Abstract
The epidermal barrier varies over the body surface to accommodate regional environmental stresses. Regional skin barrier variation is produced by site-dependent epidermal differentiation from common keratinocyte precursors and often manifests as site-specific skin disease or irritation. There is strong evidence for body-site-dependent dermal programming of epidermal differentiation in which the epidermis responds by altering expression of key barrier proteins, but the underlying mechanisms have not been defined. The LCE multigene cluster encodes barrier proteins that are differentially expressed over the body surface, and perturbation of LCE cluster expression is linked to the common regional skin disease psoriasis. LCE subclusters comprise genes expressed variably in either external barrier-forming epithelia (e.g. skin) or in internal epithelia with less stringent barriers (e.g. tongue). We demonstrate here that a complex of TALE homeobox transcription factors PBX1, PBX2 and Pknox (homologues of Drosophila Extradenticle and Homothorax) preferentially regulate external rather than internal LCE gene expression, competitively binding with SP1 and SP3. Perturbation of TALE protein expression in stratified squamous epithelia in mice produces external but not internal barrier abnormalities. We conclude that epidermal barrier genes, such as the LCE multigene cluster, are regulated by TALE homeodomain transcription factors to produce regional epidermal barriers.
Key words: Homeodomain, Transcriptional regulation, Epidermal barrier, LCE genes
Introduction
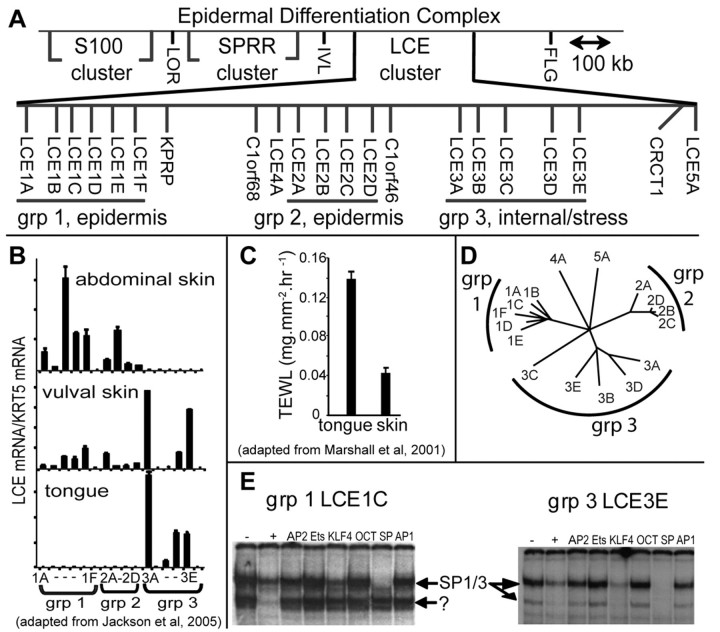
Late cornified envelope (LCE) proteins are small proline-rich proteins that form part of skin stratum corneum (Brown et al., 2007; Jackson et al., 2005; Marshall et al., 2001; Zhao and Elder, 1997). Variants within the human LCE locus at 1q21 confer susceptibility to the common skin disorder psoriasis, an inflammatory skin disease with barrier aetiology, reflecting the importance of LCE proteins for skin barrier function (de Cid et al., 2009; Liu et al., 2008; Zhang et al., 2009). It is not known why vertebrates maintain such large LCE multigene families (17 genes in human, 23 in mouse, Fig. 1A) because they encode very similar proteins (Brown et al., 2007; Jackson et al., 2005; Marshall et al., 2001; Zhao and Elder, 1997). One view is that many proteins are required to provide body-site-dependent variations in the quality of the stratum corneum or skin barrier in response to different environmental conditions (e.g. aridity, microbial, toxin and radiation exposure) at different body sites (Cabral et al., 2001; Jackson et al., 2005; Marshall et al., 2001) (Fig. 1B,C). This proposal is supported by expression data. The LCE genes form chromosomal subclusters that reflect location-dependent expression, e.g. genomic subclusters comprise genes expressed preferentially in either external barrier-forming epithelia (skin) or in internal epithelia with quite different barrier characteristics (e.g. tongue; Fig. 1B,C) (Marshall et al., 2000). Within subclusters, LCE genes are also variably expressed (Fig. 1B) and differentially responsive to barrier challenge (Jackson et al., 2005; de Cid et al., 2009).
Fig. 1.
Common and group-specific protein–DNA interactions on LCE group 1 and group 3 promoters. (A) Chromosomal arrangement of LCE genes at the epidermal differentiation complex (adapted from National Centre for Biotechnology Information, Map Viewer Build 37.1) showing clustering within groups that reflects gene similarity and expression. (B) In unstressed conditions, LCE genes are expressed in a group-specific manner. Group 1 genes are expressed most prominently in skin, whereas group 3 genes are expressed most prominently in internal stratified squamous epithelia such as the tongue. Vulval skin is intermediate, with both group 1 and 3 LCE expression (modified from Jackson et al., 2005). Expression is normalised to expression of basal cell keratin-5 (Krt5). (C) External (skin) and internal stratified squamous epithelia (tongue) have different barrier activities, measured by gravimetric transepidermal water loss (TEWL) assay (modified from Marshall et al., 2000). (D) Human LCE promoters 300 bp upstream from the transcription start site maintain group-specific similarity shown by CLUSTALW analysis and presented as a PHYLIP unrooted phylogenetic tree (The Biology Workbench) (Subramaniam, 1998). (E) EMSA competition analysis shows that SP-like factors bind the proximal region of both a group 1 and group 3 promoter, whereas unknown factors bind close by only on the group 1 promoter. −, no competition, +, self competition. CRCT, previously NICE1, newly identified cornified envelope; FLG, filaggrin; IVL, involucrin; KPRP, keratinocytes proline-rich protein; KRT5, keratin-5; LCE, late cornified envelope; LOR, loricrin; SPRR, small proline-rich, TEWL, transepidermal water loss.
The presence of so many LCE genes that are co-expressed in either external or internal stratum corneum-forming epithelia provided an opportunity to identify transcription factors binding to LCE promoters that either (1) conferred expression in all keratinocytes at similar stages of differentiation, i.e. granular layer keratinocytes; such factors should be common to all LCE genes; or (2) conferred tissue-specific expression; these factors should regulate transcription only in external or internally expressed genes.
A screen of human LCE promoters based on this rationale led to the discovery that the TALE (three amino acid loop extension) family of homeodomain transcription factors differentially regulate external and internal LCE genes. We also find that previously identified stratum corneum gene transcriptional regulators are probably TALE proteins.
The TALE transcription factor family comprises important developmental regulators that include the PBC (Drosophila Extradentical and mammalian Pbx proteins) and MEIS (Drosophila HOMOTHORAX and mammalian MEIS and PKNOX) classes of homeodomain proteins (reviewed by Laurent et al., 2008; Moens and Selleri, 2006). TALE factors co-bind promoters with HOX proteins, conferring specificity, and ablation of TALE proteins can mimic HOX defects (Manley et al., 2004; Selleri et al., 2004). In addition, TALE proteins co-bind with additional transcription factors and might have cellular roles beyond transcriptional regulation (Laurent et al., 2008).
Here we show that TALE proteins are abundant in the differentiating epidermis. They bind LCE promoters and overlap SP1 and SP3 sites, and there is a reciprocal relationship between TALE and SP binding on external (e.g. skin) and internal (e.g. tongue) LCE promoters. Perturbation of TALE proteins in transgenic mouse epidermis substantially alters external but not internal stratum corneum and produces external skin barrier defects. We propose that TALE proteins are a family of epidermal transcriptional regulators that can modulate body-site-specific epidermal barrier through regulation of LCE barrier genes and additional regionally expressed barrier genes.
Results
Common and group-specific protein binding to LCE gene promoters
Human LCE genes are co-expressed in granular layer keratinocytes, but expression levels are body-site dependent (Fig. 1B) (Jackson et al., 2005). DNA 300 bp upstream of LCE genes is conserved in a group-dependent manner, reflecting chromosomal subclustering and gene expression (Fig. 1D). We queried these regions using electrophoretic mobility shift assay (EMSA) against differentiated keratinocyte nuclear extract, searching for DNA–protein interactions that could mediate these expression patterns, i.e. common binding to all LCE promoters or group-specific binding. We used promoter proximal sequences from robustly expressed genes from skin-expressing group 1 genes (LCE1C) and internally expressing group 3 genes (LCE3E; Fig. 1B,D) (Jackson et al., 2005).
DNA between −106 and −136 from the group 1 transcription start site, and between −100 and −70 from the group 3 transcription start site prominently bound several proteins (Fig. 1E). Competitive EMSA eliminated many of the common transcription factors, but suggested that SP-like factors bound to both promoters and unknown factors bound adjacently in the group 1 promoter (Fig. 1E; note that KLF4 binds weakly to SP consensus sites explaining the weak competition). This pattern of binding fitted the criteria for both common and group-specific protein-DNA interaction, so we investigated further.
SP1 and SP3 bind external and internal promoters and overlap the binding site for an unknown factor(s) on the external promoter
We used competitive EMSA with mutated oligonucleotides spanning the protein-binding regions (supplementary material Fig. S1A for the external group 1 promoter, supplementary material Fig. S2A for the internal group 3 promoter) to map the SP and unknown factor binding sites. Supershift with SP1 and SP3 antibodies confirmed that SP1, SP3 and a truncated form of SP3 bound both promoters (supplementary material Figs S1 and S2) (Kennett et al., 1997). The unknown factors bind adjacently to SP1 and SP3, with overlap on the group 1 LCE1C promoter (Fig. 2A), suggesting competitive binding.
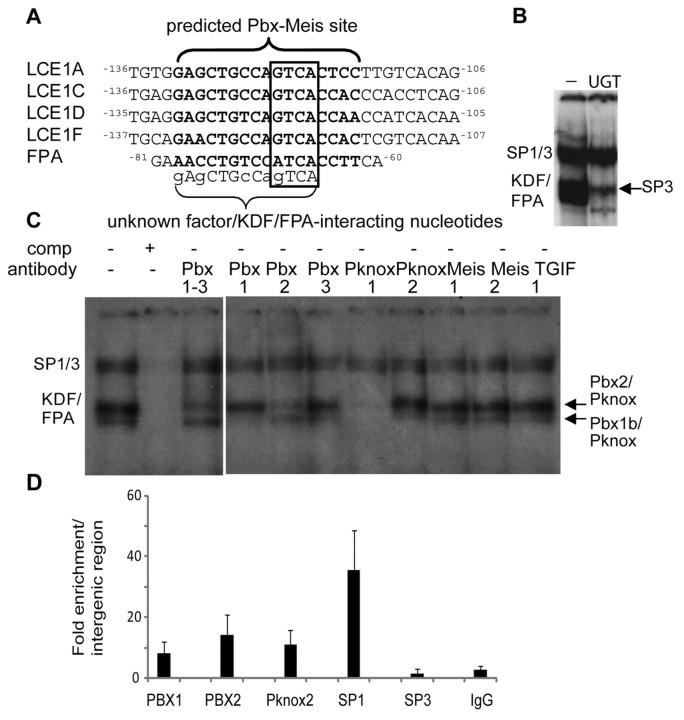
Fig. 2.
The unknown complex that binds the group 1 LCE gene overlaps an SP1–SP3 site, is similar to the involucrin-binding KDF and FPA binding factors, and binds differentially to group 1 and 3 promoters. (A) Nucleotides interacting with SP1 and SP3 in the group 1 LCE1C and group 3 LCE3E promoters were deduced as described in supplementary material Figs S1 and S2 and are shaded. Two predicted SP core binding sites are underlined. Note that the unknown factor binds the same nucleotides as SP1 and SP3. (B) Competitive EMSA of the LCE1C −136 to −106 promoter fragment with oligonucleotides from the involucrin promoter: KDF binding site (LaPres and Hudson, 1996), 1711GAAATGTGCCTGTCAGGAA1693; Footprint A or FPA (Phillips et al., 2000), 90TAGAGTGGTGAAACCTGTCCATCACCTTCA60; and H3 binding site (Lopez-Bayghen et al., 1996), 316GGAAAAATTGAGATTCAGAGCAGA292. Superscript numbers show distance from the transcription start site. Both KDF and the FPA binding protein(s) effectively and specifically compete with the lower complexes and are probably the same or a similar factors. Neither KDF nor FPA binding protein compete with SP1 or SP3 binding. Although self-fragment effectively eliminates all binding (+), neither the Oct consensus sequence nor the H3 site compete SP1 and SP3 or the lower KDF complex. (C) Competitive EMSA using the LCE1C probe shows that excess (300-fold) LCE3E −100 to −70 oligonucleotide competes both KDF/FPA and SP1 (lane 3E), despite failing to bind KDF/FPA detectably when used as an EMSA probe (e.g. Fig. 1 and supplementary material Fig. S2). The KDF/FPA binding site on the group 3 promoter was mapped using competing mutated oligonucleotides M1–M5 (see supplementary material Fig. S2) and was shown to be adjacent to and overlapping the SP1 and SP3 site, as in the group 1 promoter. (D) Sequence comparison of group 1 promoter sequences at the KDF/FPA and SP binding region shows strong sequence similarity at the KDF/FPA sites but weaker similarity at the SP region. By contrast, comparison of group 3 promoters shows conserved SP sequence and diversity at the KDF/FPA site (darker shading shows identical bases, lighter shading similar bases). (E) Competitive EMSA with LCE1C −136 to −106 probe (top panel) shows that group 1 LCE genes are more successful than group 3 LCE genes at competing with KDF/FPA (i.e. bind KDF/FPA more strongly). By contrast, group 3 LCE genes are more successful than group 1 genes at competing with the SP factors (or bind these factors more strongly). This latter result is confirmed in the bottom panel, which shows competitive EMSA with the LCE3E −100 to −70 probe. With the exception of LCE1C, group 1 promoters are less successful than group 3 promoters at competing with SP1 and SP3.
The unknown factor has previously been detected binding another stratum corneum (involucrin) promoter
A search of the literature for protein–DNA interactions associated with EDC promoters showed that the unknown complex binding site in the LCE1C promoter was similar to regulatory protein binding sites in the involucrin promoter (Fig. 2B, supplementary material Table S1). Binding sites for keratinocyte differentiation factor (KDF) (LaPres and Hudson, 1996), and additional sites identified by footprinting – FPA site (Phillips et al., 2000) and H3 site (Lopez-Bayghen et al., 1996), were tested for the ability to competitively remove the unknown factor on LCE1C. Both the KDF and the FPA sequences specifically competed with the unknown factor, but not SP1, whereas the H3 site could not compete with either factor (Fig. 2B). Therefore, it was tentatively concluded that the unknown factors were similar to KDF and FPA binding factors, and they were called KDF/FPA.
KDF/FPA factor binds strongly to LCE external promoters and weakly to internal promoters, while SP factors do the opposite
Although KDF/FPA could not be detected directly binding the group 3 promoter, competition experiments with excess group 3 probe showed that KDF/FPA bound weakly, again adjacent to and overlapping an SP site (Fig. 2C shows mapping experiment and summary findings, with mutant oligonucleotides as in supplementary material Fig. S1). To investigate whether strong and weak KDF/FPA binding were group-specific characteristics, analogous regions from additional group 1 and 3 promoters were compared (Fig. 2D).
Group 1 promoters show closest similarity over the KDF/FPA binding region, whereas group 3 promoters are quite diverse over the KDF/FPA site (Fig. 2D). By contrast, group 3 promoters show strongest similarity over the SP binding region. Group 1 promoters were clearly more efficient at competing with KDF/FPA than group 3 promoters (Fig. 2E), indicating that KDF/FPA binds most strongly to these promoters. By contrast, group 3 promoters were very efficient at competing with SP1 and SP3 binding (Fig. 2E), indicating that SP factors bind most strongly to these promoters. Group 1 promoters showed variable ability to compete with SP factors: three promoters (LCE1A, LCE1D and LCE1F) were very inefficient. These data indicate that KDF/FPA and SP factors bind LCE genes in a group-specific manner. The identification of further binding sites in the LCE and involucrin promoters provided a tool for identifying KDF/FPA binding factors.
The unknown factor (KDF/FPA) is a complex of TALE homeobox transcription factors PBX1b, PBX2 and Pknox
Because KDF/FPA bound most strongly to group 1 LCE promoters expressed in the epidermis, we asked whether KDF/FPA was enriched in epidermal keratinocytes. Promoter binding was compared in keratinocyte nuclear extracts from epidermal or oral origin (Dickson et al., 2000), HeLa epithelial cells and fibroblasts. Binding was found to be equivalent in all cell types (data not shown), indicating that KDF/FPA is not an epidermal keratinocyte-specific factor. To determine whether KDF/FPA comprised known transcription factors, the binding sites were compared with a database of known transcription factor sites (MatInspector) (Cartharius et al., 2005). Five of the eight sites identified in this work matched with a high degree of confidence a consensus matrix for the homeodomain PBX–Meis1 complex specifically at the nucleotides implicated in KDF/FPA binding (Fig. 3A).
Fig. 3.
TALE proteins PBX1b, PBX2 and Pknox bind the LCE KDF/FPA site. (A) Alignment of the KDF/FPA binding sequences from LCE genes and the involucrin promoter with a PBX–Meis1 experimentally derived matrix consensus binding sequence shown in bold (from MatInspector). PBX–Meis1 core sequences are boxed. Unknown factor(s) or KDF sites from LCE1B, LCE1E and involucrin KDF itself did not meet the stringency criterion for core (LCE1B) or matrix (LCE1E, KDF) similarity. (B) EMSA with the LCE1C promoter fragment and a well characterised PBX site from the UDP glucuronosyltransferase (UGT2B17) gene (Gregory and Mackenzie, 2002) (denoted ‘UGT’; −, no competitor) very effectively competes with KDF/FPA. Competition reveals binding by the truncated SP3 transcription factor. (C) EMSA with TALE-family-specific antibody supershift confirms that TALE family members bind the KDF/FPA site. The two KDF/FPA binding complexes are identified as upper PBX2–Pknox factor and the lower as PBX1b–Pknox factor. (D) ChIP shows that PBX1, PBX2, PKNOX2 and SP1 bind to the LCE1C site in vivo. SP3 binding was not detected. Data is presented as fold enrichment of LCE1C binding (as % of input genomic DNA) compared with enrichment of binding to an intergenic region.
The TALE (three amino acid loop extension) group of homeodomain transcription factors, including PBX, Meis, and Pknox groups, are widely expressed but have not been previously implicated in the regulation of EDC genes. To find out whether members of this family bound the KDF/FPA site, EMSA was carried out with a characterised PBX site from the UDP glucuronosyltransferase (UGT2B17) gene (Gregory and Mackenzie, 2002) (Fig. 3B) and with TALE family antibodies (Fig. 3C). The UGT2B17 Pbx oligonucleotide (UGT, Fig. 3B) very effectively outcompeted KDF/FPA from the LCE1C promoter (Fig. 3B), suggesting that the KDF site is a functional PBX site. Competitive removal of TALE factors revealed truncated SP3 binding, as expected.
TALE proteins bind DNA as heterodimers, and the consensus sequences overlap, with Meis, TGIF and Pknox member binding sites incorporating the same central TGACAG motif (Berger et al., 2008). EMSA in the presence of antibodies against TALE proteins identified complexes that bound KDF/FPA as PBX1b–Pknox and PBX2–Pknox, with Meis factors failing to bind in vitro, despite the initial bioinformatics identification of the site as a PBX–Meis site (Fig. 3C).
The KDF/FPA upper complex was supershifted with antibody crossreacting with PBX isoforms 1–3 (called PBX1–3), PBX2, Pknox1 and Pknox2 antibodies, but not PBX1, PBX3, Meis antibodies and antibodies against TGIF, which is a further TALE protein (Fig. 3C). These supershifts identified PBX2 and a Pknox protein in the upper complex. The Pknox1 polyclonal antibody used in the supershift assay can also recognise Pknox2 – its antigen is residues 26–436, which contains Pknox2 motifs and this antibody weakly detects a band co-migrating with Pknox2 on western blots (data not shown). The monoclonal Pknox2 antibody is however specific, recognising a short epitope with only moderate homology to Pknox1. Others have noted that Pknox1 antisera can compete a PBX1A–Pknox2 complex (Fognani et al., 2002). Therefore the upper complex is probably composed of PBX2–Pknox2, supported by the supershift in the presence of Pknox2-specific antibody and binding of Pknox2 in vivo (see below). However, we cannot rule out a mixture of PBX2–Pknox1 and PBX2–Pknox2.
The KDF/FPA lower complex was supershifted by PBX1, Pknox1 and Pknox2 antibodies. The PBX partner is PBX1, but the inability of PBX1–3 antibody to supershift the complex showed that it is the PBX1B isoform. PBX1–3 antibody recognises a conserved epitope near the C-terminus of PBX, which is spliced out of the B isoform.
We have shown that TALE factors PBX1b, PBX2, Pknox and SP1/3 factors can bind an overlapping region of the LCE promoter in vitro. In vivo, we confirmed that PBX1, PBX2, Pknox2 and SP1 bind the LCE1C promoter in differentiated primary human keratinocytes (Fig. 3D). SP3 binding in vivo was undetected. Weak binding or failure to detect binding might be a property of the antibodies used for ChIP. Alternatively, failure to detect SP3 binding in vivo might reflect the association of SP3 with proliferative conditions in vivo (see the Discussion).
TALE proteins are prominently expressed in epidermis and associate strongly with terminally differentiating keratinocytes
TALE family member PBX2 has been reported in the interfollicular epidermis (Selleri et al., 2004). PBX1, PBX2 and PBX3 are also found in the cytoplasm of human epidermal cells (Komuves et al., 2000) and other TALE classes have been studied in the hair follicle (Jave-Suarez and Schweizer, 2006).
We show here that TALE proteins are expressed prominently in the epidermis (Fig. 4 shows human facial skin epidermis and antibodies directed against the C-termini of TALE proteins) and can associate strongly with terminally differentiating keratinocytes (brackets and arrows, Fig. 4), which is consistent with the proposed role in regulating LCE and other EDC genes. However, irregular association of some TALE proteins with basal, proliferating keratinocytes suggests additional roles in the epidermis that are beyond the scope of this study.
Fig. 4.
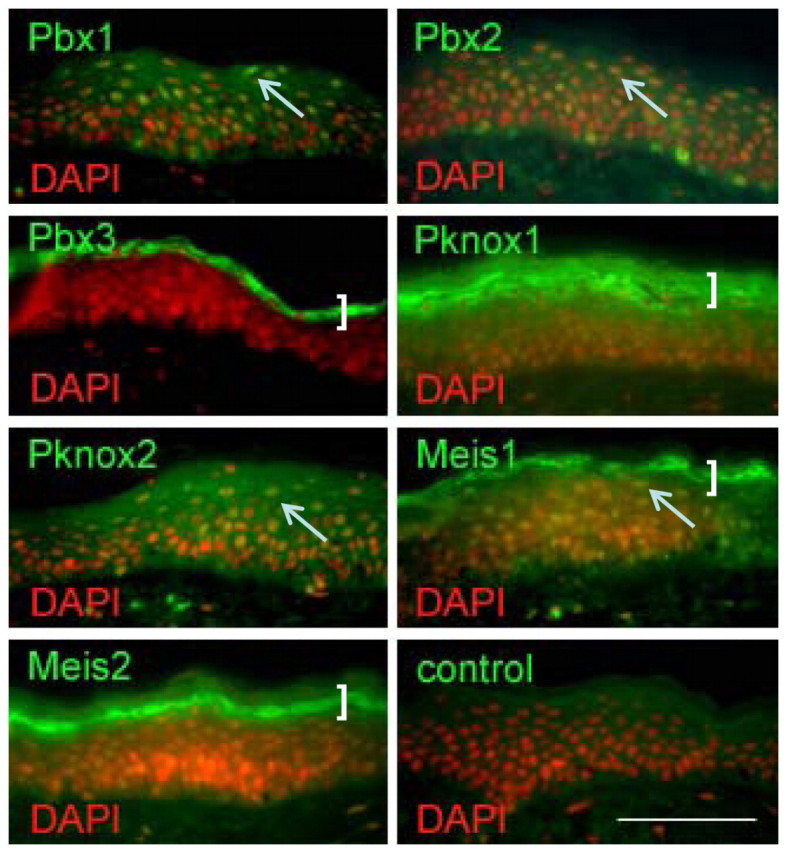
TALE factor expression in epidermis. TALE transcription factors (green) in human facial epidermis, nuclei (DAPI) are red and nuclear TALE expression yellow/orange. Arrows indicate nuclear TALE expression as the keratinocytes terminally differentiate, whereas brackets show strong granular layer expression. Control shows equivalent exposure without primary antibody. Scale bar: 50 μm.
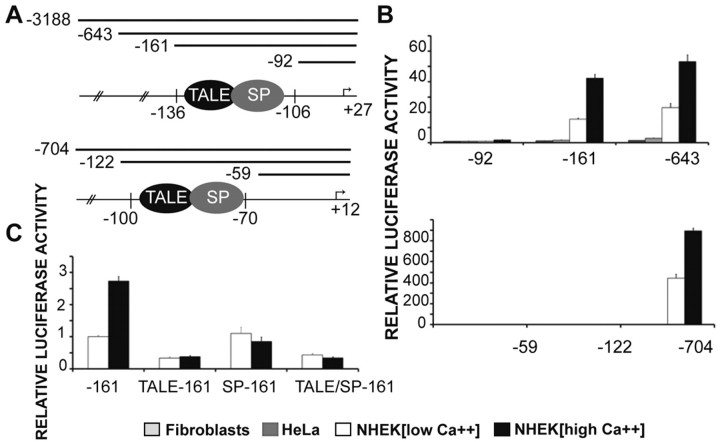
TALE factor binding is necessary for activity of the external promoter in keratinocytes
We find that the strong TALE-binding site is necessary for externally expressed LCE1C expression in Ca2+-induced (differentiated) keratinocytes using a promoter-reporter assay. A highly truncated form (−161/+27) of the externally expressed LCE1C promoter luciferase construct containing both the strong TALE-binding site and SP site (Fig. 5A) mediated strong and specific expression in primary human keratinocytes, with increased expression upon Ca2+-mediated keratinocyte differentiation. An increase in the promoter size did not increase expression further (Fig. 5A,B). Expression was specific to primary human epidermal keratinocytes and was minimal in HeLa cells and fibroblasts (Fig. 5B). Specific mutation of the TALE-binding site in the truncated LCE1C promoter construct, so that complexes can no longer bind (supplementary material Fig. S1A), caused near-complete loss of expression (Fig. 5C), demonstrating the central importance of the LCE1C TALE site. Mutation of the flanking SP site caused loss of inducibility upon Ca2+-mediated keratinocyte differentiation. Double mutation of the TALE-SP site was uninformative because of the severity of the loss of expression.
Fig. 5.
TALE factor binding is necessary for LCE1C promoter activity in vitro and differentially affects LCE1C and LCE3E promoter activity. (A) Promoter–luciferase constructs. (B) The LCE1C −161 to +27 promoter–luciferase construct containing both the strong TALE-binding site and SP site mediates Ca2+-inducible expression in normal human epidermal keratinocytes (NHEKs) without much further increase in expression with increasing promoter size. However, the LCE3E −122 to +12 construct, containing the weak TALE-binding site and strong SP site, does not promote expression; further upstream sequences are needed. (C) Mutation of the TALE-binding site in LCE1C −161 to +27 promoter–luciferase construct, so that complexes can no longer bind, causes near complete loss of expression, whereas mutation of the flanking SP site causes loss of Ca2+ inducibility. Double mutation of TALE-binding and SP sites within LCE1C is uninformative owing to the severity of the loss of expression. Firefly luciferase values were normalised against pRL-TK Renilla luciferase reporter construct. Results are shown as fold change over the LCE1C −92 to +27 or LCE3E −59 to +12 activity in each cell type. For keratinocytes, fold change is calculated against activity in keratinocytes cultured in 0.09 mM calcium (low Ca2+ medium). Error bars are ± s.e.m.
By contrast, a truncated −122/+12 internally expressed LCE 3E construct, containing the weak TALE-binding site and strong SP site (Fig. 5A) did not mediate expression, and sequences further upstream were needed for expression in normal human keratinocytes (Fig. 5B). This shows that TALE/SP binding confers different expression effects on the LCE1C (external) and LCE3E (internal) promoters.
TALE factors also bind rodent Lce promoters
Rodent (mouse and rat) Lce gene clusters have diverged from human LCE clusters, such that specific human and rodent orthologues are unrecognised (Brown et al., 2007). However, organisation into externally expressing group 1 and internally expressing group 3 genes is strongly conserved (Brown et al., 2007). Despite the human and rodent gene divergence, DNA upstream of the group 1 transcription start sites shows conserved regions (supplementary material Fig. S3A), including putative rodent TALE consensus binding sites at a similar distance from the transcription start site as the human TALE-binding region (supplementary material Fig. S3A). We used EMSA on rat Lce1q (with a TALE-binding region typical of a larger group of mouse and rat Lce genes) to show that SP1, Pbx1, Pbx2 and Pknox factors also bind in vitro (supplementary material Fig. S3B). ChIP analysis with chromatin from differentiated rat keratinocytes was used to confirm SP1, Pbx2 and Pnox2 binding in vivo (supplementary material Fig. S3C). Lack of immunoprecipitation with Pbx1a, Meis factors and SP3 is consistent with EMSA findings, but, could also reflect properties of the antibodies used for immunoprecipitation.
The extension of the analysis of TALE binding to rodent means that mouse models can be used to analyse the function of TALE proteins in epidermis in vivo.
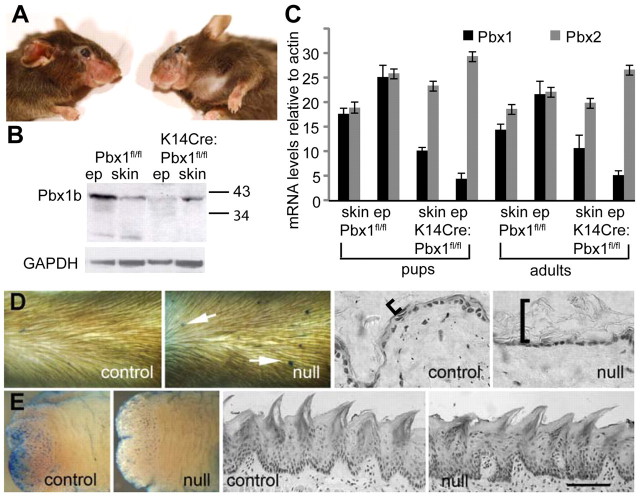
Depletion of TALE protein Pbx1 in epidermis causes stratum corneum and barrier defects in external but not internal stratum corneum
From our findings, we predict that TALE proteins selectively regulate external LCE promoters to form stringent external barriers. To test this hypothesis, we examined mice with defects in TALE expression. Mice bearing an epidermal K14-mediated conditional knockout of TALE protein Pbx1 (K14Cre:Pbx1fl/fl or K14CrePbx1fl/−) are viable, despite showing clear downregulation of epidermal PBX1 expression (Murphy et al., 2010) (Fig. 6A,B,C). However, at about 4 months of age, animals develop localised alopecia, which are suggestive of skin changes, at regions of stress such as vibrissae pads (Fig. 6A). Because TALE proteins are known to cross-regulate and we had shown small but consistent upregulation of Pbx2 mRNA in Pbx1-null epidermis (Fig. 6C), we looked for perturbation of other TALE factors in transgenic skin.
Fig. 6.
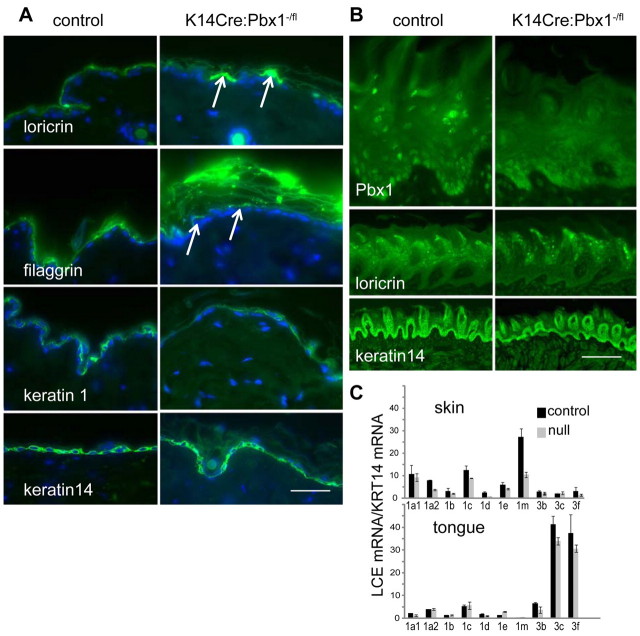
Epidermal-specific Pbx1-null mice are viable and have abnormalities in external but not internal barrier and stratum corneum. (A) Mice generated by keratin-14 Cre-driven epidermal deletion of floxed Pbx1 alleles (Murphy et al., 2010) are healthy and viable until about 4 months of age when they lose hair in stressed regions, for example, around the whisker pad. (B) Western analysis of protein from epidermis or skin (epidermis and dermis) of parental floxed Pbx1 mice (Pbx1f/f) and K14Cre-mediated epidermal Pbx1 deleted mice (K14Cre;Pbx1f/f) shows clear epidermal-specific knockdown of PBX1 protein. (C) Real-time PCR analysis of parental Pbx1f/f and K14Cre;Pbx1f/f epidermal and skin RNA from pups (day 16 to day 22) and adult (3–4 months) mice shows maintenance of epidermal-specific reduction in Pbx1 mRNA, whereas Pbx2 mRNA is slightly, but consistently upregulated. (D) Barrier assay shows lesions on the epidermis of K14Cre:Pbx1f/− mice (arrows), but not on wild-type (not shown) or control K14Cre:Pbx1f/+ mice. K14Cre:Pbx1f/− epidermal stratum corneum is thicker and noncompact (bracket) compared with control stratum corneum. (E) Pbx1-null (K14Cre:Pbx1f/−) tongue epidermis lacks lesions; ventral tongue surface is shown because it lacks blue-staining papillae except at the tip, clearly seen on the tip of control tongue. Tongue stratum corneum of Pbx1-null (K14Cre:Pbx1f/−) does not differ from control. Ep, epidermal; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Scale bars: 70 μm (D); 100 μm (E).
Immunohistochemical analysis shows widespread changes in levels and intracellular localisation of additional TALE proteins (supplementary material Fig. S4) in Pbx1-null mice. PBX2 moved to the nuclei in the epidermis of these mice, whereas other TALE proteins were downregulated, with the exception of Meis2, which appeared similar in the null and the control. These data show that the levels and cellular location of TALE proteins are interdependent in the epidermis.
Despite the viability of epidermal specific Pbx1-null (K14Cre:Pbx1f/−) mice, we showed by dye-exclusion skin barrier assay (Hardman et al., 1998) that mutant epidermis had a high frequency of tiny skin barrier defects or regions of focally enhanced permeability that were not apparent in control K14Cre:Pbx1f/+ mice or wild-type control mice (Fig. 6D). Skin barrier assay using the less-soluble hematoxylin dye permitted recognition of barrier defects after tissue sectioning (Byrne et al., 2010) (supplementary material Fig. S5). Barrier lesions can show hyperproliferation and minor hyperkeratinisation confined to the sites of increased permeability (supplementary material Fig. S5A,B) or are morphologically indistinguishable from nearby skin (supplementary material Fig. S5C,D). Epidermal hyperproliferation and hyperkeratinisation is consistent with a barrier defect (Elias, 2008). We think it improbable that scratching could cause focally enhanced permeability at multiple body sites in null mice.
To verify the skin barrier defect, we carried out a gravimetric transepidermal water loss assay on control and Pbx1-null mice. This assay is an older and established method of barrier assay (Hanley et al., 1996; Hardman et al., 1998), which can demonstrate the clear difference between external and internal stratified squamous epithelia (e.g. skin and tongue, Fig. 1C). Wild-type epidermis showed the consistent level of barrier activity expected for skin (supplementary material Fig. S5E, compare with Fig. 1C); however, TEWL values in the Pbx1-null mouse varied significantly, which is consistent with the heterogenous nature of their skin (average Pbx1-null skin TEWL>wild-type skin; P=0.027, one-tailed unpaired t-test). It is not known whether the variable TEWL activity of the Pbx1-null mutant skin is solely due to micro lesions or whether non-lesional skin also contributes.
Because the keratin-14 promoter is expressed prominently in internal as well as external stratified squamous epithelia (Vasioukhin et al., 1999) (Fig. 7B), PBX1 is also downregulated in all internal stratified squamous epithelia, including the tongue (Fig. 7B). However, mutant tongue epidermis lacked barrier defects and resembled the wild-type or control K14Cre:Pbx1f/+ tongue (Fig. 6E), showing that disruption of TALE proteins specifically affects barrier integrity in skin [note that ventral surface of tongue is shown here for clarity because the dye used for barrier assay associates with filiform and fungiform papillae on the tongue dorsal surface, making photographic representation difficult (Marshall et al., 2000); however, tongue dorsal surface also lacks barrier lesions, not shown]. These data are consistent with our finding of external, but not internal, LCE promoter dependency on a TALE site in cell culture (Fig. 5).
Fig. 7.
Differentiation is altered in epidermis, but not tongue, of keratin-14-targeted Pbx1-null mice. (A) Comparison of epidermal-specific Pbx1-null epidermis (K14Cre:Pbx1f/−) with control epidermis shows abnormal punctate expression of differentiation markers loricrin and filaggrin (arrows), whereas filaggrin retains immunoreactivity within the grossly extended mutant stratum corneum. Levels of keratin-1 and keratin-14 appear unchanged. (B) Tongue epidermis differentiates normally (keratin-14 and loricrin expression shown) despite clear knockout of Pbx1 in tongue epidermis. (C) LCE group 1 family members have reduced expression in Pbx1-null (K14Cre:Pbx1f/−) compared with control (K14Cre:Pbx1f/+) epidermis; representative experiment from two biological replicates. By contrast, expression of LCE group 3 proteins changes in Pbx1-null (K14Cre:Pbx1f/−) compared with control (K14Cre:Pbx1f/+) tongue epithelia. Note that samples with sporadic high levels of skin Lce3c, occurring with similar frequency in both transgenic and control skin samples and of unknown cause, were excluded. Values were obtained in triplicate and are normalised to levels of keratin-14. Error bars represent ± s.d. Scale bars: 50 μm (A), 400 μm (B, top panel), 50 μm (B, bottom panels).
Consistent with epidermal-specific disruption of the barrier, we show epidermal-specific disruption of the stratum corneum (Fig. 6D,E). K14Cre:Pbx1f/− mice had a loose, extended epidermal stratum corneum (Fig. 6D, brackets, see also Fig. 7A), whereas their tongue stratum corneum was indistinguishable from that in the wild-type and control mice (Fig. 6E, Fig. 7B).
Depletion of TALE protein Pbx1 in epidermis causes stratum corneum and barrier defects only in external skin
Analysis of epidermal differentiation markers showed disruption of the terminal differentiation proteins loricrin and filaggrin (Fig. 7A). Western analysis was not informative, but we showed grossly abnormal punctate expression of loricrin and filaggrin upon immunohistochemical analysis. These proteins form part of the matrix and envelope of cornified cells, and immunoreactivity to filaggrin persists in the grossly abnormal stratum corneum of the mutant mouse. The early differentiation markers keratin-1 and keratin-14 appeared unaffected. Oddly, the punctuate regions of filaggrin expression did not necessarily coincide with barrier lesions (supplementary material Fig. S5C,D). Differentiation marker expression in the tongue did not differ significantly between null and control epithelia (Fig. 7B).
Analysis of murine Lce mRNA levels (Fig. 7C) showed a reduction in group 1 Lce expression, particularly a reduction of the most prominently expresssed murine skin Lce, Lce1M, in K14CrePbx1fl/− skin. By contrast, we could not detect expression differences of murine group three Lces between the null and control tongue, consistent with a lack of tongue barrier phenotype.
Although the interdependent behavior of other TALE proteins upon Pbx1 epidermal-specific knockdown makes attribution of phenotype to a specific TALE protein impossible, these data show that disruption of TALE proteins in stratified squamous epithelia in vivo results in external skin-specific changes to the stratum corneum and epidermal barrier while leaving internal stratified squamous barrier-forming epithelia unaffected. Hence, we propose that TALE homeodomain proteins mediate a location-dependent skin barrier, acting at least partly through LCE and other stratum corneum targets.
Discussion
Here we demonstrate that TALE proteins have a key role in regulating regional epidermal barriers, at least partly through their differential regulation of LCE genes. Barrier qualities differ over the body surface, with the biggest measurable changes between internal epithelia that form the stratum corneum (e.g. tongue) and external skin (Marshall et al., 2000) (Fig. 1C). These differences probably permit the epithelia to respond appropriately to differences in aridity, microbial activity and requirement for flexibility in the different environments. However, these widely differing epithelia are derived from the same basal keratinocytes by region-specific differentiation programmes (Dhouailly et al., 2004). Based on work reported here, we propose that TALE proteins mediate these location-dependent barrier changes. In vitro, TALE proteins bind strongly to externally expressed LCE promoters but weakly to internally expressed LCE promoters. In vivo experiments confirm this result; we show that TALE perturbation in mutant mice specifically affects exterior skin, rather than the internal epithelium tongue. Taken together, these findings support the idea that the interaction of TALE proteins with LCE promoters is one mechanism by which the body develops site-specific barrier qualities from the same basal keratinocytes.
Barrier quality also differs over the exterior body surface, again to meet differing requirements for tissue sensitivity, flexibility, ultraviolet susceptibility and microbial colonisation. For example, the stratum corneum differs markedly in back skin, toe (palmar) skin and vulva, whereas less noticeable body-site differences manifest as regional susceptibility to irritation and disease. The location dependence of many of the common inflammatory skin diseases, such as psoriasis and eczema, is probably triggered by localised barrier breach (Brown and McLean, 2009; Elias and Schmuth, 2009), which might resemble the punctate regions of enhanced permeability that we found in the epidermal-specific Pbx1-null mouse. A proposal arising from this work is that TALE proteins mediate variations in the external barrier. In support of this, location-dependent change in TALE subcellular and epidermal stratum-specific location can be detected over morphologically similar body surfaces in mouse (S.J.B., unpublished results).
Regulation of location-specific keratinocyte differentiation is initiated by signalling from the underlying dermis (reviewed by Dhouailly et al., 2004). Recently a location-dependent fibroblast HOX protein code was implicated in this signalling (Rinn et al., 2008), acting through WNT intermediates (Olivera-Martinez et al., 2001; Rinn et al., 2008). How epidermis responds to the dermal signal to generate region-specific barriers is not understood and it is tempting to speculate that TALE proteins or their binding partners are responding to the dermally initiated signalling pathway.
PBX factors, as well as forming heteromeric complexes with Pknox and Meis TALE partners, form oligomeric complexes with HOX proteins, which provide HOX protein DNA-binding specificities. Several HOX proteins have been implicated by their expression in epidermis (e.g. Komuves et al., 2003; Stelnicki et al., 1998), and HOXB13 has a functional role in epidermal terminal differentiation (Mack et al., 2005). However, TALE factors are also known to form oligomeric complexes with a range of non-HOX proteins (Laurent et al., 2008). Future investigation will involve finding the partners for TALE proteins in epidermis.
We show in this work that the SP factors SP1 and SP3, as well as a truncated SP3, bind LCE promoters adjacent and, in the human promoters, overlapping the TALE site. SP3, but not SP1, can activate the p21 promoter in keratinocytes, suggesting that SP3 promotes keratinocyte proliferation over differentiation (Prowse et al., 1997). Consistent with this, SP3 can antagonise SP1 activation in a differentiation-dependent manner, with promoter activation dependent on the SP1/SP3 ratio, which favours SP1 as differentiation proceeds (Apt et al., 1996; Wong et al., 2005). Our results suggest competition between SP and TALE factors for binding, whereas our in vitro data show that although SP binding on LCE promoters is not sufficient for promoter activity, the TALE factor is crucial.
LCE genes are strongly marked by Polycomb group-mediated Histone3 trimethylation on lysine 27 (triMeK27-H3) in undifferentiated keratinocytes, which interferes with recruitment of AP1 transcription factors to Lce group 1 promoters (Ezhkova et al., 2009). Although TALE proteins show preferential association with terminally differentiated keratinocytes, they are not exclusively expressed in the upper cells where LCE expression occurs (Fig. 4) and it is tempting to speculate that epigenetic triMeK27-H3 marking could also control TALE access to Lce promoters in less well differentiated keratinocytes. Alternatively, TALE binding to LCE promoters may permit a TALE-binding partner access through restrictive chromatin. PBX1, which we show binds LCE promoters, is implicated in marking genes for activation through restrictive chromatin (Berkes et al., 2004).
Although we have emphasised the external expression of LCE group 1 genes in external stratified squamous epithelia (skin) and group 3 LCE genes in internal stratified squamous epithelia, it is worth pointing out that this applies to non-stressed skin. Minor expression of group 3 LCE genes in external epidermis can be detected (Jackson et al., 2005) and we have previously shown that a group 3 gene is massively upregulated in response to ultraviolet radiation in external skin (Jackson et al., 2005), and other group 3 genes are upregulated in response to experimental barrier loss (de Cid et al., 2009). Recent association of group 3 genes with psoriasis (de Cid et al., 2009; Zhang et al., 2009; Gudjonsson et al., 2010), a common inflammatory skin disease with underlying barrier aetiology, suggests that the group 3 genes are expressed in skin under situations of stress. It is possible that altered epidermal TALE protein activity under these conditions mediates upregulation via the weak group 3 TALE-binding site. Alternatively, stress-mediated changes in SP1 binding could make the group 3 TALE-binding site accessible, or additional upstream promoter elements that we found to increase gene expression in human cultured keratinocytes (Fig. 5) could mediate the in vivo group 3 promoter stress response.
The barrier defect in Pbx1 epidermal-specific null mice manifests as tiny regions of enhanced permeability, consistent with postnatal viability. The lack of left–right lesional symmetry suggests an environmental trigger (e.g. friction, infection) for the phenotype, and in this respect, it is similar to the skin lesions on mice where the epidermal desmosomal adhesion protein desmocollin-1 is deleted from upper layers of epidermis (Chidgey et al., 2001). It is probable that in the wild (outside the animal house), the skin would be more severely compromised. This barrier dysfunction model might mimic the skin of patients with subclinical barrier problems that lead to atopic tendencies. At the histological level, the late terminal differentiation markers, filaggrin and loricrin, are downregulated in most terminally differentiated keratinocytes of the Pbx1 epidermal-specific null mouse, but are expressed intensely in a few cells. This punctate expression does not correlate with the external lesions, and its biological basis is unknown, although it could represent a localised upregulation of genes in response to barrier loss that is not severe enough to form a lesion.
In summary, we show how location-dependent skin barriers might develop from a homogenous basal keratinocyte population through differential binding of TALE homeodomain transcription factors to the LCE barrier multigene family and other genes in the stratum corneum.
Materials and Methods
Animal work
Keratin-14 (K14) Cre recombinase transgenic mice (Vasioukhin et al., 1999) (purchased from the Jackson Laboratory, Bar Harbor, ME) were intercrossed with Pbx1-null mice (Selleri et al., 2001) to generate K14Cre:Pbx1−/+ mice. Crossing to mice harbouring a floxed Pbx1 allele (Ficara et al., 2008; Murphy et al., 2010) generated K14Cre:Pbx1f/− (mutant) and littermate K14Cre:Pbx1f/+ (control) mice. Mice were genotyped by PCR (Murphy et al., 2010) from DNA freshly isolated from ventral skin and shown to have reduced levels of epidermal Pbx1 mRNA and protein (Fig. 7, supplementary material Fig. S3). All experiments were performed with the approval of and in accordance with Stanford's administrative Panel on Laboratory Animal Care.
Cell reporter assay
LCE promoter fragments (see supplementary material Table S1 for primer sequences) were cloned into PGL3-Basic vector (Promega). Site-directed mutagenesis (see supplementary material Table S1 for primer sequences) was performed with a GeneTailer Site-Directed Mutagenesis Kit (Invitrogen). Human N-Tert keratinocytes (Dickson et al., 2000) grown in low calcium (0.4 mM) to suppress differentiation or induced to differentiate by growth in high calcium (1.2 mM) were transfected in triplicate (Lipofectamine, 2000, Invitrogen) with a Renilla firefly luciferase reporter (pRL-TK, Promega) and normalised luciferase activity determined with a Dual-Luciferase Assay kit (Promega).
Chromatin immunoprecipitation
Chromatin was extracted from neonatal human epidermal keratinocytes (Gibco) grown in Medium 154CF (Cascade Biologicals) supplemented with 0.2 mM CaCl2 and induced to differentiate by incubation for 2 days in 1.2 mM CaCl2. For rodents, ChIP chromatin was extracted from rat epidermal keratinocytes cultured in differentiating medium (Marjukka Suhonen et al., 2003; Baden and Kubilus, 1983). ChIP (Carey et al., 2009) was performed with keratinocytes crosslinked in suspension after harvesting by trypsin. Chromatin was degraded to ~0.5–1.0 kb using sonication (MSE Soniprep 150, power output 14–18 microns) for 25×15 seconds. 6–10 μg rabbit antibodies against PBX1, PBX2, SP1, SP3 (Santa Cruz Biotechnology), Pknox2 (Abnova) and keratin-1 (Covance) were used for immunoprecipitation. Primers are listed in supplementary material Table S1.
Electrophoretic mobility shift assay
EMSA was performed on radiolabelled double-stranded oligonucleotides using nuclear extracts from terminally differentiated NTERT keratinocytes (Dickson et al., 2000). Retarded complexes were supershifted with approximately 0.1–0.2 μg of antibody (rabbit antibodies against PBX1-3, PBX1, PBX2, PBX3, Pknox1 (Santa Cruz Biotechnology); rabbit anti-Meis1 and anti-Meis2 (Abcam). Competition was performed with double-stranded oligonucleotides or by using commercial oligonucleotides to transcription factor consensus binding regions (Promega) (sequences are given in supplementary material Table S1).
Immunohistochemistry
Immunohistochemistry on paraffin sections was by standard techniques. Antibodies used were: rabbit antibodies against keratin-14, keratin-1, filaggrin, loricrin and involucrin (Covance) used at 1:1000; PBX 1–3, PBX1b, PBX2, PBX3, Pknox1 (Santa Cruz Biotechnology) at 1:500; Meis1 and Meis2 (Abcam) at 1:500; mouse antibodies against Pbx1A, Pbx2 and Pbx3A (Abnova) at 1:250; PBX1B, PBX2 and PBX3 at 1:100 (Selleri et al., 2004). Antibody detection was by secondary antibodies conjugated to Alexa Fluor 488 (green) and Alexa Fluor 546 (red) (Molecular Probes). Counterstaining was with hemotoxylin and eosin for immunohistochemistry and 4′,6-diamidino-2-phenylindole (DAPI) for immunofluorescence. Images were taken with a Nikon Eclipse E600 microscope with either 20× (NA 0.4) or 60× oil immersion (NA 1.40) objectives, using a SPOT digital camera (Diagnostic Instruments) with Spot RT Software v3.0.
Real-time PCR expression analysis and western analysis
Mouse skin was frozen in liquid nitrogen and disrupted while frozen with a pestle and mortar. Epidermis was separated from dermis by incubation with trypsin at 37°C for 1 hour. RNA was isolated (Qiagen RNeasy Kit) and converted to cDNA with random primers (Iscript Select cDNA synthesis Kit, Bio-Rad). mRNA levels were monitored by real-time PCR on three biological replicates using Platinum SYBR Green qPCR mix-UDG (Invitrogen) on an Applied Biosystems 7500 Real Time PCR system. Expression was inferred from single peak dissociation curves and normalised to keratin-14. Primers (supplementary material Table S1) were confirmed for specificity by product sequencing. Western analysis was performed on skin or epidermal samples that were homogenised using a Polytron in Laemmli buffer, then loaded directly onto the gel. Antibodies used for western blot analysis were as reported previously (Murphy et al., 2010; Selleri et al., 2001; Selleri et al., 2004).
Skin barrier assay
Histological skin barrier assay (dye permeability assay, dye exclusion assay) was performed on mouse skin fixed in 4% paraformaldehyde in PBS as described previously (Hardman et al., 1998). Briefly, tissues were dehydrated with methanol (approximately 1 minute each in 25%, 50%, 75% then 100% ethanol) then rehydrated using the reverse procedure, followed by immersion in 1% toluidine blue in water, then extensive destaining in PBS. Barrier assay followed by immunohistochemistry was as described previously (Byrne et al., 2010) using hematoxylin. Transepidermal water loss (TEWL) barrier assay on excised skin (~40 mm square) was carried out as described (Hardman et al., 1998; Marshall et al., 2000) using a Cahn21 balance. Skin sample size was estimated after photography using ImageJ software.
Supplementary Material
Acknowledgments
This work was supported by Barts and The London Charity RAB05/PJ/04, Cancer Research UK C5314/A6695, British Skin Foundation 941S, and US Public Health Service CA90735. We acknowledge the assistance of Maria Ambrose in maintenance of mice and preparation of tissue, Claudia Tilli in primer design and testing and Raija Tammi for rat epidermal keratinocyte cultures.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/10/1681/DC1
References
- Apt D., Watts R. M., Suske G., Bernard H. U. (1996). High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology 224, 281-291 [DOI] [PubMed] [Google Scholar]
- Baden H. P., Kubilus J. (1983). The growth and differentiation of cultural newborn rat keratinocytes. J. Invest. Dermatol. 80, 124-130 [DOI] [PubMed] [Google Scholar]
- Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Pena-Castillo L., Alleyne T. M., Mnaimneh S., Botvinnik O. B., Chan E. T., et al. (2008). Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266-1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465-477 [DOI] [PubMed] [Google Scholar]
- Brown S. J., McLean W. H. (2009). Eczema genetics: current state of knowledge and future goals. J. Invest. Dermatol. 129, 543-552 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Tilli C. M., Jackson B., Avilion A. A., MacLeod M. C., Maltais L. J., Lovering R. C., Byrne C. (2007). Rodent Lce gene clusters; new nomenclature, gene organization, and divergence of human and rodent genes. J. Invest. Dermatol. 127, 1782-1786 [DOI] [PubMed] [Google Scholar]
- Byrne C., Avilion A. A., O'Shaughnessy R. F., Welti J. C., Hardman M. J. (2010). Whole-mount assays for gene induction and barrier formation in the developing epidermis. Methods Mol. Biol. 585, 271-286 [DOI] [PubMed] [Google Scholar]
- Cabral A., Voskamp P., Cleton-Jansen A. M., South A., Nizetic D., Backendorf C. (2001). Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J. Biol. Chem. 276, 19231-19237 [DOI] [PubMed] [Google Scholar]
- Carey M. F., Peterson C. L., Smale S. T. (2009). Chromatin Immunoprecipitation (ChIP). Cold Spring Harb. Protoc. 2009, doi:10.1101/pdb.prot5279 [DOI] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933-2942 [DOI] [PubMed] [Google Scholar]
- Chidgey M., Brakebusch C., Gustafsson E., Cruchley A., Hail C., Kirk S., Merritt A., North A., Tselepis C., Hewitt J., et al. (2001). Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. J. Cell Biol. 155, 821-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cid R., Riveira-Munoz E., Zeeuwen P. L., Robarge J., Liao W., Dannhauser E. N., Giardina E., Stuart P. E., Nair R., Helms C., et al. (2009). Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 41, 211-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D., Olivera-Martinez I., Fliniaux I., Missier S., Viallet J. P., Thelu J. (2004). Skin field formation: morphogenetic events. Int. J. Dev. Biol. 48, 85-91 [DOI] [PubMed] [Google Scholar]
- Dickson M. A., Hahn W. C., Ino Y., Ronfard V., Wu J. Y., Weinberg R. A., Louis D. N., Li F. P., Rheinwald J. G. (2000). Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436-1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P. M. (2008). Skin barrier function. Curr. Allergy Asthma Rep. 8, 299-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias P. M., Schmuth M. (2009). Abnormal skin barrier in the etiopathogenesis of atopic dermatitis. Curr. Opin. Allergy Clin. Immunol. 9, 437-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E. (2009). Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficara F., Murphy M. J., Lin M., Cleary M. L. (2008). Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell 2, 484-496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fognani C., Kilstrup-Nielsen C., Berthelsen J., Ferretti E., Zappavigna V., Blasi F. (2002). Characterization of PREP2, a paralog of PREP1, which defines a novel sub-family of the MEINOX TALE homeodomain transcription factors. Nucleic Acids Res. 30, 2043-2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P. A., Mackenzie P. I. (2002). The homeodomain Pbx2-Prep1 complex modulates hepatocyte nuclear factor 1alpha-mediated activation of the UDP-glucuronosyltransferase 2B17 gene. Mol. Pharmacol. 62, 154-161 [DOI] [PubMed] [Google Scholar]
- Gudjonsson J. E., Ding J., Johnston A., Tejasvi T., Guzman A. M., Nair R. P., Voorhees J. J., Abecasis G. R., Elder J. T. (2010). Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 130, 1829-1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley K., Rassner U., Elias P. M., Williams M. L., Feingold K. R. (1996). Epidermal barrier ontogenesis: maturation in serum-free media and acceleration by glucocorticoids and thyroid hormone but not selected growth factors. J. Invest. Dermatol. 106, 404-411 [DOI] [PubMed] [Google Scholar]
- Hardman M. J., Sisi P., Banbury D. N., Byrne C. (1998). Patterned acquisition of skin barrier function during development. Development 125, 1541-1552 [DOI] [PubMed] [Google Scholar]
- Jackson B., Tilli C. M., Hardman M. J., Avilion A. A., MacLeod M. C., Ashcroft G. S., Byrne C. (2005). Late cornified envelope family in differentiating epithelia-response to calcium and ultraviolet irradiation. J. Invest. Dermatol. 124, 1062-1070 [DOI] [PubMed] [Google Scholar]
- Jave-Suarez L. F., Schweizer J. (2006). The HOXC13-controlled expression of early hair keratin genes in the human hair follicle does not involve TALE proteins MEIS and PREP as cofactors. Arch. Dermatol. Res. 297, 372-376 [DOI] [PubMed] [Google Scholar]
- Kennett S. B., Udvadia A. J., Horowitz J. M. (1997). Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 25, 3110-3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuves L. G., Shen W. F., Kwong A., Stelnicki E., Rozenfeld S., Oda Y., Blink A., Krishnan K., Lau B., Mauro T., et al. (2000). Changes in HOXB6 homeodomain protein structure and localization during human epidermal development and differentiation. Dev. Dyn. 218, 636-647 [DOI] [PubMed] [Google Scholar]
- Komuves L. G., Ma X. K., Stelnicki E., Rozenfeld S., Oda Y., Largman C. (2003). HOXB13 homeodomain protein is cytoplasmic throughout fetal skin development. Dev. Dyn. 227, 192-202 [DOI] [PubMed] [Google Scholar]
- LaPres J. J., Hudson L. G. (1996). Identification of a functional determinant of differentiation-dependent expression in the involucrin gene. J. Biol. Chem. 271, 23154-23160 [DOI] [PubMed] [Google Scholar]
- Laurent A., Bihan R., Omilli F., Deschamps S., Pellerin I. (2008). PBX proteins: much more than Hox cofactors. Int. J. Dev. Biol. 52, 9-20 [DOI] [PubMed] [Google Scholar]
- Liu Y., Helms C., Liao W., Zaba L. C., Duan S., Gardner J., Wise C., Miner A., Malloy M. J., Pullinger C. R., et al. (2008). A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041, doi:10.1371/journal.pgen.1000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bayghen E., Vega A., Cadena A., Granados S. E., Jave L. F., Gariglio P., Alvarez-Salas L. M. (1996). Transcriptional analysis of the 5′-noncoding region of the human involucrin gene. J. Biol. Chem. 271, 512-520 [DOI] [PubMed] [Google Scholar]
- Mack J. A., Li L., Sato N., Hascall V. C., Maytin E. V. (2005). Hoxb13 up-regulates transglutaminase activity and drives terminal differentiation in an epidermal organotypic model. J. Biol. Chem. 280, 29904-29911 [DOI] [PubMed] [Google Scholar]
- Manley N. R., Selleri L., Brendolan A., Gordon J., Cleary M. L. (2004). Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 276, 301-312 [DOI] [PubMed] [Google Scholar]
- Marjukka Suhonen T., Pasonen-Seppanen S., Kirjavainen M., Tammi M., Tammi R., Urtti A. (2003). Epidermal cell culture model derived from rat keratinocytes with permeability characteristics comparable to human cadaver skin. Eur. J. Pharm. Sci. 20, 107-113 [DOI] [PubMed] [Google Scholar]
- Marshall D., Hardman M. J., Byrne C. (2000). SPRR1 gene induction and barrier formation occur as coordinated moving fronts in terminally differentiating epithelia. J. Invest. Dermatol. 114, 967-975 [DOI] [PubMed] [Google Scholar]
- Marshall D., Hardman M. J., Nield K. M., Byrne C. (2001). Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl. Acad. Sci. USA 98, 13031-13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens C. B., Selleri L. (2006). Hox cofactors in vertebrate development. Dev. Biol. 291, 193-206 [DOI] [PubMed] [Google Scholar]
- Murphy M. J., Polok B. K., Schorderet D. F., Cleary M. L. (2010). Essential role for Pbx1 in corneal morphogenesis. Invest. Ophthalmol. Vis. Sci. 51, 795-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera-Martinez I., Thelu J., Teillet M. A., Dhouailly D. (2001). Dorsal dermis development depends on a signal from the dorsal neural tube, which can be substituted by Wnt-1. Mech. Dev. 100, 233-244 [DOI] [PubMed] [Google Scholar]
- Phillips M. A., Qin Q., Rice R. H. (2000). Identification of an involucrin promoter transcriptional response element with activity restricted to keratinocytes. Biochem. J. 1, 45-53 [PMC free article] [PubMed] [Google Scholar]
- Prowse D. M., Bolgan L., Molnar A., Dotto G. P. (1997). Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 272, 1308-1314 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Wang J. K., Allen N., Brugmann S. A., Mikels A. J., Liu H., Ridky T. W., Stadler H. S., Nusse R., Helms J. A., et al. (2008). A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 22, 303-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L., Depew M. J., Jacobs Y., Chanda S. K., Tsang K. Y., Cheah K. S., Rubenstein J. L., O'Gorman S., Cleary M. L. (2001). Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128, 3543-3557 [DOI] [PubMed] [Google Scholar]
- Selleri L., DiMartino J., van Deursen J., Brendolan A., Sanyal M., Boon E., Capellini T., Smith K. S., Rhee J., Popperl H., et al. (2004). The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell. Biol. 24, 5324-5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelnicki E. J., Komuves L. G., Kwong A. O., Holmes D., Klein P., Rozenfeld S., Lawrence H. J., Adzick N. S., Harrison M., Largman C. (1998). HOX homeobox genes exhibit spatial and temporal changes in expression during human skin development. J. Invest. Dermatol. 110, 110-115 [DOI] [PubMed] [Google Scholar]
- Subramaniam S. (1998). The biology workbench – a seamless database and analysis environment for the biologist. Proteins 32, 1-2 [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein L., Wise B., Fuchs E. (1999). The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA 96, 8551-8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. F., Barnes L. M., Dahler A. L., Smith L., Popa C., Serewko-Auret M. M., Saunders N. A. (2005). E2F suppression and Sp1 overexpression are sufficient to induce the differentiation-specific marker, transglutaminase type 1, in a squamous cell carcinoma cell line. Oncogene 24, 3525-3534 [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Huang W., Yang S., Sun L. D., Zhang F. Y., Zhu Q. X., Zhang F. R., Zhang C., Du W. H., Pu X. M., et al. (2009). Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205-210 [DOI] [PubMed] [Google Scholar]
- Zhao X. P., Elder J. T. (1997). Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics 45, 250-258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.