Abstract
A fungal extract (MSX 63619), from the Mycosynthetix library of over 50,000 fungi, displayed promising cytotoxicity against a human tumor cell panel. Bioactivity-directed fractionation led to the isolation of an o-pyranonaphthoquinone decaketide, which we termed obionin B (1). The structure of 1 was deduced via spectroscopic and spectrometric techniques. The IC50 value of 1 was moderate, ranging from 3 to 13 μM, depending on the cell line tested.
Keywords: Polyketide, Cytotoxicity, Pleosporales, o-Naphthoquinone
Nature, in general, and fungi, specifically, have and continue to be a valuable source of new drug leads.1–3 For example, two of the most well known drug classes of the 20th century, the antibiotics and the cholesterol lowering agents, both originated as fungal secondary metabolites (e.g. penicillin and compactin). More recently, in 2010 the U.S. FDA approved a new treatment for multiple sclerosis, fingolimod (Gilenya), which derived from the fungal secondary metabolite, myriocin.4 Yet, despite these successes, it has been postulated that fewer than 10% of the estimated 1 to 1.5 million species of fungi in the world have been identified taxonomically, with possibly an even smaller percentage being studied for bioactive leads.5 Thus, given the under explored biodiversity, and the aforementioned successes as inspiration, our research team has been probing the Mycosynthetix library of filamentous fungi, representing over 50,000 isolates, for new anticancer drug leads.6
An extract of the filamentous fungus MSX 636197 exhibited promising cytotoxic activity, as evidenced by less than 20% survival of human tumor cells when treated with 20 μg/mL of crude extract. The 1:1 chloroform/methanol extract of the solid fermentation of MSX 63619 was subjected to bioactivity-directed fractionation using flash chromatography on silica gel followed by preparative RP-HPLC on C18.8 This resulted in the isolation (>95% purity according to HPLC) and characterization of a new decaketide, which was named obionin B (1). This compound was evaluated against the human tumor panel and a pair of antibacterial assays.
Compound 1,9 which was a deep purple color, displayed HRESIMS data of m/z 355.1541 in the negative mode (corresponding to C21H24O5 - H, calcd for 355.1545 [M - H]−), indicating an index of hydrogen deficiency of ten. The 1H- and COSY-NMR data showed signals consistent with a straight saturated alkyl chain from δH 0.8–2.3. The rest of the signals were singlets, starting with a three-proton singlet at δH 3.81, suggesting a methoxy group. A two-proton singlet was present at δH 5.16, followed by one-proton singlets at δH 5.56, 6.26, 6.30, and 12.31, the latter consistent with an intramolecular hydrogen bonded phenol. The 13C-NMR data showed 21 peaks, consistent with the HRMS data, including seven peaks upfield of δC 35, six of which were methylenes and the seventh a methyl according to the multiplicity edited HSQC experiment. These data indicated that the straight saturated alkyl chain was n-heptyl. The only other signals upfield of δC 100 were a methylene at δC 63.1 that correlated with the two-proton singlet at δH 5.16, and the methoxy signal at δC 55.9. The remaining signals were for sp2 carbons, with five between δC 100–120, five between δC 130–170 and two between δC 175–180 (see Supporting Information for the 1H- and 13C-NMR spectra and Table S1).
Upon purification of 1, the diode-array detector on the HPLC displayed UV maxima at 243, 297, and 464 nm. A search of the Dictionary of Natural Products for the formula C21H24O5 and a UV range of 460–468 nm returned a single hit; obionin A.10 The downfield 1H- and 13C-NMR signals of 1 were in close agreement with the NMR data for the o-pyranonaphthoquinone portion of obionin A (2), except for the H-11 resonance, which was split into two one-proton doublets due to the presence of chiral centers in the side chain of 2; this was a two-proton singlet (δH 5.16) in achiral 1. However, the data for the upfield region were significantly different. Obionin A (2) has a branched saturated alkyl chain (Figure 1) instead of the n-heptyl chain of 1. The HMBC spectrum showed that the n-heptyl group of 1 was attached to the o-pyranonaphthoquinone at the same position (C-9) as the branched saturated chain in obionin A (Figure 2). To verify that the quinone was ortho instead of para, key HMBC correlations were observed from H-6 to C-4, and H-4 to both C-6 and C-2 (Figure 2). The remaining HMBC correlations (Figure 2) confirmed the structure of 1, which was termed obionin B in deference to the earlier studies on obionin A.10
Figure 1.
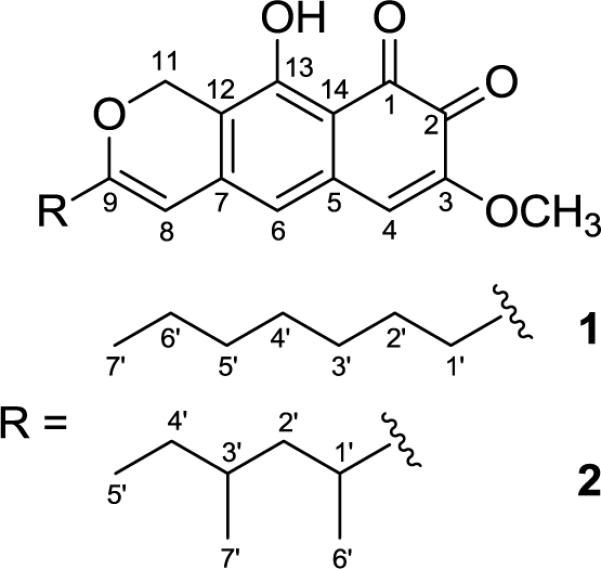
Structures of obionin A (2) and obionin B (1)
Figure 2.
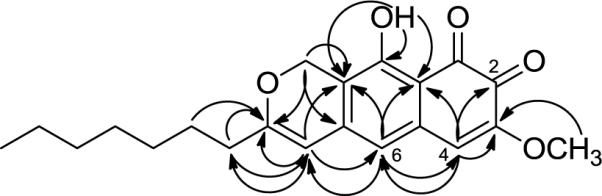
HMBC correlations of obionin B (1)
Compound 1 was assayed against several human cancer cell lines using methods described previously,11 including the MCF-7 (breast carcinoma), NCI-H460 (large cell lung carcinoma), SF-268 (astrocytoma), HT-29 (colorectal adenocarcinoma), and MDA-MB-435 (melanoma) cell lines. The IC50 values (Table 1) indicated that obionin B was moderately cytotoxic. Compound 1 was inactive (MIC values > 500 μg/mL; data not shown) when tested for antimicrobial activity using methods described previously11 against Escherichia coli and Bacillus subtilis. Previously, obionin A (2) was isolated based on the brine shrimp toxicity of the fungal extract,10 although the authors stated that 2 did “not account for that activity;” to the best of our knowledge, 2 has not been examined for cytotoxicity. A related pair of o-pyranonaphthoquinones, the laccaridiones, were reported as promising antimycotic leads due to inhibition of Candida albicans adhesion to epithelial and endothelial cells, as well as the ability to reduce the release and inhibit the catalytic activity of secreted aspartic proteases.12 These effects resulted in reduced virulence properties (e.g. colonization and penetration of host tissues) without being fungistatic or fungicidal.12 However, only laccaridione B has been investigated for cytotoxicity towards human tumor cells, with IC50 values ranging from 4.5 μM in the K-562 human erythroleukemia cell line to 34 μM in HeLa cervical carcinoma cells.13 In comparison, obionin B (1) exhibited broad potency on the lower end of this range (Table 1).
Table 1.
Cytotoxicity of compound 1 against a panel of human tumor cell lines.
| IC50 values in μMa | |||||
|---|---|---|---|---|---|
| Compound | MCF-7 | H460 | SF-268 | HT-29 | MDA-MB-435 |
| obionin B (1) | 11.1 | 7.6 | 13.2 | 3.1 | 7.3 |
| camptothecinb | 0.03 | 0.005 | 0.13 | nt | nt |
| silvestrolb | ntc | nt | nt | 0.004 | 0.006 |
IC50 values are determined as the concentration required to reduce cellular staining with sulforhodamine B by 50% relative to untreated controls following 72 h of continuous exposure.11
Positive controls.
Indicates `not tested'
In conclusion, an o-pyranonaphthoquinone decaketide (obionin B, 1) has been isolated and characterized from a terrestrial fungus from the Mycosynthetix library of filamentous fungi; 1 exhibited moderate cytotoxicity toward a panel of five human tumor cell lines (Table 1). Compound 1 is structurally related to the known nonaketides obionin A,10 leptosphaerodione,14 and the laccaridiones.13 Interestingly, the former two were both isolated from marine-derived fungi of the same Order (Pleosporales) as the terrestrial fungus (MSX 63619)7 investigated herein; this is the second time we have reported a correlation between secondary metabolites isolated from fungi of both terrestrial and marine origin.15
Supplementary Material
Acknowledgments
This research was supported by P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. The Golden LEAF Foundation (Rocky Mount, NC) provided partial support to D. J. K. Mycology technical support was provided by Maurica Lawrence. The authors thank Mingming Su of the David H. Murdock Research Institute, Kannapolis, NC, for mass spectrometry data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material Supplementary data (1H- and 13C-NMR spectra and data) associated with this article can be found, in the online version, at (doi # to be filled in by publisher).
References and notes
- (1).Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- (2).Pearce C, Eckard P, Gruen-Wollny I, Hanske FG. In: Natural Product Chemistry for Drug Discovery. Buss AD, Butler MS, editors. The Royal Society of Chemistry; Cambridge: 2010. pp. 215–244. [Google Scholar]
- (3).Butler MS. Nat. Prod. Rep. 2008;25:475–516. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- (4).Strader CR, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:900–907. doi: 10.1021/np2000528. [DOI] [PubMed] [Google Scholar]
- (5).Hawksworth DL, Rossman AY. Phytopathology. 1997;87:888–891. doi: 10.1094/PHYTO.1997.87.9.888. [DOI] [PubMed] [Google Scholar]
- (6).Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. In: Bioactive Compounds from Natural Sources. Second Edition: Natural Products as Lead Compounds in Drug Discovery. Tringali C, editor. Taylor & Francis; London: 2011. pp. 37–63. [Google Scholar]
- (7).Mycosynthetix fungal strain 63619 was isolated in June, 1992 by Dr. Barry Katz of MYCOsearch from woody plant material. DNA analysis was performed by MIDI Labs, Inc. (Newark, DE), and the D2 variable region of the Large Subunit (LSU) rRNA was sequenced and compared to their database; the closest match could only determine that this fungus was of the Order Pleosporales; these data were deposited in Genbank (accession No. JN032133). The culture was stored on a malt extract slant and was transferred periodically. A fresh culture was grown on a similar slant, and a piece was transferred to a medium containing 2% soy peptone, 2% dextrose, and 1% yeast extract (YESD media). Following incubation (7 d) at 22 °C with agitation, the culture was used to inoculate 50 mL of a rice medium, prepared using rice to which was added a vitamin solution and twice the volume of rice with H2O, in a 250 mL Erlenmeyer flask. This was incubated at 22 °C until the culture showed good growth (approximately 14 d).
- (8).The 1:1 CHCl3/MeOH extract of MSX 63619 (351 mg) was eluted at 30 mL/min on a RediSep Rf silica gel column (12 g) using a Teledyne ISCO CombiFlash Rf. The solvent gradient was 100% hexanes to 100% CHCl3 over 15 column volumes (CV), 100% CHCl3 for 9 CV, then from 100:0 to 80:20 CHCl3/MeOH over 30 CV. The material eluting from 0–2% MeOH was active in the cytotoxicity assay and pooled (96.42 mg). This fraction was subjected to preparative HPLC (Phenomenex Gemini C18, 250 × 21.2 mm, 5 μm, 15 mL/min, 20–100% CH3CN in H2O over 30 min, hold 100% CH3CN for 10 min). Compound 1 was isolated at >95% purity from the 100% CH3CN fraction (9.96 mg).
- (9).Obionin B (1): dark purple solid: IR (diamond) νmax 2955, 2928, 2857, 1686, 1598, 1557, 1336, 1097, 877 cm−1; UV (MeOH) λmax (log ε), 242 (4.12), 294 (3.84) 465 (3.80) nm; 1H-and 13C-NMR (see Supporting Information); HRESIMS m/z 355.1541 [M - H]− (calcd for C21H24O5 - H, 355.1545).
- (10).Poch GK, Gloer JB. Tetrahedron Lett. 1989;30:3483–3486. [Google Scholar]
- (11).Ayers D, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Falkensammer B, Pleyer L, Ressler S, Berg A, Zepelin MBV, Nagl M, Lass-Florl C, Speth C, Dierich MP, Wurzner R. Mycoses. 2008;51:505–514. doi: 10.1111/j.1439-0507.2008.01515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Berg A, Reiber K, Dorfelt H, Walther G, Schlegel B, Grafe U. J. Antibiot. 2000;53:1313–1316. doi: 10.7164/antibiotics.53.1313. [DOI] [PubMed] [Google Scholar]
- (14).Guerriero A, Dambrosio M, Cuomo V, Pietra F. Helv. Chim. Acta. 1991;74:1445–1450. [Google Scholar]
- (15).Sy-Cordero AA, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. doi: 10.1021/np2004243. In Revision, submitted 05/19/2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


