rates of molecular evolution and levels of polymorphism among genes has been a major challenge in evolutionary biology (Larracuente et al., 2008; Bustamante et al., 2005). One proposal is that the intrinsic properties of enzymes and their role and position in functional pathways have direct effects on the molecular evolution of the participating genes. This view has taken the study of molecular evolution and adaptation at the genic level to the level of pathways, systems and networks of genes (Eanes, 1999; Jovelin and Phillips, 2009; Wagner, 2005). Clearly, one network of special interest is that of the energy-producing pathways and, in particular, the central pathway of glycolysis. Over the past twenty years, our laboratory has studied in Drosophila the genetic variation and population genetics of the glycolytic pathway and its side branches. Our long-term interest in this particular pathway has both historical links and a practical side; what the glycolytic pathway lacks in size, it more than makes up for in its information or knowledge base.
Certainly, with respect to many features, the glycolytic pathway is among the best understood in biology. Despite encoding members of a single functioning pathway, the glycolytic genes vary by an order of magnitude in levels of polymorphism and divergence. In Drosophila, as in other organisms, the expression levels also vary by an order of magnitude among steps, with the traditional non-equilibrium enzymes possessing the lowest expression levels and genes at the bottom of the pathway possessing the highest levels. Of major importance is that the individual enzymes have been extensively studied by enzymologists with respect to catalytic mechanism, and that some, such as hexokinase (HEX) and triosephosphate isomerase (TPI), represent textbook cases. Furthermore, physiologists have focused many questions on the glycolytic pathway from the standpoint of optimal design for regulation and function (Hochochka and Somero, 2002). The protein structures and structure-function relationships are well known for most members and this allows the placement of individual mutations in a functional context. As in much of population genetics, as well as in investigations into the molecular variation in this pathway, progress has been linked to a progression of methodological opportunities.
The first opportunity to explore genetic variation in the glycolytic pathway emerged from the many allozyme studies of the 1970s and 1980s. Because they exploited staining methods that were coupled to energy-state co-factors such as NADH and NADPH, these studies were focused, albeit unintentionally, on polymorphism of genes in the energy-producing pathways (Mitton, 1997). The introduction of PCR in the late 1980s and early 1990s, allowed assessing variation directly in DNA sequences of any gene, and at both the intra- and interspecific levels. Moreover, aside from simply getting a better handle on the underlying molecular nature of polymorphism and divergence, this new data opened the door to the use of genealogical models and historical interference to assess the action of natural selection (Eanes, 1999). Nevertheless, the emphasis still remained on the single gene because each study required an initial discovery (cloning) and characterization of a gene's sequence to set the process in motion. The availability of the fully annotated genome sequence of Drosophila melanogaster at the turn of the current century (Adams et al., 2000) enormously changed the landscape by removing the time-consuming and rate-limiting requirement of individual gene discovery. This has now permitted the study of gene evolution in the larger context of their roles and positions in pathways and interaction networks (Greenberg et al., 2008).
Before the first D. melanogaster full genome sequence was unveiled, our laboratory had been moving towards a pathway-centered view of selection on glycolysis and its immediate branches. Along with discoveries from other laboratories, we had begun a project using PCR-cloning strategies to recover the primary gene sequences for most of the pathway members (Verrelli and Eanes, 2000; Eanes, 1999; Duvernell and Eanes, 2000; Eanes et al., 1993; Hasson et al., 1998; Merritt et al., 2005). Of course, this approach became irrelevant when the genome sequence of D. melanogaster became available. The opportunity to study many problems using the population genetics of D. melanogaster has been advanced by the plan to produce full sequences of 192 Drosophila genomes from a single population from Raleigh, NC, USA, and the recent release of 38 genome sequences from this collection by the Drosophila Population Genomics Project (http://www.DPGP.org).
For decades, D. melanogaster has been the main population genetic model in biology (Powell, 1997). Most of the ideas of how to study selection using DNA sequence variation have originated from Drosophila studies and the DNA sequences (including some of ours) collected through the 1990s. Drosophila melanogaster's history of worldwide colonization is well understood. The species has an Afrotropical origin and has spread over most of the world as a human commensal on fermenting resources. Its colonization of the Western Hemisphere and Australia is as recent as the last 200 years (David and Capy, 1988). There is a growing appreciation that this has resulted in geographical selection for many traits, especially those associated with the ability to withstand the stresses associated with overwintering in temperate populations (Schmidt et al., 2005; Schmidt and Paaby, 2008; Schmidt et al., 2008; Reaume and Sokolowski, 2006; Mitrovski and Hoffmann, 2001; Hoffmann et al., 2001; Umina et al., 2005). From an experimental standpoint, the genetic tools of D. melanogaster are unparalleled and methods are available to permanently knockout or modify activity in many genes (Bellen et al., 2004; Venken and Bellen, 2005) and to control metabolic function (Merritt et al., 2005; Merritt et al., 2006; Eanes et al., 2006; Eanes et al., 2008; Merritt et al., 2009). In addition, D. melanogaster has been a focal model in the study of energy partitioning and aging, where differential expression of the energy-producing and energy-consuming pathways has come under intense scrutiny as a response to dietary restriction (Gershman et al., 2007; Baker and Thummel, 2007). In this review, I will be discussing work emphasizing Drosophila as a model organism. To best study selection on metabolic pathways requires using a species model in which metabolic pressures can be associated with agents of selection in natural populations. We believe D. melanogaster is uniquely positioned to study natural selection on metabolic tradeoffs, as they translate into selection on single genes and pathways.
Metabolic control and selection
To understand how natural selection will impact the evolution of molecular variation in energy-producing pathways requires that we address the genotype-phenotype relationship. A universal phenomenon of genetic systems is the strong hyperbolic relationship between gene dosage and phenotype – the observation that defines genetic dominance. The intrinsic cause of genetic dominance has been debated since the pioneering work of Wright, Fisher and Haldane (Wright, 1929; Wright, 1934; Haldane, 1939). This problem was later taken up by Kacser and Burns (Kacser and Burns, 1981) from the perspective of flux through a biochemical pathway. They concluded that flux is a system-wide feature of the interactions across the total pathway and, as a consequence, individual steps exercise little control; in other words, genetic dominance is an inherent consequence of this system-wide control and this explains the hyperbolic relationship at most steps. This does not reject some uneven distribution of control, but the causes of inequity are hard to predict. Elements of their hypothesis are not without critics, but their paper clearly identified the metabolic control of enzymes as a key property in understanding levels of molecular variation among genes (Fell, 1997; Watt and Dean, 2000; Eanes, 1999; Wright and Rausher, 2010).
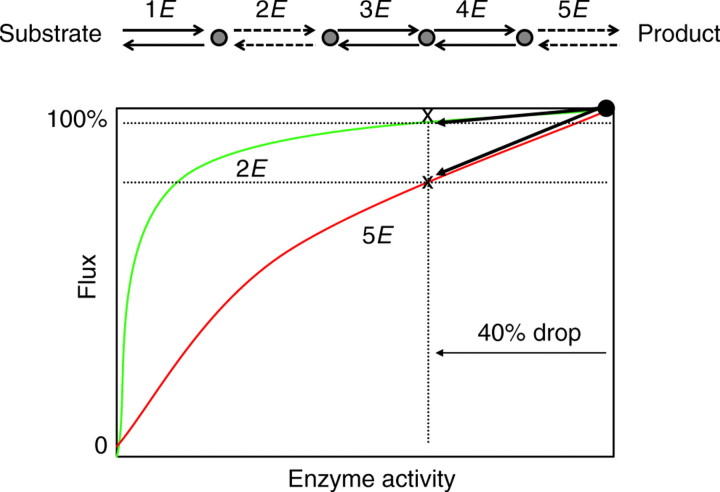
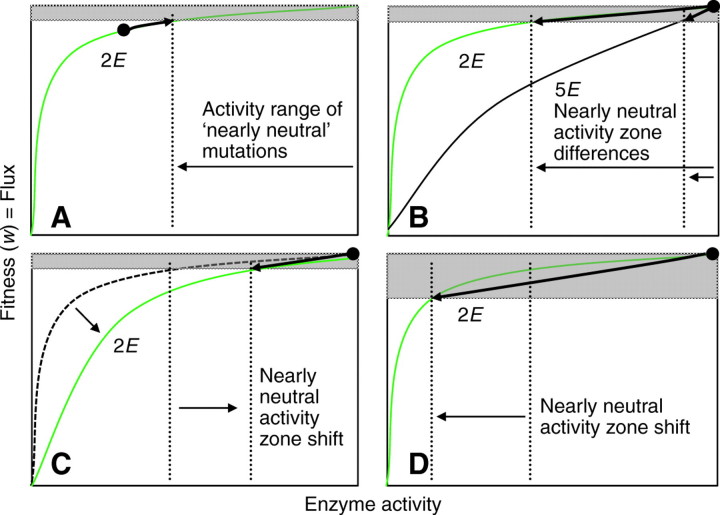
How do we connect this important property of pathway steps with its effect on molecular evolution and polymorphism? A hypothetical pathway of five steps in which metabolic control (or the sensitivity of flux to perturbation) varies at steps 2E and 5E is shown in Fig. 1. The same mutation (causing a 40% reduction in expression level) in 2E and 5E will have very different impacts on flux. For example, the mutation in enzyme 2E results in a <5% reduction in flux, but results in a 20% reduction in flux for enzyme 5E (Fig. 1). We need next to consider for enzymes imbedded in a pathway how enzyme-specific changes in control and the influence of population size translate into different levels of segregating genetic variation in natural populations and/or rates of divergence between species. This involves the effect of diminishing fitness gains and their interaction with the stochastic effects imposed by finite population size or genetic drift. It is often assumed that fitness is a monotonically increasing function of flux. Hartl et al. (Hartl et al., 1985) discussed the reasons why, if increasing activity is associated with increasing fitness, enzymes do not continue to accumulate mutations that increase function and activity level. They proposed that if fitness is positively associated with increasing flux, then many mutations that increase activity (through catalytic improvement, enhanced transcription, increased translational efficiency, or increased stability and shorter half-lives) will be fixed by selection and the population mean activity will mutation-by-mutation incrementally climb the fitness-activity hyperbolic curve (Fig. 2A, upper left). This improvement will not continue indefinitely because of the ever-diminishing return associated with the hyperbolic function, but will reach a point at which the associated incremental gains in fitness for a new mutation will be matched by the random noise of genetic drift. At this point, the substitution of new mutations via a selection-driven process stalls and the genes or enzymes begin to evolve under a ‘nearly neutral’ process (Ohta, 1992). Fig. 2B (upper right) shows how this proposal predicts very different impacts on the range of activity function detectable by selection for two enzymes with very different control functions. Viewed from the standpoint of the neutral activity zone and the action of purifying selection, gene or enzyme 2E will accommodate higher rates of neutral amino acid substitution and polymorphism than gene or enzyme 5E. Conversely, under diversifying or balancing selection that favors variation in flux for a pathway, we might expect 5E to accumulate more mutations. Therefore, depending on one's view of the nature of selection acting on a pathway, metabolic control will influence molecular evolution. This set of proposals serves as a useful working hypothesis and establishes basic expectations about how metabolic control differences might lead to differences in genetic variation and divergence among genes.
Fig. 1.
An example of a five-step pathway (enzymes 1E to 5E) in which two of the member enzymes, 2E and 5E, possess different metabolic control. Formally, the control is defined as the slope of the function at any point, but practically speaking the point of mean population activity is the reference. It shows that similar proportional drops in activity (40%) caused by mutations have a much greater impact on pathway flux for enzyme 5E than for enzyme 2E.
Fig. 2.
The evolution and consequences of metabolic control, see text. (A) When fitness is proportional to flux the mean activity will increase through mutation until the incremental advance in fitness, Δw, is of the order of the reciprocal of the effective population size, 1/Ne. At this point the mutation substitution rate converges on the neutral substitution rate and mean activity stalls. (B) Two different enzymes with different control functions and, as a consequence, different activity ranges over which mutational reductions qualify as neutral. Gene 2E will accommodate more neutral mutation than gene 5E. (C) The effect of changing the metabolic control function for 2E. In this case mutations, formerly with neutral fitness effects, will be purged by natural selection. (D) The effect of decreasing population size. This lowers the neutral fitness threshold and increases the activity range over which mutations are effectively neutral. The rate of neutral mutation substitution will increase, as will polymorphism.
So far, these proposals assume no contextual changes to the ‘system’, yet they are expected to have significant consequences for the selection pressures on genes by shifting the function–activity zone boundary for the nearly neutral mutation. These contextual changes would arise as the result of: (1) a change in the functional context of the pathway, branch or enzyme (either through epistasis or the environment); or (2) a change in the effective population size of the species. In the former case, interactions with other genes or a change in flux demand in competing branches may redistribute pathway control and consequently alter the shape of the hyperbolic function for a particular step (Bagheri et al., 2003; Bagheri and Wagner, 2004) (Fig. 2C, bottom left). In the case of population size fluctuations, the step control does not change, but the population size change shifts the boundary of the nearly neutral fitness range and therefore the activity–function zone over which mutation is detectable to selection (Fig. 2D, bottom right). This idea that the efficiency of natural selection in removing or favoring mutations is dependent on the effective population size and genetic drift was first introduced by Ohta (Ohta, 1992) and has taken a firm hold in population genetics and genome evolution, where it appears to be useful in explaining many patterns in molecular data that appear to have a dependence on population size (Kreitman, 1996; Lynch, 2007). Because of both of these phenomenon, the intensity of both positive and negative selection on the components of a pathway is likely to fluctuate through time, and could possibly include rounds of compensatory evolution in which mutations that possess lower activity and were fixed at earlier times are replaced by mutations of higher function at later intervals (Kulathinal et al., 2004; Hartl and Taubes, 1996). These consequences will depend on the intrinsic level of metabolic control for particular steps and again emphasize the importance of this property.
If metabolic control is not shared by all steps, are there intrinsic or contextual properties of enzymes that might generally set the expected level of their individual control? This is a long debated and unresolved question (Fell, 1997). The classic view is that some enzymes have inherent control by virtue of their regulatory potential (Hochachka and Somero, 2002). Most notable in the glycolytic pathway are the textbook examples of hexokinase and phosphofructokinase. These enzymes are assumed to operate in vivo far from thermodynamic equilibrium and this feature allows them to sharply ramp up and down activity in response to allosteric energy-state signals, such as AMP and NADH levels. Their allosteric response to these co-factors is an undeniable fact, but their contribution to control, as defined here, is unclear. The second property potentially determining the level of step importance in control is pathway topology, in particular the location and presence of branches that appear to be points of increased control (LaPorte et al., 1984; Stephanopoulos and Vallino, 1991). In metabolic pathways, branches also partition resources to energy-storage pools, such as glycogen, lipids and trehalose. This is also important because it is expected that storage pools will come under natural selection where we see life history tradeoffs (Chippindale et al., 1996). Wright and Rausher (Wright and Rausher, 2010) recently carried out an extensive series of theoretical simulations to address the question of where under specified selection a model's control will evolve in a linear pathway, and how this distribution in control might affect the acquisition and accumulation of adaptive mutations among steps. In general, they conclude that, in a largely irreversible pathway, flux control will evolve to be unequally distributed and vested in the top steps. Under natural selection and depending on the proximity to an adaptive optimum, these top steps will experience the greatest molecular evolution in the sense of allele substitution. Their study did not examine the more complex influence of branch points. From these conclusions it appears that uncovering the rules for differential control at branches is central to understanding metabolic adaptation.
Assessing natural selection
The first strategy we have taken to explore these ideas is to examine the population genetic evidence for natural selection across the glycolytic pathway. Natural selection acts on new mutations in two ways. It can remove all variation incompatible with the optimal function of an enzyme or it can fix activity-modifying mutations in response to diversifying selection acting within and among populations. It is safely assumed that nearly all spontaneous mutation suffers the first fate. In the second case, positive selection will favor and substitute non-coding, amino acid replacement, and even synonymous mutations that sufficiently impact function (at a rate exceeding a neutral drift rate). It appears this is not uncommon in Drosophila genes, where it is estimated that 20 to 40% of amino acid differences between species have been positively selected (Smith and Eyre-Walker, 2002; Shapiro et al., 2007). Finally, selection may also maintain polymorphisms via any number of potential mechanisms that together encompass balancing selection and local adaptation. It is possible to explore both these possibilities in population genetic data and to look at the distribution of evidence for one or the other form of selection across the genes of the glycolytic pathway. There are a number of approaches that have been introduced to search among genes for evidence of these so-called ‘signatures’ of natural selection (Vasemagi and Primmer, 2005). The two approaches we have used in our work are: (1) screening for evidence of short-term contemporary selection presented as geographical variation in allele frequencies in the form of latitudinal clines; and (2) testing for patterns of longer-term positive selection that have resulted in apparent excess amino acid substitution between species. It should be emphasized that a lack of evidence here does not preclude other types of selection acting on a gene as, depending on the form, strength and past timing of selection, not all such events will leave an imprint in DNA sequences (Nielsen, 2005).
Clines
In D. melanogaster, numerous clines with latitude are known for inversions, microsatellites, morphological traits, and life-history-associated genes (Schmidt et al., 2005; James et al., 1997; Gockel et al., 2001; Schmidt et al., 2000; Schmidt and Paaby, 2008; Schmidt et al., 2008; Fry et al., 2008). In addition, many clines in allozyme genes have been described, the most famous being in genes for alcohol dehydrogenase (Adh) and glycerol 3-phosphate dehydrogenase (Gpdh) (Oakeshott et al., 1981; Oakeshott et al., 1982; Oakeshott et al., 1983; Berry and Kreitman, 1993). These clines can reflect a long-term balance between selection and migration, or cases of recent adaptation in the spreading source population. For example, Oakeshott et al. (Oakeshott et al., 1983) described latitudinal clines in the phosphogluconate dehydrogenase (Pgd) Fast (F) allele in Europe, North America and Australia. The F allele was fixed in Europe. Recently, Glinka et al. (Glinka et al., 2003) and Beisswanger et al. (Beisswanger et al., 2006), in a study of European and African populations for evidence of adaptive sweeps, identified an X chromosome region bearing the footprint of very low heterozygosity. The F allele of Pgd was found in the center of the inferred sweep region (which spanned less than 60 kilobases). They estimated that this selection event (and presumably the associated cline in the F allele) occurred less than 6000 years ago, which would be consistent with the colonization of Europe by D. melanogaster following the introduction of agriculture and fermentation (their approach belies the future of detecting natural selection in genomes). Generally, clines have been studied in the context of a single gene, and here we begin to apply it across a pathway and branches.
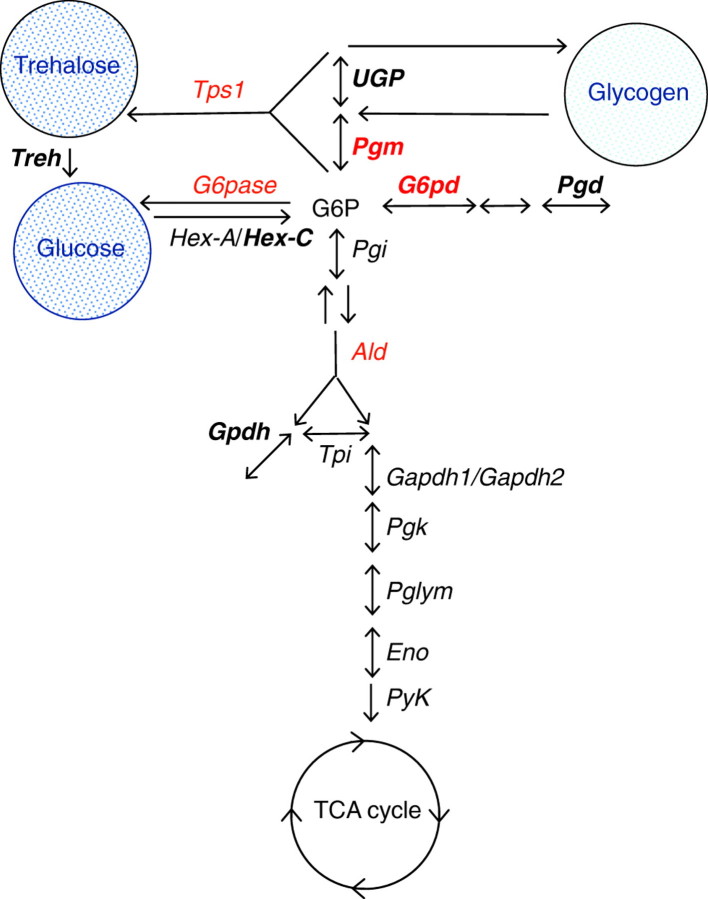
In 2004, we published the results of clinal variation for ten populations of D. melanogaster collected throughout the Eastern US from Florida to Vermont in 1997 (Sezgin et al., 2004). We assembled data on 12 new genes (23 single-nucleotide polymorphisms, SNPs) and combined these with earlier reports to establish the incidence of clinal variation in 20 metabolic genes. Of the 12 new genes, three – those encoding glutamate dehydrogenase (Gdh), trehalase (Treh) and glucose 1-phosphate uridylyltransferase (UGP) – showed significant allele frequency clines. These combined with the earlier reports showed that, with respect to just the glycolytic pathway and its immediate branches, seven of the 14 genes studied possess one or more SNPs that are clinal (Fig. 3). The pathway and branch genes hexokinase C (Hex-C), phosphoglucomutase (Pgm), glucose 6-phosphate dehydrogenase (G6pd), Treh, Pgd, UGP and Gpdh all possess SNPs with clines. How is the evidence for clines distributed across the pathway? Our results show a high incidence of clines for genes at the top of the pathway. The most enlightening observation, one that also emphasizes the enhanced information content of sequence data per se, is the number of cases (six out of seven) in which the derived (non-ancestral) amino acid change is the allele that increases with the colonization of temperate environments in the North. Because D. melanogaster is a tropical species that has invaded temperate regions, this observation is consistent with the acquisition of new mutations (or derived rare mutations in Africa) being adaptive responses to this new climatic challenge.
Fig. 3.
Summary for the studied enzymes of the glycolytic pathway in Drosophila. The results of geographical clinal variation in SNP polymorphism and of the McDonald–Keitman (M–K) tests are combined to assess adaptive amino acid substitutions. Three genes (Ald, G6Pase and Tps1) show significant M–K tests (red letters). Five genes (Treh, UGP, Pgd, Hex-C and Gpdh) show significant variation that is clinal (bold). Two genes (Pgm and G6pd) show both significant clines and significant rejection in the M–K tests (red and bold). Note, G6Pase, Ald and Hex-A were not studied in the cline report. See text for details.
McDonald–Kreitman tests
Our second approach focused on the longer timescale associated with amino acid substitutions between species. It applies the classic expectation of the neutral theory of molecular evolution of Kimura (Kimura, 1983) that predicts a correlation between polymorphism and divergence. Under the neutral model, the ratio of polymorphisms for nonsynonymous and synonymous sites should be the same as the ratio of fixed differences between species for both types of sites (McDonald and Kreitman, 1991). Assuming the absence of selection on synonymous sites, excesses of amino acid polymorphism or divergence are consistent with adaptive amino acid changes (tested by a G-test of the two-by-two table). Deviations revealed by the McDonald–Kreitman (M–K) test are generally interpreted as excesses of amino acid substitutions (Smith and Eyre-Walker, 2002), although excesses of polymorphism are often seen in samples of random genes from D. melanogaster (Shapiro et al., 2007). The introduction of polymorphism data greatly enhances our ability to infer the action of selection and makes the M–K test (as well as the Bayesian estimation below) preferable over using the dN/dS ratio in species such as Drosophila, which are outbred and might possess equilibrium structures closer to those assumed in neutral models (see Li et al., 2008).
We carried out M–K tests for 17 genes in the glycolytic pathway and its immediate branches (Flowers et al., 2007). The test can be carried out using different partitions of the data. In the case of a two-species data set (as here with D. melanogaster and D. simulans), the polymorphism data can come from one or the other species separately, or combined. By using a third closely related species as an ‘outgroup’ (D. yakuba here), the divergence can be partitioned by the D. melanogaster or the D. simulans lineage as well, depending on one's interest in lineage-specific effects. When we apply this test throughout the pathway we see that, similar to our observations on clines and pathway position, most of the evidence indicates positive selection at the top of the pathway surrounding glucose 6-phosphate (G6P). This is the point of energy apportioning to glycolysis, glycogen, trehalose, and the pentose shunt. The M–K tests indicated that glucose 6-phosphatase (G6pase), G6pd, trehalose 6-phosphate synthase 1 (Tps1) and aldolase (Ald) individually deviate from neutrality in the direction attributed to an excess of amino acid fixation, whereas Pgm showed a significant excess of amino acid polymorphism. When fixations are assigned to the D. melanogaster or D. simulans lineages, the departures from neutrality are consistent with balancing selection in D. melanogaster at Pgm (Verrelli and Eanes, 2000), positive selection at Tps1, G6pase and Ald in the D. simulans lineage, and positive selection in both lineages at G6pd (Eanes et al., 1993; Eanes et al., 1996). When considered with respect to pathway position, four out of the five genes that deviate from neutrality (Tps1, G6pase, G6pd, Pgm) code for enzymes at the G6P crossroads (Fig. 3). With the exception of Ald, neutrality could not be rejected for the remaining 14 genes in lineage-specific tests, or when polymorphisms and fixations were combined across lineages.
It should be emphasized that the magnitude of selection associated with these amino acid substitution events need not be large. Estimates of the average coefficient of selection (s) based on the numbers of observed amino acid replacements support our assertion that natural selection has operated on Pgm, G6pd, Tps1, G6pase and Ald. The Bayesian method we employed to determine the strength of selection uses the joint nucleotide configurations of the M–K table to estimate the posterior probability distribution of the scaled selection coefficient, 2Nes, where N is the population size (Bustamante et al., 2002). Estimates of positive coefficients are highest for Tps1, G6pd, G6pase and Ald, and the 97.5% quantiles of the posterior probability distribution of 2Nes for these loci are greater than zero. This suggests that amino acid substitutions at these loci have been driven to fixation by positive selection, and, if we use the commonly assumed heuristic estimate of Ne=∼106 as the effective population size in D. melanogaster, we see that values of s as small as 10–5 are sufficient to be associated with the fixations observed here. With regard to this observation, Beisswanger et al. (Beisswanger et al., 2006) also estimated the selection coefficient associated with the fixation of the Pgd Fast allele in Europe to be s=10–3. Clearly, these fitness differences, although sufficient to drive significant evolutionary change, are well below a level that is experimentally detectable.
Curiously, at the opposite end of the scale, Pgm shows a highly significant negative value of 2Nes (Flowers et al., 2007), which reflects an excess of amino acid polymorphism in D. melanogaster (Verrelli and Eanes, 2000). The Poisson random field model used here to fit the data does not include the possibility of balancing selection favoring the retention of amino acid polymorphisms (Sawyer and Hartl, 1992). Therefore, the interpretation of a negative selection coefficient for Pgm is unclear. In D. melanogaster, there are 16 replacement polymorphisms in our sample and no amino acid fixations since the common ancestor with D. simulans. This excess of replacement polymorphism at Pgm is also consistent with a recent relaxation of functional constraints, the segregation of slightly deleterious mutations, or balancing selection. With respect to this, the clinal distribution of major amino acid Pgm haplotypes in the Eastern US (Verrelli and Eanes, 2001a) and the existence of many intermediate frequency amino acid polymorphisms provide evidence for the action of balancing selection. However, the level of silent polymorphism in Pgm is not high for D. melanogaster, nor is there any detectable flatness in the frequency spectrum of linked synonymous mutations (as might be expected for sites flanking an ancient balanced polymorphism). By contrast, in studies of long-term selection for metabolic response to aging, Pgm has surfaced as a gene of significance in Drosophila (Teotonio et al., 2009).
The combined evidence using both geographical variation and the M–K test shows that amino acid variation for enzymes at this intersection may be involved in metabolic adaptation in Drosophila (note, G6Pase, Hex-A and Ald were not included in the cline study). A possible explanation for adaptive selection on these branch point enzymes might be the localization of flux control at this junction. G6P is an allosteric effector of enzymes lying in branching pathways that lead to the storage, mobilization, transport, and breakdown of carbohydrate. In D. melanogaster, there is direct evidence that G6P branch point enzymes are capable of controlling flux allocation. Flux apportionment to the pentose shunt is extremely sensitive to variation at G6pd (Labate and Eanes, 1992) and Pgd (Cavener and Clegg, 1981), and glycogen storage levels have been reported to depend on Pgm genotype (Verrelli and Eanes, 2001b). In addition, overexpression of Tps1 in D. melanogaster increases trehalose content (Chen et al., 2002), suggesting that this enzyme may significantly control flux in the trehalose pathway branch.
Some closing thoughts
There are a number of questions that emerge from this study as it stands at this point. These questions involve: (1) the generality of our observation; (2) the complexities of assessing metabolic control in an experimental context in Drosophila; and (3) the identification of the agents of selection acting across the pathway and its branches.
Of course, it is important to establish the generality of our observation that there is the strongest evidence for adaptive selection at the intersection with G6P. We have taken the theoretical prediction that branch points possess novel control and posited that with increased control comes the opportunity for selection to act. We have not actually demonstrated greater control at these points or established causality. The question of the novelty of branch points per se can be retested in the larger metabolic network (see Greenberg et al., 2008), but for glycolysis there is a limited number of member enzymes and, thus, the generality cannot be further tested just by adding more genes. Further examination of this question will require the sequencing of more full genomes and collection of the necessary population level data in other species. Working with other Drosophila species is obvious, because we already have a number of full genomes sequenced (Drosophila 12 Genomes Consortium 2007), but it would be of interest to extend this broad pathway-based approach to a diversity of species with different life histories and life-styles, in particular, to microbes, marine species and plants. Another issue is that the glycolytic pathway differs from top to bottom with respect to other properties that should exert strong influences on the distribution of control and evolution of the pathway genes (Wright and Rausher, 2010). The ‘bottom’ enzymes have much higher expression levels (with the associated high codon bias), and as a group are considered to operate closer to equilibrium (Hochachka et al., 1998). They also must accommodate twice the flux rate because of the split of hexose 6-phosphate into triose phosphate. This has lead to the proposal that these downstream enzymes have evolved colocalization and possible channeling to facilitate flux under conditions of high metabolic demand (Suarez, 2003). The implications of these other properties to metabolic control and the evolution of the enzymes across the pathway are unclear.
The arguments I have presented here are simple, but the world is more complex. The basic metabolic control arguments may be more testable in microbes in which growth rate is closely connected to flux (Dykhuizen and Dean, 1990); the study of this problem in Drosophila (or for that matter in any other metazoan species) with its multiple tissues, each with differing physiological demands and required control, presented special concerns. The need for different controls in different tissues is reflected in the appearance of tissue-specific gene families. In our studies of D. melanogaster, the case of hexokinase is a point in hand; there are four genes with homology to hexokinases. These hexokinases have very different roles and patterns of evolution: the Hex-C gene is most active in the fat body (Moser et al., 1980), shows an amino acid polymorphism cline and is much less conserved across species than Hex-A, which is limited to nervous system and muscle (Duvernell and Eanes, 2000). This complexity can to some extent be addressed by experimentation. In D. melanogaster, the genetic tools are available to directly perturb enzyme dose and assess control over certain phenotypes. Our own projects have examined the impact of controlled modification of flight metabolism (Merritt et al., 2006). So far, we see that, with respect to the traditionally viewed regulatory step, HEX-A, there is a complete loss of flight ability for genotypes in the range of 27 to 50% of normal activity. By contrast, the classic equilibrium enzymes, phosphoglucose isomerse (PGI) and TPI, show no apparent reduction in flight performance at much lower levels (Eanes et al., 2006), even down to only 10% of normal levels. This type of direct experimentation has the promise of overcoming some of these complexities and marrying the power of genetic models to testing long-running hypotheses in metabolic physiology.
Lastly, one important goal is to understand the agents or the cause of selection that acts on points in the pathway. Are our observations really about controlling flux apportionment at the top of the pathway? In this regard, a complicating issue is that some enzymes and pathways may have tangential functions that are not coupled to the immediate role of simply turning over substrate. Many metabolic enzymes have been implicated in other capacities and the list is growing (Kim and Dang, 2005). In particular, they are involved in energy-sensing roles that direct downstream signaling (Wellen et al., 2009; Rathmell and Newgard, 2009; Zhao et al., 2009). Hexokinase is called a Jack-of-all-trades because of its role in energy-state sensing in both animals and plants (Moore et al., 2003). PGI has been shown to be a cytokine (Sun et al., 1999), and there are likely to be other as yet undiscovered roles for other enzymes in the energy-producing pathways. It may be that selection acts through these roles to generate the evolutionary patterns we observe.
Acknowledgements
Many individuals contributed to the work primarily reviewed in this paper. They especially include Brian Verrelli, Luciano Matzkin, Estaban Hasson, Paul Schmidt, Jon Flowers, Efe Sezgin, Chen-Tseh Zhu, Thomas Merritt, Dave Duvernell, Seiji Kumagai and Yihao Duan. I would like to thank Matthew Talbert and Dan Dykhuizen, as well as two anonymous reviewers, for commenting on and significantly improving earlier versions of the manuscript.
Footnotes
This study was supported by US Public Health Service Grant GM-45247 to W.F.E. and is contribution number 1197 from the Graduate Program in Ecology and Evolution, State University of New York, Stony Brook, New York. Deposited in PMC for release after 12 months.
References
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., Scherer S. E., Li P. W., Hoskins R. A., Galle R. F., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185-2195 [DOI] [PubMed] [Google Scholar]
- Bagheri H. C., Wagner G. P. (2004). Evolution of dominance in metabolic pathways. Genetics 168, 1713-1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri H. C., Hermisson J., Vaisnys J. R., Wagner G. P. (2003). Effects of epistasis on phenotypic robustness in metabolic pathways. Math. Biosci. 184, 27-51 [DOI] [PubMed] [Google Scholar]
- Baker K. D., Thummel C. S. (2007). Diabetic larvae and obese flies – emerging studies of metabolism in Drosophila. Cell Metab. 6, 257-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswanger S., Stephan W., De Lorenzo D. (2006). Evidence for a selective sweep in the wapl region of Drosophila melanogaster. Genetics 172, 265-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., Tsang G., Evans-Holm M., Hiesinger P. R., Schulze K. L., Rubin G. M., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A., Kreitman M. (1993). Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134, 869-893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Sawyer S. A., Olsen K. M., Purugganan M. D., Hartl D. L. (2002). The cost of inbreeding in Arabidopsis. Nature 416, 531-534 [DOI] [PubMed] [Google Scholar]
- Bustamante C. D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M. T., Glanowski S., Tanenbaum D. M., White T. J., Sninsky J. J., Hernandez R. D., et al. (2005). Natural selection on protein-coding genes in the human genome. Nature 437, 1153-1157 [DOI] [PubMed] [Google Scholar]
- Cavener D. R., Clegg M. T. (1981). Evidence for biochemical and physiological differences between genotypes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 78, 4444-4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ma E., Behar K. L., Xu T., Haddad G. G. (2002). Role of trehalose phosphate synthase in anoxia tolerance and development in Drosophila melanogaster. J. Biol. Chem. 277, 3274-3279 [DOI] [PubMed] [Google Scholar]
- Chippindale A. K., Chu T. J. F., Rose M. R. (1996). Complex trade-offs and the evolution of starvation resistance in Drosophila melanogaster. Evolution 50, 753-766 [DOI] [PubMed] [Google Scholar]
- David J. R., Capy P. (1988). Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4, 106-111 [DOI] [PubMed] [Google Scholar]
- Duvernell D. D., Eanes W. F. (2000). Contrasting molecular population genetics of four hexokinases in Drosophila melanogaster, D. simulans and D. yakuba. Genetics 156, 1191-1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D. E., Dean A. M. (1990). Enzyme activity and fitness: evolution in solution. Trends Ecol. Evol. 5, 257-262 [DOI] [PubMed] [Google Scholar]
- Eanes W. F. (1999). Analysis of selection on enzyme polymorphisms. Annu. Rev. Ecol. Syst. 30, 301-326 [Google Scholar]
- Eanes W. F., Kirchner M., Yoon J. (1993). Evidence for adaptive evolution of the G6PD gene in Drosophila melanogaster and D. simulans lineages. Proc. Natl. Acad. Sci. USA 90, 7475-7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes W. F., Kirchner M., Yoon J., Biermann C. H., Wang I. N., McCartney M. A., Verrelli B. C. (1996). Historical selection, amino acid polymorphism and lineage-specific divergence at the G6pd locus in Drosophila melanogaster and D. simulans. Genetics 144, 1027-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes W. F., Merritt T. J., Flowers J. M., Kumagai S., Sezgin E., Zhu C. T. (2006). Flux control and excess capacity in the enzymes of glycolysis and their relationship to flight metabolism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103, 19413-19418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes W. F., Merritt T. J., Flowers J. M., Kumagai S., Zhu C. T. (2008). Direct evidence that genetic variation in glycerol-3-phosphate and malate dehydrogenase genes (Gpdh and Mdh1) impacts adult ethanol tolerance in Drosophila melanogaster. Genetics 181, 607-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell D. A. (1997). Understanding the Control of Metabolism. London: Portland Press; [Google Scholar]
- Flowers J., Sezgin E., Kumagai S., Duvernell D., Matzkin L., Schmidt P., Eanes W. (2007). Adaptive evolution of metabolic pathways in Drosophila. Mol. Biol. Evol. 24, 1347-1354 [DOI] [PubMed] [Google Scholar]
- Fry J. D., Donlon K., Saweikis M. (2008). A worldwide polymorphism in aldehyde dehydrogenase in Drosophila melanogaster: evidence for selection mediated by dietary ethanol. Evolution 62, 66-75 [DOI] [PubMed] [Google Scholar]
- Gershman B., Puig O., Hang L., Peitzsch R. M., Tatar M., Garofalo R. S. (2007). High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol. Genomics 29, 24-34 [DOI] [PubMed] [Google Scholar]
- Glinka S., Ometto L., Mousset S., Stephan W., De Lorenzo D. (2003). Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165, 1269-1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockel J., Kennington W. J., Hoffmann A., Goldstein D. B., Partridge L. (2001). Nonclinality of molecular variation implicates selection in maintaining a morphological cline of Drosophila melanogaster. Genetics 158, 319-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A. J., Stockwell S. R., Clark A. G. (2008). Evolutionary constraint and adaptation in the metabolic network of Drosophila. Mol. Biol. Evol. 25, 2537-2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S. (1939). The theory of the evolution of dominance. J. Genet. 37, 365-374 [Google Scholar]
- Hartl D. L., Taubes C. H. (1996). Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182, 303-309 [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Dykhuizen D. E., Dean A. M. (1985). Limits of adaptation: the evolution of selective neutrality. Genetics 111, 655-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson E., Wang I. N., Zeng L. W., Kreitman M., Eanes W. F. (1998). Nucleotide variation in the triosephosphate isomerase (Tpi) locus of Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 15, 756-769 [DOI] [PubMed] [Google Scholar]
- Hochachka P. W., Somero G. N. (2002). Biochemical Adaptation. New York: Oxford University Press; [Google Scholar]
- Hochachka P. W., McClelland G. B., Burness G. P., Staples J. F., Suarez R. K. (1998). Integating metabolic pathway fluxes with gene-to-enzyme expression rates. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 120, 17-26 [Google Scholar]
- Hoffmann A. A., Hallas R., Sinclair C., Mitrovski P. (2001). Levels of variation in stress resistance in Drosophila among strains, local populations, and geographic regions: patterns for desiccation, starvation, cold resistance, and associated traits. Evolution 55, 1621-1630 [DOI] [PubMed] [Google Scholar]
- James A. C., Azevedo R. B. R., Partridge L. (1997). Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics 146, 881-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R., Phillips P. C. (2009). Evolutionary rates and centrality in the yeast gene regulatory network. Genome Biol. 10, R35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H., Burns J. A. (1981). The molecular basis of dominance. Genetics 97, 639-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. W., Dang C. V. (2005). Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 30, 142-150 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1983). The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; [Google Scholar]
- Kreitman M. (1996). The neutral theory is dead. Long live the neutral theory. BioEssays 18, 678-683 [DOI] [PubMed] [Google Scholar]
- Kulathinal R. J., Bettencourt B. R., Hartl D. L. (2004). Compensated deleterious mutations in insect genomes. Science 306, 1553-1554 [DOI] [PubMed] [Google Scholar]
- Labate J., Eanes W. F. (1992). Direct measurement of in vivo flux differences between electrophoretic variants of G6PD in Drosophila melanogaster. Genetics 132, 783-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte D. C., Walsh K., Koshland D. E. (1984). The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J. Biol. Chem. 259, 14068-14075 [PubMed] [Google Scholar]
- Larracuente A. M., Sackton T. B., Greenberg A. J., Wong A., Singh N. D., Sturgill D., Zhang Y., Oliver B., Clark A. G. (2008). Evolution of protein-coding genes in Drosophila. Trends Genet. 24, 114-123 [DOI] [PubMed] [Google Scholar]
- Li Y. F., Costello J. C., Holloway A. K., Hahn M. W. (2008). “Reverse ecology” and the power of population genomics. Evolution 62, 2984-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. (2007). The Origins of Genome Architecture. Sunderland, MA: Sinauer Associates; [Google Scholar]
- McDonald J. H., Kreitman M. K. (1991). Adaptive evolution at the Adh locus in Drosophila. Nature 351, 652-654 [DOI] [PubMed] [Google Scholar]
- Merritt T. J., Duvernell D., Eanes W. F. (2005). Natural and synthetic alleles provide complementary insights into the nature of selection acting on the Men polymorphism of Drosophila melanogaster. Genetics 171, 1707-1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt T. J., Sezgin E., Zhu C. T., Eanes W. F. (2006). Triglyceride pools, flight and activity variation at the Gpdh locus in Drosophila melanogaster. Genetics 172, 293-304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt T. J., Kuczynski C., Sezgin E., Zhu C. T., Kumagai S., Eanes W. F. (2009). Quantifying interactions within the NADP(H) enzyme network in Drosophila melanogaster. Genetics 182, 565-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovski P., Hoffmann A. A. (2001). Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc. R. Soc. Lond. B. Biol. Sci. 268, 2163-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton J. B. (1997). Selection in Natural Populations. New York Oxford: Oxford University Press; [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W. H., Liu Y. X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332-336 [DOI] [PubMed] [Google Scholar]
- Moser D., Johnson L., Lee C.-Y. (1980). Multiple forms of Drosophila hexokinases. Purification, biochemical and immunological characterization. J. Biol. Chem. 255, 4673-4679 [PubMed] [Google Scholar]
- Nielsen R. (2005). Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197-218 [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Chambers G. K., Gibson J. B., Willcocks D. A. (1981). Latitudinal relationships of esterase-6 and phosphoglucomutase gene frequencies in Drosophila melanogaster. Heredity 47, 385-396 [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Gibson J. B., Anderson P. R., Knibb W. R., Chambers G. K. (1982). Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution 36, 86-96 [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Chambers G. K., Gibson J. B., Eanes W. F., Willcocks D. A. (1983). Geographic variation in G6pd and Pgd allele frequencies in Drosophila melanogaster. Heredity 50, 67-72 [DOI] [PubMed] [Google Scholar]
- Ohta T. (1992). The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23, 263-286 [Google Scholar]
- Powell J. R. (1997). Progress and Prospects in Evolutionary Biology: The Drosophila Model. New York: Oxford University Press; [Google Scholar]
- Rathmell J. C., Newgard C. B. (2009). A glucose-to-gene link. Science 324, 1021-1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume C. J., Sokolowski M. B. (2006). The nature of Drosophila melanogaster. Curr. Biol. 16, R623-R628 [DOI] [PubMed] [Google Scholar]
- Sawyer S. A., Hartl D. L. (1992). Population genetics of polymorphism and divergence. Genetics 132, 1161-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Paaby A. B. (2008). Reproductive diapause and life-history clines in north american populations of Drosophila melanogaster. Evolution 62, 1204-1215 [DOI] [PubMed] [Google Scholar]
- Schmidt P. S., Duvernell D. D., Eanes W. F. (2000). Adaptive evolution of a candidate gene for aging in Drosophila. Proc. Natl. Acad. Sci. USA 97, 10861-10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. S., Matzkin L., Ippolito M., Eanes W. F. (2005). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59, 1721-1732 [PubMed] [Google Scholar]
- Schmidt P. S., Zhu C. T., Das J., Batavia M., Yang L., Eanes W. F. (2008). An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105, 16207-16211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E., Duvernell D. D., Matzkin L. M., Duan Y., Zhu C. T., Verrelli B. C., Eanes W. F. (2004). Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168, 923-931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., Turissini D. A., Fang S., Wang H. Y., Hudson R. R., Nielsen R., et al. (2007). Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104, 2271-2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. G., Eyre-Walker A. (2002). Adaptive protein evolution in Drosophila. Nature 415, 1022-1024 [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G., Vallino J. J. (1991). Network rigidity and metabolic engineering in metabolite overproduction. Science 252, 1675-1681 [DOI] [PubMed] [Google Scholar]
- Suarez R. K. (2003). Shaken and stirred: muscle structure and metabolism. J. Exp. Biol. 206, 2021-2029 [DOI] [PubMed] [Google Scholar]
- Sun Y. J., Chou C. C., Chen W. S., Wu R. T., Meng M., Hsiao C. D. (1999). The crystal structure of a multifunctional protein: Phosphoglucose isomerase/autocrine motility factor/neuroleukin. Proc. Natl. Acad. Sci. USA 96, 5412-5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotonio H., Chelo I. M., Bradic M., Rose M. R., Long A. D. (2009). Experimental evolution reveals natural selection on standing genetic variation. Nat. Genet. 41, 251-257 [DOI] [PubMed] [Google Scholar]
- Umina P. A., Weeks A. R., Kearney M. R., Mckechnie S. W., Hoffmann A. A. (2005). A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691-693 [DOI] [PubMed] [Google Scholar]
- Vasemagi A., Primmer C. R. (2005). Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol. Ecol. 14, 3623-3642 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J. (2005). Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6, 167-178 [DOI] [PubMed] [Google Scholar]
- Verrelli B. C., Eanes W. F. (2000). Extensive amino acid polymorphism at the Pgm locus is consistent with adaptive protein evolution in Drosophila melanogaster. Genetics 156, 1737-1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli B. C., Eanes W. F. (2001a). Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster. Genetics 157, 1649-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli B. C., Eanes W. F. (2001b). The functional impact of Pgm amino acid polymorphism on glycogen content in Drosophila melanogaster. Genetics 159, 201-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. (2005). Robustness and Evolvability in Living Systems. Princeton and Oxford: Princeton University Press; [Google Scholar]
- Watt W. B., Dean A. M. (2000). Molecular-functional studies of adaptive genetic variation in prokaryotes and eukaryotes. Annu. Rev. Genet. 34, 593-622 [DOI] [PubMed] [Google Scholar]
- Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076-1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. M., Rausher M. D. (2010). The evolution of control and distribution of adaptive mutations in a metabolic pathway. Genetics 184, 483-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. (1929). Fisher’s theory of dominance. Am. Nat. 63, 274-279 [Google Scholar]
- Wright S. (1934). Physiological and evolutionary theories of dominance. Am. Nat. 68, 24-53 [Google Scholar]
- Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., et al. (2009). Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324, 261-265 [DOI] [PMC free article] [PubMed] [Google Scholar]