Abstract
We present a novel algorithm to accelerate feature based registration, and demonstrate the utility of the algorithm for the alignment of large transmission electron microscopy (TEM) images to create 3D images of neural ultrastructure. In contrast to the most similar algorithms, which achieve small computation times by truncated search, our algorithm uses a novel randomized projection to accelerate feature comparison and to enable global search. Further, we demonstrate robust estimation of non-rigid transformations with a novel probabilistic correspondence framework, that enables large TEM images to be rapidly brought into alignment, removing characteristic distortions of the tissue fixation and imaging process. We analyze the impact of randomized projections upon correspondence detection, and upon transformation accuracy, and demonstrate that accuracy is maintained. We provide experimental results that demonstrate significant reduction in computation time and successful alignment of TEM images.
Keywords: Image Registration, Electron Microscopy
I. INTRODUCTION
Image registration is a fundamental process in medical imaging applications aimed at establishing spatial correspondences between images [1]. The algorithm presented here was developed in order to enable the rapid and accurate creation of 3D volumetric images of neural ultrastructure by the alignment of transmission electron microscopy (TEM) serial sections with very large fields of view at very high spatial resolution. Recent advances in TEM imaging now provide the capability to image the ultrastructure of the brain at unprecedented resolution with extremely large volumes of interest ([2], [3], [4], [5]). Determining the detailed connections of brain circuits is an essential unsolved problem in neuroscience. Analysis of volumetric TEM images will provide critical insight making it possible to uncover the connectivity of brain circuits. However, the volumetric reconstruction of neural circuitry from series of EM images pose several unique challenges, posed by high spatial resolution (order of nanometers) which makes apparent the large number of scales at which interesting biological structure exists, the very large size of the data (terabytes to petabytes) which necessitates efficient algorithms, and the distortion of tissue due to the section handling and imaging processes [6], [7], [8]. The volumetric construction of neural circuitry from TEM remains a substantial and challenging problem [9]. 1
A. Electron Microscopy
The reconstruction of three-dimensional neural tissue with conventional serial electron microscopy is not a new endeavor. Twenty five years ago, White et al. [10] imaged the entire nervous system of the nematode C. elegans down to the last neuron, dendrite, and synapse. Yet the technique classically was done by hand, with photographic techniques and manual tracing. Currently, there are several types of serial section EM providing three dimensional (3D) volumetric resolution over regions of up to a cubic millimeter. The approaches differ mainly by the sectioning process, the image acquisition and their alignment [3]. The first is SSTEM (Serial Section Transmission EM, [11], [12]) which involves slicing the tissue to ultrathin (50nm) sections, where each section is cut from the tissue block by a diamond knife, creating ribbons of thin sections. Following the embedding sectioning and staining processes the sections are imaged with TEM where a broad beam of electrons is directed at the thin sections allowing a substantial fraction of the electrons to pass through the tissue and then be focused onto a detector. The second of these high-throughput techniques is the Serial Block-Face Scanning EM (SBFSEM) [2]. In SBFSEM a tissue block is processed in the manner typical for TEM; however, instead of slicing thin sections for TEM imaging, the block is mounted in the vacuum chamber of a specially modified scanning EM (SEM). Within the SEM, the top-most layer of tissue from the block is successively sliced off after it is imaged, which is then simply discarded. The freshly exposed blockface is scanned with an SEM electron beam and the backscattered electrons are recorded. This way, 3D data set can be collected with no manual involvement. The advantage of SBFSEM is that the backscattering from the solid block requires no slice registration. However, the signal-to-noise ratio and the in-plane resolution are generally lower than what can be obtained using SSTEM. SBFSEM can provide sections as thin as 20-30 nm, but is limited to 20-30 nm per pixel resolution insection ([11],[3]). Although SSTEM lead to larger sections thickness (of ≈ 40 nm), it offers significantly better in plane resolution (2-4 nm) and relatively higher signal-to-noise ratio [6]. A third alternative is Serial Section Electron Tomography (SSET) which is based on reconstruction from 2D projections taken at different angles [13], [14]. The in-plane resolution of this method is similar to that of SSTEM and the axial resolution can be far better, but it has not proven a practical method for large-scale reconstructions of neuronal circuits. Finally, Focused Ion Beam Scanning EM (FIB-SEM) [15] offers excellent in-plane and axial resolution, but the volumes that can currently be acquired with this method are too small for large-scale reconstruction.
B. Related Work
A significant literature has developed addressing the special requirements of microscopy images for alignment. Algorithms for 3D reconstruction from 2D images have extensively utilized feature based approaches due to their superior performance, and have included the block matching strategy proposed by [16], where the local displacements were utilized to robustly estimate a global rigid transformation. Dauguet et al. [17] developed a solution for 3D reconstruction of a series of TEM images based on the finite support properties of the cubic B-splines, where the initial estimate for the affine registration was based on the block matching technique described in [16]. Koshevoy et al. [6] addressed the section to section matching as part of a complete algorithm for assembling 3D volumes from EM data. Their approach first identifies Scale Invariant Feature Transform (SIFT)[18] feature descriptors and then exploits these features to match adjacent slices, where the transformation is parameterized by Legendre polynomials. Thus the algorithm relies on the assumption that the tissue undergoes only smooth and continuous deformations, and does not account for discontinuities such as tears or folding.
Kaying et al. [7] presented an automatic calibration and stitching approach that can correct for non-linear distortions caused by the electromagnetic lenses. SIFT features [18] of local image patches were used, and the correspondences were determined based on the Euclidean distance of the features. However rather than estimating the transformation independently for each pair of images, the approach maximizes the similarity between all overlapping pairs jointly and therefore corrects only for electron microscope distortions that are shared by all images.
The large size of the images requires extreme computational efficiency consideration. Accordingly, there have been several attempts to automate and accelerate the collection of three-dimensional EM data. Serial Block-Face Scanning EM (SBFSEM) was utilized by [2] to study 3D tissue ultrastructure. However, the image resolution obtained was not sufficient to enable identification of neurobiological structures, such as synaptic contacts and gap junctions, which requires a minimum of 2 (nm/pixel) [12]. Therefore, although the SBFSEM technique avoids the distortion caused by slicing tissue, it limits the achievable resolution to lower than we need to achieve for reconstruction of three-dimensional neural ultrastructure.
C. Summary of Nonrigid Registration Methods
Bringing a pair of TEM images, I and J, into alignment requires the identification of a nonrigid transformation. Let us denote a transformation by T , and define a dissimilarity function D(I, J, T) that describes the differences between images I and J under the transform T . Image I and J are aligned, by a transformation T, when D(I, J, T) is minimized.
Cachier et al. [19] developed a classification of nonrigid registration algorithms based on two axes, the first consisting of the transformation model, and the second consisting of the image features used to guide the search for the transformation that aligns the images. The transformation model was categorized by identifying three ways in which transformations were regularized: parametric models, competitive regularization, and incremental regularization.
Parametric models referred to transformations T characterized by a small number of parameters, which were estimated by minimizing the dissimilarity between images I and J when aligned according to transform T:
| (1) |
The registration that results is intrinsically regularized by the small number of parameters used to represent the transformation T .
Competitive regularization referred to algorithms based on the following minimization problem:
| (2) |
In these approaches the minimization of the image dissimilarity function is explicitly regularized by a function that depends on the transformation T.
Incremental regularization algorithms differ from competitive regularization algorithms by considering a sequence of transformation estimates, and carrying out regularization of a function of the sequence of transformations (such as the change in transform from iteration to iteration) rather than the transformation itself:
| (3) |
Cachier et al. [19] further described registration algorithms by identifying categories of geometric feature based (GFB) registration, intensity based registration (IB), and introduced a new category of iconic feature based (IFB) algorithms that utilize both geometric distance and intensity information. Standard intensity based algorithms are based directly on the image grey values measured at each voxel and utilize an intensity based similarity function and an optimizer that finds a local optimum of the objective function ([20],[21],[22]). GFB alignment approaches are commonly based on features such as salient points, surfaces or segmented structures in the image. A fundamental algorithm in this context is the Iterative Closest Point (ICP) [23] and its popular extensions ([24] [25]). The new category of IFB algorithms seek to identify features that should be aligned, search for correspondences based on their intensity similarity measure and find an optimal geometric transformation that brings the corresponding features into alignment.
Cachier et al. [19] demonstrated that it was valuable to consider correspondences, denoted C, identified between two images, separately from the transformation T that brings the two images into alignment. Representing C and T by vector fields, [19] proposed a registration energy function using competitive regularization:
| (4) |
where σ and λ are scalar parameters, and demonstrated an iterative alternating minimization strategy in which correspondences were first identified by solving:
| (5) |
and then the transformation was estimated by minimizing:
| (6) |
This was further extended to a mixed competitive and incremental regularization function of the form:
| (7) |
where w ∈ [0, 1] determines the balance between the competitive and incremental regularization schemes. Experiments demonstrated alternated minimization was an effective and robust strategy for achieving registration of the images I and J.
D. Similarity measures
A broad range of image similarity or dissimilarity measures have been investigated in the literature. Three of the most commonly used are sum of squared differences (SSD), cross-correlation (CC) and mutual information (MI), each of which can be computed either globally or locally.
Hermosillo et al. [26] investigated dissimilarity measures of images I and J at scale σ, with the following SSD measure as an example of the type:
| (8) |
and demonstrated that minimization of dissimilarity measures of this type through calculus of variations leads to iterative minimization by gradient descent. This leads to update equations with terms of the form [26], [27]:
| (9) |
We can immediately notice that only regions of image J (the image undergoing transformation) with a significant gradient magnitude drive changes through the iterative minimization procedure. Homogeneous regions, away from regions with large gradient magnitude, do not contribute to the energy function, and do not influence the minimization. Symmetric similarity and inverse consistent registration techniques aim to ensure that the role of image I and image J is equivalent in estimating the transformation [28], but maintain the property that regions with large gradient magnitude are primarily important to the registration that is achieved. Hermosillo et al. [26] demonstrate this is also the case for dissimilarity functions of the form of local and global cross-correlation, and local and global mutual information. These observations provide further motivation for the use of regularization in order to constrain the transformation in regions away from large signal intensity gradients, and demonstrate that the force driving the minimization may be distributed sparsely across the image even when a dense transformation is being estimated.
It has been argued that local dissimilarity measures have a number of advantages over global dissimilarity measures ([16], [19], [26], [29], [30], [31]). Global dissimilarity measures, including mutual information, assume that the link between images I and J is the same over the entire image. That is, that the image statistics are stationary, whereas local similarity measures are effective even in the presence of non-stationarity. Further, it has been demonstrated that global computation of mutual information actually reduces the quality of image alignment achieved in comparison to computation of image statistics only in the regions closest to edges [30], due to a combination of non-stationarity of image statistics and a reduction in the quality of mutual information estimates from including voxels away from regions of significant gradient magnitude that do not drive the minimization.
While SSD is appropriate for single-modality images, and mutual information can be challenging to compute from local regions due to the smaller number of samples available to estimate the required probability density functions [32], local cross-correlation is effective for both single-modality and multi-modality image registration. As demonstrated first by [31] and confirmed by [29],[16], local cross-correlation is effective for multi-modality registration because it exploits the affine relationship between signal intensities of the local regions of the images, a relationship which is most apparent in regions of significant gradient magnitude.
E. Search for Correspondences
Both GFB and IFB registration methods proceed to search for correspondences between features. For IFB algorithms, the search problem is usually solved by assuming a best corresponding feature will be within a certain search range, and then brute force evaluation of all potential correspondences within the search region. Recent examples such as [33] have shown the efficacy of local cross correlation for such truncated searches, but can fail to identify the best correspondences due to the truncated search region, become slower as the local region size must scale with the biological features of interest, and are driven by a single best correspondence pair at each voxel. For GFB algorithms, such as ICP [23], a brute-force approach which involves computing the distances between the query point and all the points in the neighboring image can be used.
F. Our Contribution
We propose a new registration algorithm in the category of iconic feature based registration algorithms [19]. As with previously described such algorithms, our algorithm identifies correspondences between pairs of images at scale σ, and computes a nonrigid transformation from the correspondences. The algorithm is suitable for any model of alignment transformation, and transformations may be regularized with competitive and/or incremental regularization.
The first algorithmic contribution is a novel and efficient search strategy that dramatically accelerates feature based registration. We recast the conventional search for corresponding iconic features as a near neighbor search. As the dimensionality of the search space is very high, this has not previously been regarded as tractable. We demonstrate the search can be made extremely efficient while preserving accuracy by projecting to a low-dimensional search space using randomized projections, and then using efficient low-dimensional search. The key result enabling this is the seminal result of Johnson and Lindenstrauss (JL) [34] demonstrating the remarkable distance preserving property of this type of projection, which shows that points close together in high dimensional space are almost certainly very close together when projected into low dimensional space. This enables us to achieve a dramatic acceleration of the identification of correspondences. Our second contribution is a novel algorithm for efficient and robust estimation of the alignment transformation from a set of exact or probabilistic correspondences.
An early version of this work has appeared in [35], in which efficacy of randomized projections for correspondence detection was examined. This paper extends that previous work to iconic feature based registration with arbitrary transformation models, utilizes a multiscale pyramid, provides a detailed analysis of the impact of dimensionality reduction on the transformation estimation, and has an extensive experimental evaluation with larger TEM images.
The remainder of the paper is organized as follows. Section 2 introduces the main steps of our registration algorithm. Section 3 provides a set experiments and results evaluating the registration algorithm on TEM images and the conclusions are presented in Section 4.
II. MATERIAL AND METHODS
A. Theory
In this section we outline the main components of the proposed registration algorithm. Volumetric images of neural ultrastructure are constructed by pairwise alignment of consecutive images of thin slices of tissue. The key registration challenge to solve is the alignment of consecutive 2D images. We describe the alignment algorithm in this context, but the algorithm is independent of the spatial dimension of the data, and can be used for 3D and 4D registration as well.
Given a fixed image and a moving image, our aim is to find the transformation T aligning the moving image with the fixed image.
The algorithm can be decomposed into three main steps. In the first step, a scale containing key biological features is selected and each image is divided into multiple image patches. The patches, of size d pixels, may be circular, square or rectangular subregions (or of arbitrary shape), may overlap each other, and may be taken from the original image or an intensity transformation of the image (such as the image after Gaussian convolution, or the image after convolution with a derivative of a Gaussian). Each patch is normalized to have zero mean and unit magnitude. A low dimensional representation of each patch is then computed, by projection of the patch through a random matrix that meets the requirements of the Johnson-Lindenstrauss Lemma [34] for distance preservation.
In the second step the projected patches are compared. We measure the dissimilarity between projected patches of the moving and fixed images, using Euclidean distance. This is equivalent to local normalized cross correlation [36].
Finally, we construct two sets of geometric points based on the center coordinates of the projected patches. A regularized nonrigid transformation is then estimated to bring the images into alignment, by estimating the correspondences of the patches and the transformation that is most consistent with the estimated correspondences. Table 1 presents an outline of the algorithm.
TABLE I.
Evaluating the difference between the transformation obtained by our automatic algorithm to the manual transformation on the set of TEM images. The high accuracy achieved is obtained both with and without the JL projection.
| Dimension | Scale of (1000 × 1000 voxels) | Scale of (5000 × 5000 voxels) |
|---|---|---|
| k = 30 (with JL projection) | 3.62 ± 2.32 | 3.18 ± 1.80 |
| d = 10000 (without JL projection) | 3.38 ± 1.85 | 3.03 ± 1.30 |
B. Similarity measure
We consider two images Iσ = I* Gσ and Jσ = J* Gσ , resulting from the convolution of each image with a Gaussian kernel of standard deviation σ. We select the dissimilarity measure of Euclidean distance of the zero mean unit magnitude image patches:
| (10) |
where represents a patch of image Iσ centered at i, from which the image intensity mean has been subtracted, and normalized to unit length, and similarly for J. D(Iσ, Jσ, Cn,i,j) represents the dissimilarity between the images Iσ, Jσ, at patch locations i and j where the sum is over a local neighborhood x defined by the patch geometry.
C. Distance Preserving Random Projections to a Low Dimensional Space
The Johnson-Lindenstrauss Lemma [34] asserts that any set of n feature points in d-dimensional Euclidean space can be projected down to k-dimensional Euclidean space, where k = O(∈−2 log n), while maintaining pairwise distances with a bounded distortion (of at most ∈). Recent work has identified very efficient projection operators, such as the projection proposed by [37].
A number of different random projections meet the requirements of the Johnson-Lindenstrauss Lemma for distance preservation. One such projection can be constructed as follows. Let R be a k × d random matrix with R(i, j) = rij, where the independent random variables rij are from either one of the following two probability distributions:
| (11) |
| (12) |
Consider the set of n patches of d pixels from image Iσ, in Rd, represented as an n × d matrix, where each patch is represented by a row. The random projection can be performed by constructing a k × d random matrix; by employing this approach, projection of each point requires a matrix-vector multiply taking O(dk) operations. Recent theoretic work suggests that even more efficient projections are possible, with [38] proposing an algorithm to project from dimension d to dimension k with O(d) operations.
Each of these projections can be precomputed for each image. Computing and storing these projections is time and memory efficient. The key point of carrying out this projection is that evaluation of the image dissimilarity D(Iσ, Jσ, Cn,i,j) provides distances between the original patches from Rd and the projected patches from Rk, that are almost unchanged by the projection, but the evaluation is dramatically more efficient for k dimensional patches. The image dissimilarity calculation is accelerated by a factor of d/k.
In contrast with JL projection, Principal Component Analysis (PCA) is aimed at finding an orthogonal basis of vectors that accounts best for the variability in the data. Compression based projections, such as PCA, do not put a bound on the distance between two patches after projection [39]. In addition, in a data driven compression scheme such as PCA, it is necessary also to communicate either the basis functions, or to agree ahead of time on what the best basis functions are. The need to have the same basis function reduces the overall efficiency of PCA and generating an adaptive basis function which relies on the data information adds a significant computational cost in cases of large set of points and dimensions.
D. Search for Correspondences
We consider first defining Cn,i,j ∈ [0, 1] as variables that indicate whether or not patch i and j correspond. The most straightforward identification of correspondences Cn,i,j can be obtained by directly minimizing the image dissimilarity function:
| (13) |
subject to the constraint , which leads to the correspondence solution of Cn,i,j = 1 when patch i and patch j have the smallest dissimilarity, Cn,i,j = 0 otherwise. The usual implementation of this type of search requires the evaluation of the dissimilarity between each pair of patches, and we refer to this as brute-force search. We also consider an acceleration of this search using a kd-tree data structure [40], [41], which can reduce the number of comparisons that must be made to obtain the same result.
E. Transformation Estimation
While the transformation can be directly estimated from the correspondences computed above, we have found it more effective to relax the requirement that Cn,i,j ∈ [0, 1] and to consider probabilistic correspondences of the form Cn,i,j ∈ R while maintaining the constraint that . We describe in this section how this is done.
Let si indicate the location of the patch i in the fixed image Iσ ∈ R2 and mj the location of patch in the moving image Jσ ∈ R2, with ns and nm denoting the number of patches of each. Let T denote the transformation from the fixed image to the moving image.
We consider the geometric alignment of patch pairs (si, mj) under the transformation T, and define a probability based on the closeness of the patches. In the case of homogeneous isotropic Gaussian measurement error the probability is given by:
| (14) |
where σn represents the uncertainty in the localization. With this model, considering each pair of patches, we can compute the probability of observing a particular alignment of patches under a given transform T.
Further, if we consider the pairwise correspondences of patches as a random variable Cn,i,j and T as a parameter, then we can formulate the problem of estimating these using Expectation-Maximization.
| (15) |
The optimization starts by initialization of the transformation T, and repeats until convergence of the two EM steps. In the E-step, T is fixed and the probability of matches is computed as follows:
| (16) |
The prior probability of the matches is determined by the intensity dissimilarity of the patches. Hence, the prior is based on the normalized dissimilarity:
| (17) |
The prior is used to initialize the probabilistic correspondences C0, and thus to determine an initial transformation T0.
In the M-step, Cn is fixed and the value of the parameter T is maximized:
| (18) |
Accordingly, the criterion optimized by the algorithm yields
| (19) |
subject to a constraint imposed by the form of the regularization of the transformation represented by log P (T).
F. Model of Transformation
The above procedure for estimation of a transformation is suitable for rigid, affine, or nonrigid transformations. Some nonrigid transformations, such as thin plate spline representations, can be computed directly in closed form due to the analytic form of their derivatives [25], [42]. Arbitrary nonrigid transformations can also be modelled by a spatial composition of local rigid or affine transformations [43].
In this section, we demonstrate how to achieve a local symmetric affine transformation that brings a sub-region of the images into alignment, based on the approach presented in [44].
First a whitened version of the moving and fixed scene patch coordinates is formed, by subtracting the mean ( respectively) and multiplying the result by the covariance matrix raised to the −1/2. The whitening operators Ws, Wm applied to the fixed and moving patch coordinates respectively, lead to a new set of points si′ = Ws* si, mj′ = Wm* mj with zero mean and an identity covariance matrix. Then a rigid transformation is applied to the new whitened set si′, mj′ together with unrolling the whitening transformation. This sequence of operations is equivalent to computing an affine transformation on the original patch sets si, mj and provides an efficient means of estimating the desired affine transformation.
| (20) |
Further, it is valuable to note that the M-step transformation optimization with probabilistic correspondences can be obtained by extension of the unit quaternion method [45] and by simplifying Equation 19. By defining as the barycenter of the mj weighted by , the effective term maximized is equal to
| (21) |
Since the second term in Equation 21 is constant during the M-Step, the maximization to obtain T̂ requires consideration only of the term .
A Block-Affine Nonrigid Transform
A nonrigid transformation that accounts for local distortions, and discontinuities such as tears and folding, can be computed by spatial decomposition of the images into multiple regions, and computing an optimal affine transformation for each region. Such a block-affine transformation is based on the classical approach of modelling a number N of affine transformations Ti, corresponding to local regions with center position ci. A family of nonrigid transformations can be formed from the set of local affine or rigid transformations [43], by associated a non-negative weight function wi(x) with each image location x. Different weight functions can be defined, but a common choice is a Gaussian wi(x) = G(μl, σl)(x) which determines the region of influence based on the mean and standard deviation μl,σl.
G. Multiscale Registration Scheme
Registration is achieved with a standard multiscale scheme to further improve the robustness and reduce the computational time. A Gaussian pyramid is generated from the input images, and registration is performed over the scale space from coarse to fine. Three pyramid levels were used. The multiscale framework is designed to utilize the transformation computed in the coarser scale to initialize the transformation estimation process at the next finer scale, where the coarsest scale relies on the priors for initialization.
Brute-force Rotation Initialization
SSTEM images are imaged at an arbitrary orientation, due to the process of section acquisition. Consecutive images can exhibit a large rotational difference. At the coarsest scale of the multi-resolution pyramid, the patches are rotated by multiple angles (10 different angles, equally distributed between 0 to 360 were tested) and the patch correspondence is evaluated with these different starting conditions. The angles which provide best pairwise matches in the coarse scale, determine the orientation initialization. Additionally, to account for artifacts such as tears and folds which commonly occur in TEM images, the coarse sections were divided to four subparts, and the angles corresponding to the minimum dissimilarity after excluding the subparts with artifacts manually, determined the initial orientation for each section.
H. Data Acquisition
The algorithm was evaluated on TEM images of the lateral geniculate nucleus of a ferret. A young adult ferret was transcardially perfused using a 2% paraformaldehyde, 2.5% glutaraldehyde fixative mixture. Brain sections 300 micrometers thick were cut using a vibratome and immersed in 2.5% glutaraldehyde overnight at 4°. They were then washed with Sorenson's buffer and incubated for at 4° in a 1% osmium tetroxide/ 1.5% potassium ferrocyanide solution. The sections were then washed in maleate buffer and incubated in 1% uranyl acetate solution. Then they were washed in maleate buffer again and dyhydrated by immersion in a series of increasingly concentrated ethanol solutions. The sections were then incubated in a series of 100% propylene oxide, 50%/50% propylene oxide/ Eponate 812 resin mixture, and 100% Eponate. Finally resin-embedded sections were cured for 2 days in a 60° oven. The polymerized blocks were sectioned using an EMS-Diasum diamond knife and a Leica ultramicrotome. Sections were picked up on pioloform-coated slot grids and imaged at 80 kV in an FEI T12 BioTwin electron microscope. The algorithm was tested on a set of 160 sections, where each image is about 10000 × 10000 pixels in size with a pixel resolution of 3nm and a slice thickness of 60nm. ”Blendmont”, a utility that is part of the IMOD package (http://bio3d.colorado.edu/imod/) was used to reconstruct the large field of view image from the 5 × 5 mosaics of smaller images coming from the camera.
Reference standard alignments of these images were obtained by manual alignment. Manual alignment is a time consuming and difficult process, which generates a reference standard 3D volume in which some registration artifacts may remain. The manual registration denoted as (T*), was performed by manually selecting corresponding points in a pair of consecutive images and computing an optimal pairwise transformation based on Horn's method [45].
III. EXPERIMENTS AND RESULTS
A. Effectiveness of the Dissimilarity Measure
To evaluate the effectiveness of our proposed patch definition and dissimilarity measure for identifying correspondences for various types of neurobiological objects, patch dissimilarity was measured for different neural ultrastructure including myelinated white matter, dendrites, synapses, and microtubules. The dissimilarity function was computed between the features visible in the image, with patches within the same image and with patches in a successive image. Figure 2 provides an illustration of the result, showing the inverse of the dissimilarity function, demonstrating that the feature is a local maxima of the function on both the original slices from where the patch was extracted and in the consecutive slice. This demonstrates that the dissimilarity measure is effective in identifying correct matches throughout the slices.
Fig. 2.
Effectiveness of similarity measure for identifying correspondences. The fusion of the similarity map computed for the patch (a) with the successive slice (b) and with the original slice, from which the patch was extracted (c). The maxima regions in both slices (b,c) respectively, are highlighted in red rectangles and enlarged. The red maxima values in the fusion image correspond to higher similarity values, showing that the features are a sharp local maxima of the function.
B. Impact of the Dimensionality Reduction on the Transformation Accuracy
Figure 3 (a) shows how the average error in Euclidean distance between patch pairs increases as the projection dimension k decreases. This increase in distance distortion leads to an increase in the number of poor correspondence estimates as k decreases. Despite the increasing number of poor matches, the transformation estimation procedure maintains robustness and is able to accurately estimate the transformation despite the increasing number of poor correspondence estimates. Figure 3(b) illustrates that the algorithm can identify the accurate transformation even for small dimensionality projections (e.g. k = 30).
Fig. 3.
Impact of dimensionality k on registration accuracy. (a) Illustrates the decrease in Euclidean distance error as k increases. (b) Shows the decrease in the transformation estimation error as k increases until reaching a value for which it is small and stable. It can be seen that for k 30 the transformation is identified accurately.
C. Transform Estimation
We further examined the ability of our proposed registration algorithm to accurately estimate an alignment transformation T. We sought to compare the accuracy achieved using randomized projection with that achieved using the full dimensionality features.
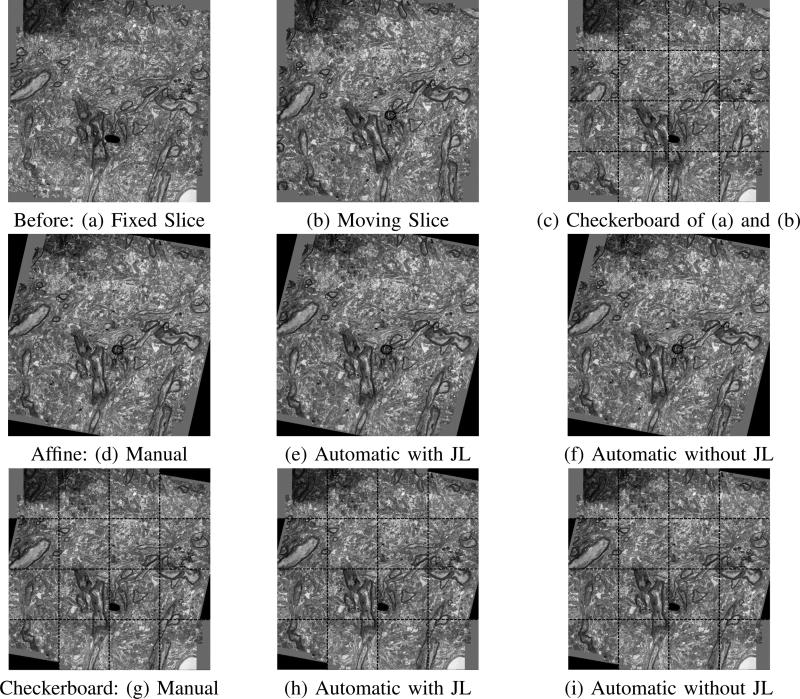
The search for correspondences in both cases was done using both a brute force approach and a kd-tree data structure, and the computation time needed to achieve registration was recorded. The kd-tree data structure retrieved the three closest neighbors with the smallest Euclidean distance for each query patch and the transformation estimation was based on the 1000 patch pairs that obtained the minimal Euclidean distance. Figure 4, illustrates the volume constructed with and without the randomized projection, compared to the manual registration. The three manual reconstruction presented were obtained based on Horn's method [45] with a different manually selected set of points. The Frobenius norm of the difference between the transformations obtained by the registration algorithm in both and the manual reference standard alignment (T*) was computed. Table I presents these norms and shows that both the full patch information and projected patches provide accurate transformation estimates. We conclude that the projection to low dimensional space does not reduce the ability to recognize sufficient corresponding patches. The Frobenius norm is shown averaged across 160 slices of the data, except for the brute-force approach on the full dimensional data, where we tested the approach on only 16 slices, due to the large computation time required.
Fig. 4.
An orthogonal view of the reconstructed volume performed (a-c) Manually with three different sets of points (d) Automatically with d = 10000 (e) Automatically with k = 30 .
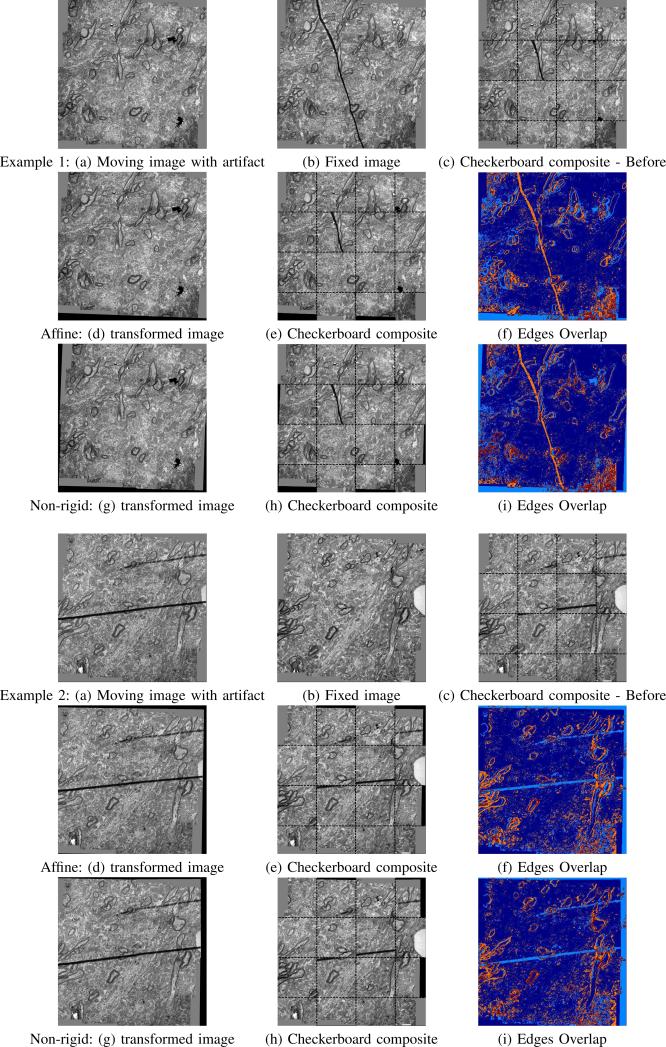
The non-rigid approach was tested with N = 4 blocks on images of size ≈ 1000 × 1000 voxels. The Gaussian standard deviation parameter was set to σl = 40 equally for all regions. The results show that the due to non-rigid distortions in the images, the sections cannot be matched with an affine transformation alone. The checkerboard composite and the overlap of the edges demonstrate that the proposed non-rigid transformation with a Gaussian regularization obtains finer local alignment especially in cases of distortions.
D. Speed and Computational Complexity
The computational complexity of the entire process is determined by the number of images in the data set, the image size n and the patch size d. Our preliminary results were performed on slice sections of n ≈ 108 = 104 × 104, which were downsampled using Gaussian smoothing plus bi-linear interpolation to obtain three scales with sizes of n ≈ 5000 × 5000, n ≈ 106 = 1000 × 1000, n 104 = 100 × 100. Improved TEM techniques utilizing multiple camera arrays have lead to datasets of n = 1010 = 105 × 105. The size of a typical local image patch is d = 104 = (100 × 100), and employing JL lemma and randomized projection turns the initial n × d image patch matrix into an n × k projected patch matrix (where k = 30 in our experiments).
The running time is composed of the preprocessing time required to project each image patch, the time to search for correspondences between the images and the time to perform transformation estimation. As noted in Sec. II-C, the projection scheme currently requires O(dk), but it has been suggested effective projection can be carried out in O(d) operations [38].
The brute force search involves comparing each projected patch to all the projected patches in the neighboring image. The complexity of this search time per query is O(dn). Thus, the overall complexity for using the full d-dimensional patches is O(n2d). Hence, our randomized projection alone reduces the complexity to O(n2k) ≈ O(n2 log n) saving O(d − k) operations (104 operations per search in our experiments).
Additional significant acceleration may be obtained by employing recent advances in approximation algorithms for performing approximate near neighbor (ANN) search. Relying on the algorithm presented by [46], the speedup between the naive and accelerated searches is O(n1−1/c2) and for c = 2 this becomes O(n3/4) which for recent TEM data is O(107.5). We also note that the query time quoted here assumes the need to project from high dimension to low dimension before doing the search. However, since our patches participate as both search and target regions, we can benefit from pre-computing the projection of all the patches ahead of time, which further reduces the search cost by O(d). Recall d = 104 in our case, so this is a substantial improvement.
Projection of 160000 patches from one section required 842 sec ≈ 14 min on average. As described above, the time to project each patch to low dimension is a small fraction of the overall running time which is determined by the search for correspondences. Therefore, Table II focuses on the most expensive component of the algorithm which is the search for correspondences and demonstrates the efficiency of the algorithm.
TABLE II.
Comparison of the computation times of the different approaches.
| Search per n queries | Brute force | Brute force + JL | JL + kd-trees | ANN-LSH |
|---|---|---|---|---|
| Theoretical | O(n2d) | O(n2k) | O(ndn1/4) | |
| Practical n = 42336 | 283400sec = 3.3days | 3614sec = 60min | 420sec = 7min | – |
| Practical n = 160000 | 4.4 × 106 sec ≈ 51 days1 | 53506sec = 892min | 6300sec = 105min | – |
Table II presents results for testing with patches of dimension d = 10000 for n = 42336 and n = 160000 per section. The brute force computation time required 283400 sec = 3.3 days and 4400000 sec≈ 51 days respectively 2. In theory by utilizing the full ANN approach we expect to obtain savings of O(n3/4), leading to a factor of 8000 for n = 160000. Currently, in our experiments we have found that the JL projection alone provides a saving factor of ≈80, 110 for n = 42336, n = 160000 respectively and that the kd-tree search data structure provides an additional saving factor of 8.5 leading to ≈ 800−fold speed up for these n values. The significant speed-up achieved (e.g. on n = 42336 the algorithm requires several minutes instead of more than three days), is currently based on the randomized projection from d to k and search using the kd-tree data structure.
IV. DISCUSSION AND CONCLUSION
We described a novel algorithm for iconic feature based alignment and demonstrated its successful application to large TEM images. The registration algorithm achieves a significantly accelerated search by forming a low dimensional representation of the features by randomized projections. We demonstrated that the accelerated search still achieves high robustness and accuracy of the alignment. In this work we have demonstrated these contributions on TEM data. We analyzed the impact of dimensionality reduction on the transformation accuracy and demonstrated for the first time the feasibility and effectiveness of this approach, showing that the projected features are as effective for registration as the full-dimensionality features. Future work will evaluate alternative data structures for search strategies for the acceleration of accurate correspondence estimation. This will include further development and evaluation of data structures that support ANN search strategies, such as locality sensitive hashing (LSH) and tree search and assessment of faster variants of projection algorithms to improve performance efficiency of the projection into k-dimensional space.
Fig. 1.
Schematic outline of the alignment algorithm.
Fig. 5.
Successive pair of slices before and after alignment. (a) and (b) present the fixed and moving image before alignment and (c) shows their checkerboard composite. (d),(e),(f) show the alignments results of the manual, and automatic algorithms with JL projection and without projection respectively. (g),(h),(i) demonstrate the checkerboard of the fixed image in (a) and the aligned moving image in (d), (e) and (f) respectively.
Fig. 6.
Two examples comparing the transformation results of the affine and non-rigid approach. The checkerboard composite and the overlap of the edges demonstrate that the proposed approach using a Gaussian regularization of a block-rigid transformation to estimate the non-linear deformation field improves the accuracy of the alignment especially in cases of distortions.
V. ACKNOWLEDGMENT
This investigation was supported in part by NIH grants R01 RR021885, R01 EB008015, R03 EB008680, R01 LM010033 and P30 HD018655.
Footnotes
Personal use of this material is permitted. However, permission to use this material for any other purposes must be obtained from the IEEE by sending a request to pubs-permissions@ieee.org.
The brute force computation time for n = 160000 was estimated by utilizing the kd-tree search on the high dimensional data multiplied by the saving factor calculated for the kd-trees compared to the brute-force approach on the projected data.
References
- 1.Maintz J, Viergever M. A survey of medical image registration. Medical Image Analysis. 1998;2(1):1–16. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- 2.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biology. 2004;2:329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggman K, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Current Opinion in Neurobiology. 2006;16(5):562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Blow N. Following the wires. Nature Methods. 2007;4(11):975–980. [Google Scholar]
- 5.Micheva K, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55(1):25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koshevoy P, Tasdizen T, Whitaker R, Jones B, Marc R. Assembly of large three-dimensional volumes from serial-section transmission electron microscopy. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2006 [Google Scholar]
- 7.Kaynig V, Fischer B, Mller E, Buhmann J. Fully automatic stitching and distortion correction of transmission electron microscope images. Journal of Structural Biology. 2010;171(2):163–73. doi: 10.1016/j.jsb.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Jain V, Murray J, Roth F, Turaga S, Zhigulin V, Briggman K, Helmstaedter M, Denk W, Seung H. Supervised learning of image restoration with convolutional networks. ICCV. 2007 [Google Scholar]
- 9.Smith SJ. Circuit reconstruction tools today. Current Opinion in Neurobiology. 2007;17(5) doi: 10.1016/j.conb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 11.Mishchenko Y. Automation of 3D reconstruction of neural tissue from large volume of conventional serial section transmission electron micrographs. Journal of Neuroscience Methods. 2009;176(2):276–289. doi: 10.1016/j.jneumeth.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson J, Jones B, Yang J-H, Shaw M, Watt C, Koshevoy P, Spaltenstein J, Jurrus E, Kannan U, Whitaker R, Mastronarde D, Tasdizen T, Marc R. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biology. 2009;7(3):493–512. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonic S, Thvenaz P, Sorzano C, Unser M. Biophotonics for life sciences and medicine. Fontis Media; 2005. ch. Spline-based 3D-to-2D image registration for image-guided surgery and 3D electron microscopy.
- 14.Carazo J, Herman G, Sorzano C, Marabini R. Electron Tomography: methods for three-dimensional visualization of structures in the cell. 2nd ed. Springer; 2006. ch. Algorithms for three-dimensional reconstruction from imperfect projection data provided by electron microscopy.
- 15.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. The Journal of Neuroscience. 2008;28(12):2959. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ourselin S, Roche A, Subsol G, Pennec X, Ayache N. Reconstructing a 3D structure from serial histological sections. Image and Vision Computing. 2001;19:25–31. [Google Scholar]
- 17.Dauguet J, Bock D, Reid CR, Warfield SK. Alignment of large image series using cubic b-splines tessellation: Application to transmission electron microscopy data. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2006 doi: 10.1007/978-3-540-75759-7_86. [DOI] [PubMed] [Google Scholar]
- 18.Lowe D. Distinctive image features from scale-invariant key-points. International Journal of Computer vision. 2004;60:91–110. [Google Scholar]
- 19.Cachier P, Bardinet E, Dormont D, Pennec X, Ayache N. Iconic feature based nonrigid registration: the pasha algorithm. Computer Vision and Image Understanding. 2003;89:272–298. [Google Scholar]
- 20.Collignon A, Maes F, Delaere D, Vandermeulen D, Suethens P, Marchal G. Information Processing in Medical Imaging. Kluwer Academic Publishers Dordrecht; 1995. pp. 263–274. ch. Automated multi-modality image registration based on information theory.
- 21.Viola P, Wells WM., III Alignment by maximization of mutual information. Proceedings of the Fifth International Conference on Computer Vision. 1995:16–23. [Google Scholar]
- 22.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE transactions on Medical Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 23.Besl P, McKay N. A method for registration of 3-d shapes. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1992;14(2):239–256. [Google Scholar]
- 24.Granger S, Pennec X. Multi-scale EM-ICP: A fast and robust approach for surface registration. ECCV. 2002;4:418–432. [Google Scholar]
- 25.Chui H, Rangarajan A. A new point matching for non-rigid registration. Computer Vision and Image Understanding. 2003;89:114–141. [Google Scholar]
- 26.Hermosillo G, d'Hotel CC, Faugeras O. Variational methods for multimodal image matching. International Journal of Computer Vision. 2002;50(3):329–343. [Google Scholar]
- 27.Pennec X, Cachier P, Ayache N. Medical Image Computing and Computer Assisted Intervention (MICCAI) LNCS; 1999. Understanding the demon's algorithm: 3d non-rigid registration by gradient descent; pp. 597–606. [Google Scholar]
- 28.Johnson H, Christensen G. Consistent landmark and intensity-based image registration. IEEE Transactions on Medical Imaging. 2002;21(5):450–461. doi: 10.1109/TMI.2002.1009381. [DOI] [PubMed] [Google Scholar]
- 29.Ourselin S. PhD thesis. Universite de Nice Sophia-Antipolis; 2002. Recalage d'images medicales par appariement de regions, application a la construction d'atlas histologiques. [Google Scholar]
- 30.Freiman M, Werman M, Joskowicz L. A curvelet-based patient-specific prior for accurate multi-modal brain image rigid registration. Medical Image Analysis. 2010 doi: 10.1016/j.media.2010.08.004. in press. [DOI] [PubMed] [Google Scholar]
- 31.Weese J, Rosch P, Blaffert T, Quist M. Medical Image Computing and Computer Assisted Intervention(MICCAI) LNCS; 1999. Gray-value based registration of ct and mr images by maximization of local correlation. pp. 656–664. [Google Scholar]
- 32.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Transactions on Medical Imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 33.Avants B, Tustison N, Song G, Cook P, Klein A, Gee J. A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage. 2010;54(3):2033–44. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson W, Lindenstrauss J. Extensions of lipschitz mapping into a hilbert space. Contemp. Math. 1984;26:189–206. [Google Scholar]
- 35.Akselrod-Ballin A, Bock D, Reid RC, Warfield SK. Accelerating feature based registration using the johnson-lindenstrauss lemma. Medical Image Computing and Computer Assisted Intervention (MICCAI) 2009 doi: 10.1007/978-3-642-04268-3_78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballard D, Brown C. Computer Vision. Prentice-Hall; Englewood Cliffs, NJ: 1982. [Google Scholar]
- 37.Achlioptas D. Database-friendly random projections: Johnson-lindenstrauss with binary coins. Journal of Computer and System Sciences. 2003;66:671–687. [Google Scholar]
- 38.Liberty E, Ailon N, Singer A. Dense fast random projections and lean walsh transforms. Lectures in Computer Science. 2008:512–522. [Google Scholar]
- 39.Ailon N, Chazelle B. Faster dimension reduction. communications of the ACM. 2010;53:97–104. [Google Scholar]
- 40.Friedman JH, Bentley JL, Finkel RA. An algorithm for finding best matches in logarithmic expected time. ACM Transactions on Mathematical Software. 1977;3(3):209–226. [Google Scholar]
- 41.Arya S, Mount D, Netanyahu N, Silverman R, Wu A. An optimal algorithm for approximate nearest neighbor searching in fixed dimensions. Journal of the ACM. 1998;45(6):891–923. source code available from www.cs.umd.edu/mount/ANN/ [Google Scholar]
- 42.Hufnagel H, Pennec NAX, Ehrhardt J, Handels H. Generation of a statistical shape model with probabilistic point correspondences and EM-ICP. International Journal for Computer Assisted Radiology and Surgery (IJCARS) 2008;2(5):265–273. [Google Scholar]
- 43.Arsigny V, Commowick O, Pennec X, Ayache N. A fast and log-euclidean polyaffine framework for locally affine registration. Journal of Mathematical Imaging and Vision. 2009;33(2):222–238. [Google Scholar]
- 44.Ho J, Yang M, Rangarajan A, Vemuri B. A new affine registration algorithm for matching 2D point sets. IEEE Workshop on Applications of Computer Vision (WACV ’07) 2007 [Google Scholar]
- 45.Horn BKP. Closed-form solution of absolute orientation using unit quaternions. Journal of the Optical Society of America. 1987;4:629–642. [Google Scholar]
- 46.Andoni A, Indyk P. Near-optimal hashing algorithms for approximate nearest neighbor in high dimensions. Communications of the ACM. 2008:117–122. [Google Scholar]