Abstract
Strong statistical associations between polymorphisms in HIV-1 population sequences and carriage of HLA class I alleles have been widely used to identify possible sites of CD8 T cell immune selection in vivo. However, there have been few attempts to prospectively and systematically test these genetic “hypotheses” arising from population-based studies at a cellular, functional level.
We assayed CD8 T cell epitope-specific IFNγ responses in 290 individuals from the same cohort which gave rise to 874 HLA-HIV associations in genetic analyses, taking into account autologous viral sequences and individual HLA genotypes. We found immunological evidence for 58% of 374 associations tested as sites of primary immune selection and identified up to 50 novel HIV-1 epitopes using this “reverse genomics” approach. Many HLA adapted epitopes elicited equivalent or higher magnitude IFNγ responses than the non-adapted epitopes, particularly in Nef. At a population level, inclusion of all the immunoreactive variant CD8 T cell epitopes in Gag, Pol, Nef and Env suggested that HIV adaptation leads to an inflation of Nef-directed immune responses relative to other proteins.
We conclude that HLA-HIV associations do mark viral epitopes subject to CD8 T cell selection. These results can be used to guide functional studies of specific epitopes and escape mutations as well as test, train and evaluate analytical models of viral escape and fitness. The inflation of Nef and HLA adapted variant responses may have negative effects on natural and vaccine immunity against HIV, and therefore has implications for diversity coverage approaches in HIV vaccine design.
Introduction
The dual challenges of HIV-1 diversity and evasion of human immunity have concentrated efforts in the vaccine field to optimise diversity coverage in vaccines on the one hand (1, 2), and distinguish protective from non-protective immune responses on the other (3). With respect to CD8 T cell immunity, diversity and immunogenicity considerations may well intersect if specific, predictable genetic variations in HIV-1 have important functional consequences for prevalent epitope-specific responses. HIV-1 mutational escape from cellular immune responses generated in acute and chronic infection contributes to HIV-1 diversity at the population level. In particular, HLA restricted CD8 CTL responses are sufficiently suppressive to exert selection pressure on HIV quasispecies, however in most individuals, ongoing viral replication allows the eventual outgrowth of CTL adapted viruses (4, 5). Such variations therefore have functional implications for immunogenicity, and if present in a vaccine immunogen, would effectively be “pre-adapted” to certain HLA types. Furthermore, presence of escape mutations in a vaccine immunogen may influence the immunodominance of vaccine-induced CTL responses, as suggested by significant changes in immunodominance hierarchies that follow early viral evolution and diversification in natural infection (6). Understanding the immunological consequences of specific HIV variations may become increasingly important as more are incorporated into polyvalent vaccines designed to optimise population diversity coverage (1, 7, 8).
Though information on specific variations can be derived from a number of observational CTL escape studies (5, 9-11), the breadth of HLA backgrounds and viral mutations examined in these studies are narrow, relative to the great breadth of HLA genotypes and HIV-1 diversity present in human populations. Since the first population based HLA-HIV association study in 2002 (12), several large scale studies have identified natural HIV-1 polymorphisms and networks of polymorphisms which appear to be significantly HLA allele-specific across the full HIV-1 subtype B and C proteomes, after accounting for viral phylogeny and linkage disequilibria in the MHC (13-19). These associations are not a functional demonstration of immune escape but rather, may be considered individual hypotheses, based on a statistical association, of an in vivo biologic interaction between a HLA class I molecule and the viral epitope spanning the polymorphism or distant epitopes linked functionally to the polymorphic site. Though recent approaches have also sought to identify whole mutational networks involving multiple viral codons into the analyses (16), it is not possible to prove the order of consecutive changes by these analyses alone. That is, it is possible that residues co-vary because of compensatory fitness balancing interactions between viral residues or because of co-dominant targeting by the same HLA restricted CTLs. These studies have used published existing CTL epitope and escape data and known compensatory patterns to validate associations, however the repertoire of confirmed, published epitopes are not complete, particularly for less common HLA alleles, alleles associated with non-white Caucasian populations and HLA-C in general. HLA-C-restricted responses may be particularly important in view of recent evidence linking levels of HLA-C cellular expression to better immunological control (20). There is even less viral escape data to validate the functional effect of all polymorphisms observed in vivo. We therefore sought to use population derived HLA-HIV associations as starting hypotheses and systematically characterise the epitope-specific CD8 T cell responses that may account for them in vivo, as well as determine the functional effects of HLA associated variations on T cell reactivity in individuals and in a population. Those HLA-HIV associations for which no evidence for direct influence on viral epitope T cell interactions could be found after systematic testing would also increase the likelihood of them being driven by compensatory interactions or networks within the HIV proteome. We used a previously published dataset of genome wide HLA-HIV-1 associations derived from a large diverse population from the United States of America (USA) to predict the epitopic targets of prevalent CTL responses (19), and assayed these responses ex vivo in the same population. For each individual, we tested known and predicted “non-adapted” or immune susceptible HIV-1 epitopes along with the paired adapted epitope sequence relevant to their own HLA-A, -B and -C alleles and autologous viral epitope sequences. We primarily aimed to determine the proportion of HLA-HIV genetic associations that could be additionally explained or supported by T cell epitope data gained as a result of this systematic testing, compared with just using published epitope information. Having carried out large-scale population-based cellular testing, we aimed to generally characterise the distribution of these prevalent T cell responses across the HIV proteome, their response rates and magnitude. We also aimed to analyse how immune reactivity is influenced by the strength of the epitope predication value, the autologous virus sequence and clinical indices. Finally we sought to determine the changes to reactivity caused by HLA-driven polymorphism on individual epitopes and overall patterns of immune reactivity at the population level that could impact vaccine design considerations.
Materials and Methods
Study cohort and samples
The cohort of individuals examined in this study (n = 414) were a subset of the 555 individuals with chronic HIV-1 infection who were co-enrolled in the Adult AIDS clinical trials group (AACTG) studies A5142 and A5128 from the USA. The AACTG A5142 was a randomised clinical trial comparing three first-line antiretroviral drug regimens in individuals with no previous antiretroviral therapy and a viral load of greater than or equal to 2000 copies/mL plasma (21). There was no inclusion/exclusion criteria based on CD4 T cell counts. Subjects were recruited from 55 centres across the USA between 2003 and 2004, and were co-enrolled in A5128 if they provided consent for inclusion in the ACTG human DNA bank (22). Baseline pre-treatment viral load measurements were available. All participants provided written informed consent to these investigations and the study was approved by the Institutional Review Board governing the AACTG prior to commencement.
The subset of 414 individuals had HIV-1 sequencing, HLA class I genotyping resolved to four-digit types in all but three cases, and participated in a previous population analysis involving 800 individuals which generated a dataset of 874 HLA allele associated HIV-1 genome-wide subtype B polymorphisms (19). These study participants were selected based on availability of cryopreserved PBMCs for immunological studies. PBMCs obtained from baseline visit time points in the trial and before commencement of antiretroviral therapy had been cryopreserved in central AACTG facilities between 2003 and 2004, and transported to the Centre for Clinical Immunology and Biomedical Statistics (CCIBS), Perth, Western Australia in 2008.
Formulation of HLA based peptide sets
For every one of 874 HLA associations identified in the previous genetic analysis involving the AACTG 5142/5128 cohort (19), we applied the Epipred T cell epitope prediction program (23; http://atom.research.microsoft.com/bio/epipred.aspx) to a sequence window of 13 amino acid residues flanking either side of the HLA associated site in the population consensus sequence, to score the probability of CD8 T cell epitopes with a matching HLA allelic restriction. Scores were generated for sequence containing the adapted amino acid as well as the non-adapted amino acids to predict the effect of the polymorphism on immune reactivity. The Epipred prediction algorithm was trained on characteristics of known CD8 T cell epitopes including HLA-specific peptide binding motifs, TCR contact residues, epitope length and flanking sequences to generate a probability score for predicted epitopes relative to known, published epitopes assigned a score of 1. Epipred used Bayes rule to compute the posterior probability that a viral sequence contains an epitope assuming a prior probability of 10%. A detailed example of an Epipred calculation for a single input HLA allele-peptide sequence is provided in supplementary material. All epitope sequences with a score ≥0.4 (representing at least 40% positive predictive value of being a true epitope flanking an association, and a four-fold increase from prior probability) were considered putative epitopes for immunological testing, even if they contained the HLA adapted polymorphism. Peptides representing the paired HLA adapted (resistant/escaped) or non-adapted (susceptible/wildtype) sequences were synthesized and tested to confirm HLA restricted immune reactivity to the non-adapted epitope, and loss or reduction of reactivity due to specified HLA associated epitope variations from that epitope.
Additional epitopes (n = 137) that were not spanning any HLA-HIV polymorphism associations in the genetic analysis (19) but were in the “A” (optimally defined/confirmed) or “B” (not optimally defined) lists of defined CD8 T cell epitopes published in the January 2009 update of the Los Alamos National Laboratory (LANL) HIV immunology database (http://www.hiv.lanl.gov/content/immunology) were added to the testing protocol to act as positive controls where possible (identified as “A and B list epitopes without HLA-HIV associations”).
Epitope selection was predicated by the HLA genotype of the subject. However, the number of predictions that were finally tested was constrained by the numbers of PBMC available. For this reason, epitopes for each individual were ranked in order of preference for testing based firstly on being possible novel epitopes, secondly on Epipred score, and thirdly on sequence match to the autologous viral sequence. Ranked lists of epitopes for every individual in the cohort were generated electronically using an in-house database. PBMCs were thawed, rested overnight in 10% heat inactivated FCS and RPMI (R10) and the number of cells ascertained using a Vi-Cell XR (Beckman Coulter, Australia) as previously described (24). Epitopes were then selected for testing for each individual from the ranked lists based on the number of cells.
IFNγ ELISpot assays
IFNγ responses to HIV-1 derived epitopes were quantified using Mabtech reagents in 96-well nitrocellulose-backed plates (Millipore, USA). Plates coated with 2 μg/mL of anti-IFNγ antibody were blocked with R10 for a minimum of 30 minutes, washed using an ELx 405 washer (BioTek, USA) after which 30,000 to 50,000 PBMCs, along with anti-CD28 antibody (Pharmingen, Australia) at a final concentration of 1 μg/mL, were added to each well (24). Lyophilised peptides (Invitrogen, Australia) were reconstituted to 10 mg/mL in DMSO, from which 1 mg/mL aliquots were made and stored at -20°C before use. The 1 mg/mL peptide stocks were further diluted to 50 μg/mL in R10 and tested in single or duplicate wells at a final concentration of 5 μg/mL. Where possible, triplicate wells of media alone served as negative controls while anti-CD3 antibody was used as a positive control either in single or duplicate wells. After addition of cells, peptides and anti-CD3 antibody, the ELISpot plates were incubated overnight at 37°C. Plates were then washed and IFNγ spots were developed with biotinylated antibody and streptavidin horse-radish peroxidise according to the manufacturer's instructions. IFNγ spots were detected using 3, 3′, 5, 5′-tetrametylbenzidine (24).
The large number of peptides, PBMC samples and individualised testing required use of a previously described automated system (24), in which the electronically generated peptide lists for each individual were integrated with the Biomek FX automated sample handling platform (Beckman Coulter, Australia); with software developed to electronically track the locations and volumes of all reagents including peptides and PBMCs on the 96-well nitrocellulose plate. Databases were created in-house to track reagent stock volumes, the number of freeze/thaw cycles of peptide stocks and document experimental procedures and results. Once optimised, epitope-specific IFNγ responses were investigated in a maximum of 30 individuals in one day (24). The plates were read on an AID plate reader (Automimmun Diagnostika GmbH, Germany) and the average count for the background was subtracted from all wells. Positive responses were defined as greater than twice the mean of the background and greater than or equal to 100 SFUs/106 PBMCs (25). Very high spot counts for nine epitopes which could not be enumerated by the AID plate reader and were designated “too numerous to count” (TNTC) were assigned a value of 15,000 SFUs/106 PBMCs for all quantitative analyses, based on the uppermost limit of values actually enumerated in the study.
Statistical analyses
A number of predicted epitopes had more than one possible HLA restriction and in some cases, individuals carried two or more of the associated HLA alleles. In this case, Epipred scores were used to identify the most likely responding peptide-HLA combination in the individual. IFNγ responses were therefore inherently more likely to be attributed to putative epitopes with high scores or known epitopes (where the Epipred score was assigned as 1) over putative epitopes with low scores as the most conservative approach to the analyses.
For each epitope the proportion of responders was calculated as the proportion of individuals tested who had a response ≥100 SFUs/106 PBMCs (25). Selected analyses involving comparisons of relative magnitude of responses included all non-zero responses to account for the use of a pre-defined cut-off for positivity, and to avoid a zero-inflated distribution of responses given that non-zero responses were normally distributed on the log scale. Mann-Whitney tests were utilized for evaluation of group epitope-specific differences, Spearman correlations for assessment of correlations with Epipred scores and generalized linear mixed models for assessing individual-specific associations using TIBCO Spotfire S+ 8.2 for Windows. All other analyses were performed using Prism 5.02 (GraphPad).
Results
Predicted CD8 T cell epitopes spanning HLA associations
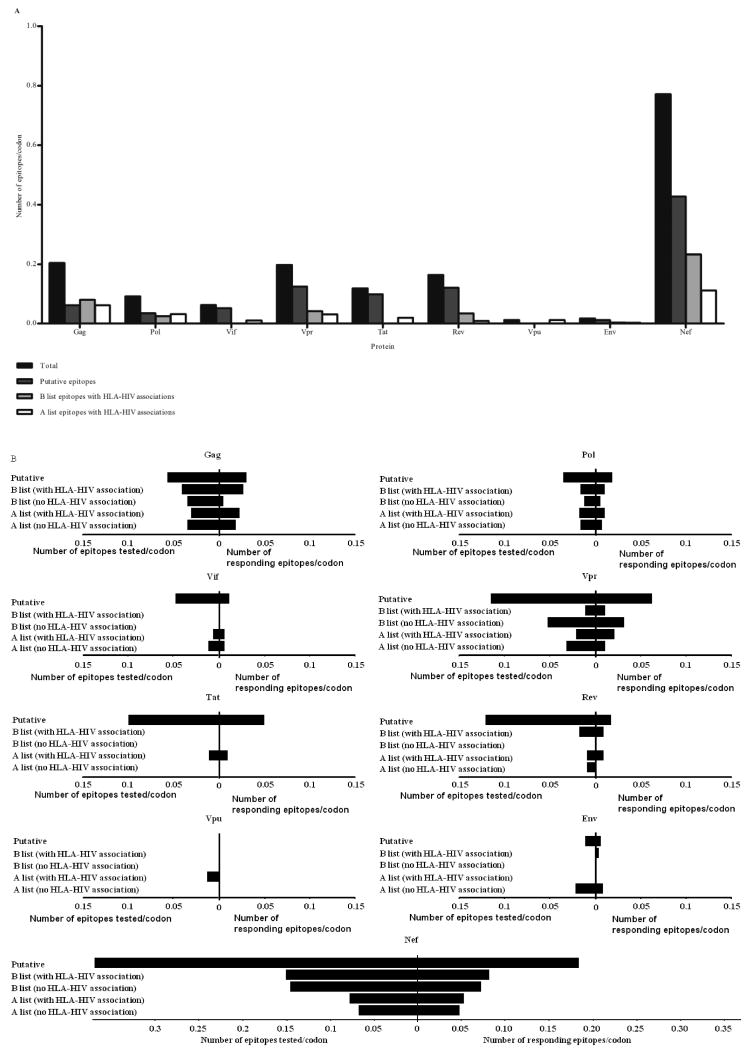
There were 221 already known CD8 T cell epitopes shown at or near sites of HLA-HIV associations with matching HLA restriction. There were a further 53 epitopes which had minor variations in length or sequence to known epitopes but these changes were not at sites of HLA associated polymorphism. The remaining 157 epitopes appeared completely novel giving a total of 431 epitopes with unique HLA restrictions and Epipred scores between 0.4 and 1 spanning 367 (of 874 total) HLA associations (Figure 1). There were 507 HLA allele specific polymorphisms, all with q-values (false-discovery rates (26)) <0.2 following phylogenetic correction, for which no epitope sequence with predictive scores greater than 0.4 were detected within a 26 codon sequence spanning the association. Among the 431 epitopes associated with the HLA allele-specific polymorphisms, there was a markedly higher proportion of epitopes in Nef (37% of all predicted epitopes; 0.77 epitopes per codon) compared to all other proteins (Figure 2A).
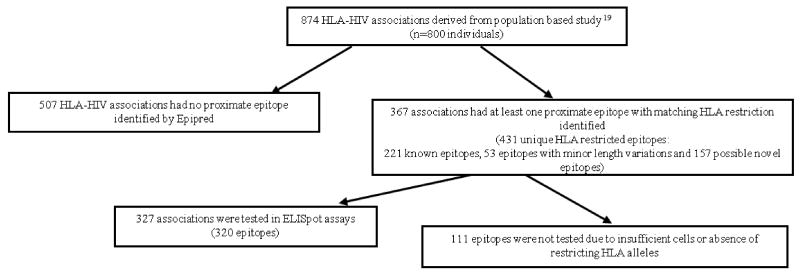
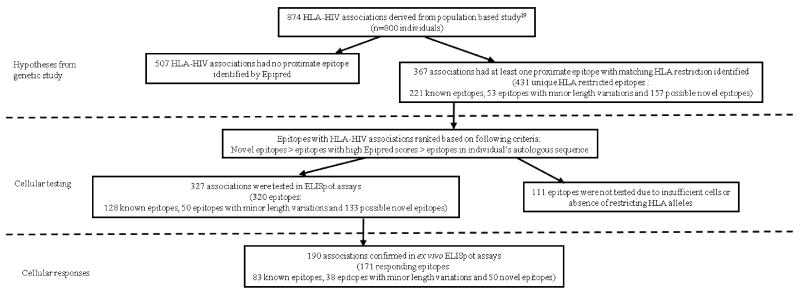
Figure 1. Summary of immunological investigations of 874 HLA-HIV associations in the study.
Figure 2. Distribution of epitopes predicted (A), tested and targeted (B) across the HIV-1 proteome.
(A)Numbers of putative and known epitopes around sites of HLA associated HIV adaptation are shown for each protein. (B)The distribution of epitopes tested and eliciting positive IFNγ responses spanning the nine viral proteins is shown. The numbers of epitopes are adjusted for varying lengths of different proteins by dividing them by the number of codons in each protein.
For 52 epitopes, the presence of the HLA associated substitution in the epitope changed the Epipred score of the epitope from greater than 0.4 to less than 0.4, predicting reduced or lost immune reactivity and in keeping with CTL escape in vivo. However we detected Epipred scores above 0.4 for 131 adapted epitopes, of which 25 had higher scores relative to the non-adapted epitope. In these cases, we presume that the HLA adapted variant sequence retained characteristics of an epitope still predicted to elicit a T cell response by the Epipred programme.
IFNγ T cell responses to predicted epitopes spanning HLA associations
We then sought to test these predictions in assays of ex vivo epitope-specific T cell responses using the IFNγ ELISpot assay. Of the 414 patient-specific PBMC samples thawed and enumerated, 290 had cell counts greater than 1.5 × 105 cells/mL; with an average viability of 82% (range = 33 to 100%) after thawing and these were used in subsequent immunological investigations. Using the known HLA class I alleles carried by individuals in the cohort with sufficient PBMCs available for testing (n = 290), we generated a list of known and putative CD8 T cell epitopes unique to each individual in the study. Of all 431 potential HLA/epitope combinations for testing arising from our genetic analysis, there were 320 (spanning 327 HLA-HIV associations) which were ultimately tested, because for the remainder there were insufficient numbers of subjects with the relevant HLA or less commonly, problems in synthesising the peptide. Of these 327 HLA-HIV associations, only 35% were proximate to well characterised, published CD8 T cell epitopes with the relevant HLA restriction.
CD8 T cell responses to these epitopes together with A and B list epitopes without HLA-HIV associations were investigated across 94 HLA-individualised 96-well plates for 290 individuals with on average, 13 epitopes tested per individual (range = 1 to 56 epitopes). At least one positive IFNγ response was elicited by 51% of the epitopes tested, and in 140 of the 290 individuals investigated. The number of responses per individual ranged from none to 33 with an average of two epitope-specific responses. Among individuals who mounted positive responses, the median magnitude of their IFNγ responses was 590 SFUs/106 PBMCs (inter-quartile range = 280 to 1440 SFUs/106 PBMCs). 128 individuals did not respond to any tested peptides and 22 individuals failed to elicit a response to the anti-CD3 antibody positive control. These 22 individuals who did not respond to the positive control had an average cell viability of 67% compared with 84.1% in the remainder of the cohort (p < 0.0001, Mann-Whitney test).
Protein distribution of prevalent detected IFNγ T cell responses
The protein distribution of responses was similar to the distribution of predicted epitopes, with epitopes in Nef eliciting the largest proportion of responses overall (38% of all epitope-specific responses) and had the highest number of responding epitopes per codon compared with all other proteins (Figure 2B), though the overall average magnitude of all responses were not significantly different across Gag, Nef, Tat, Pol and Env (Figure 3). Notably no IFNγ responses were detected against Vpu epitopes, including the known Vpu epitope ER9 (EYRKILRQR) (27) though the genetic analyses identified the E29Q, I33L and R37K mutations within the epitope associated with carriage of HLA-A*33 in this cohort (19).
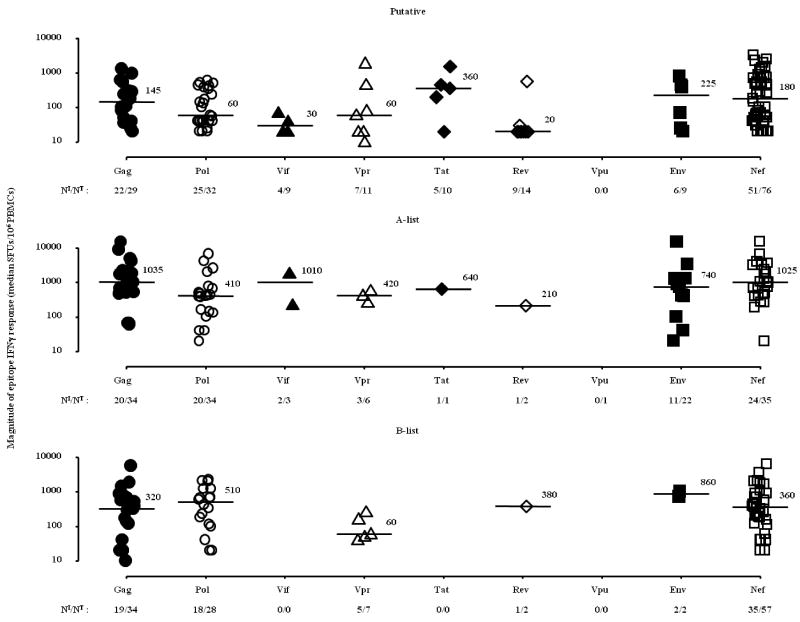
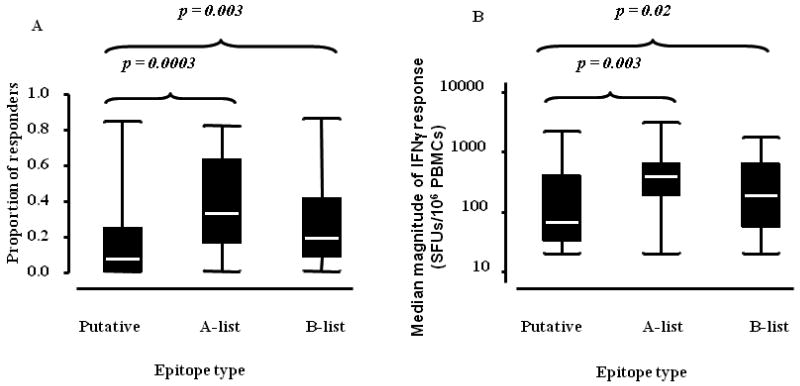
Figure 3. Magnitude of epitope-specific IFNγ responses (median) across all HIV-1 proteins for putative, A-list and B-list epitopes.
The plots display the median magnitude of all non-zero IFNγ responses for each epitope. Protein medians are annotated; NI/NT ≡ number epitopes/number epitopes tested. No responses were detected against the four VPU peptides tested.
On a within-individual basis, Nef-derived epitopes elicited the highest magnitude response more commonly (n = 74 epitopes, median = 1780 SFUs/106 PBMCs; interquartile range = 750 to 4600 SFUs/106 PBMCs) while there were 31 epitopes in Gag (median = 980 SFUs/106 PBMCs; interquartile range = 300 to 4000 SFUs/106 PBMCs) and 26 epitopes in Pol (median = 680 SFUs/106 PBMCs; interquartile range = 360 to 1130 SFUs/106 PBMCs) that accounted for the highest magnitude response in responding individuals. These patterns of reactivity both at the population and individual level largely reflected the distribution of HLA associations and epitope predictions as there were a greater number of epitopes from Nef firstly predicted, (Figure 2A) and tested (Figure 2B). In a mixed model regression analysis which takes the numbers of epitopes tested into account, Nef epitopes were more likely to mount positive responses compared to Env (p = 0.02) but not compared to epitopes in Gag (p > 0.9) and Pol (p = 0.1). A slight majority (57%) of these “highest magnitude per individual” responses targeted known epitopes while the remaining IFNγ responses were directed against putative epitopes and minor variants of known epitopes. Of note the number of individuals tested for each epitope was also a function of the prevalence of the restricting HLA allele, such that epitopes associated with rare alleles were tested less frequently. We identified a group of 33 epitopes which were tested in at least five individuals and elicted positive responses in at least 40% of those individuals tested. In this group of prevalent “responding” epitopes, 61% were clustered in Nef (Figure 4). We did not detect any statistically significant differences in the distribution of HLA restrictions between putative versus A or B list epitopes (data not shown).
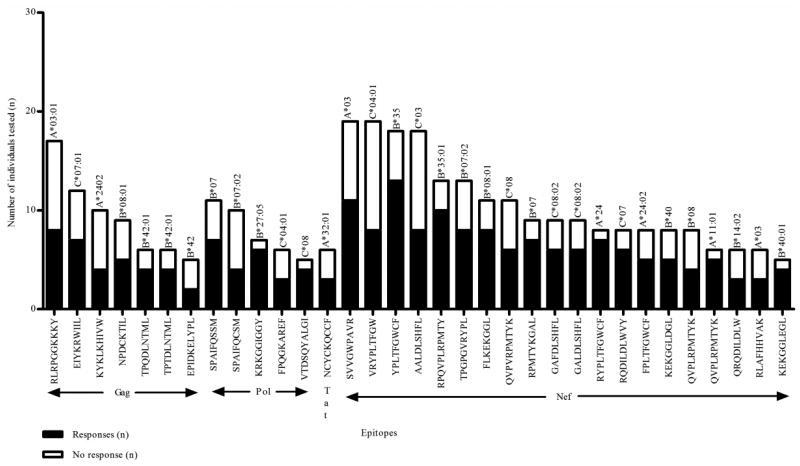
Figure 4. Immunogenic epitopes identified in IFNγ ELISpot assays.
Epitopes that were tested in five or more individuals and had a positive response rate of at least 40% are shown on the x-axis with the number of responders (■) and non-responders (□) shown on the y-axis. The restricting HLA allele is shown above each bar. The majority of epitopes were identified from the central region of Nef.
HLA associations marking novel CD8 T cell epitopes
In the study cohort overall, positive IFNγ responses were directed against a total of 143 known epitopes drawn from those associated with the HLA associations in the original genetic analyses or those added from the 2009 LANL update and not associated with HLA-driven polymorphism. Of these, 73 A list epitopes and 70 B list epitopes elicited at least one IFNγ response in this cohort. Known epitopes in general had an average response rate of 33% with 122 known epitopes eliciting no responses at all. There were consistent responses against nine novel epitopes in individuals carrying the HLA allele predicted to restrict the epitope (Table I). These nine epitopes were considered “high probability” novel epitopes because they were not listed in A or B lists of the 2009 LANL update (http://www.hiv.lanl.gov/content/immunology), there was common carriage of only one HLA allele predicted to bind the epitope, there were at least five individuals tested, and the response rate among those tested was at least 40% and therefore comparable to the mean response rate (33%) seen for known epitopes. For example, the HLA association studies identified HLA-C*04:01-driven polymorphism within FF9 (FPQGKAREF) in the Gag/Pol transframe region restricted by HLA-C*04:01. This epitope elicited responses in three of six individuals with carriage of HLA-C*04:01 tested (median = 380 SFUs/106 PBMCs, range = 340 to 1100 SFUs/106 PBMCs).
Table I. High probability novel epitopes.
| Protein | HLA | Epitope | Number of individuals tested | Positive responses (%) |
|---|---|---|---|---|
| Gag | C*07:01 | EIYKRWIIL | 12 | 58 |
| Gag | B*42 | EPIDKELYPL | 5 | 40 |
| Pol | B*27:05 | KRKGGIGGY1 | 7 | 86 |
| Pol | C*04:01 | FPQGKAREF | 6 | 50 |
| Tat | A*32:01 | NCYCKQCCF | 6 | 50 |
| Nef | A*03 | SVVGWPAVR | 19 | 58 |
| Nef | C*08 | QVPVRPMTYK | 11 | 55 |
| Nef | B*14:02 | QRQDILDLW | 6 | 50 |
| Nef | C*04:01 | VRYPLTFGW | 19 | 42 |
High probability novel epitopes were those for which the sequence or the HLA-restriction was not published as at 2009 LANL update; were tested in five or more individuals and had a positive response rate of greater than 40%.
This epitope was not listed in the January 2009 update of A or B-list epitopes but has been described in reference 28.
There were a further 41 “possible” novel epitopes which were also not listed in the LANL A or B lists and elicited at least one positive IFNγ response in the study, however there were either less than five individuals tested or the response rate was less than 40% (Table II). For example, HIV adaptation to HLA-B*14:02 was associated with a change from tyrosine (Y) at position 133 in Nef and was predicted to lie within the TW9 (TRYPLTFGW) epitope. IFNγ responses were investigated in eight individuals with HLA-B*14:02 and there were three responders (median magnitude = 480 SFUs/106 PBMCs; range = 400 to 740 SFUs/106 PBMCs).
Table II. Possible novel epitopes.
| Protein | Novel HLA-restrictions | Epitope | Number of individuals tested | Positive responses (%) | Known alternative HLA-restriction |
|---|---|---|---|---|---|
| Gag | B*57 | HQAISPRTL | 1 | 100 | B*1515, B*151029 |
| Gag | B*14:01 | DRWEKIRLR | 2 | 50 | None |
| Gag | B*15:01 | RLRPGGK(R)KKY | 13 (15) | 38 (7) | A*0330, A*030131 |
| Gag | B*45:01 | AEQASQDVKNW | 5 | 20 | B*4432, B*440232 |
| Gag | C*03 | RLRPGGKKKY | 7 | 14 | A*0330, A*030131 |
| Gag | A*03:01 | RAPRKKGCWK | 27 | 4 | None |
| Gag | A*31:01 | TVKCFNCGK | 6 | 17 | None |
| Pol | B*35 | FPQGKARE(K)F | 37 (37) | 27 (8) | None |
| Pol | B*35:12 | VPLTEEAEL | 13 | 15 | None |
| Pol | A*68 | LVDFRELNK | 13 | 15 | None |
| Pol | B*08:01 | QVRDQAEHL | 14 | 14 | None |
| Pol | A*02:01 | IIKIQNFRV | 38 | 5 | None |
| Pol | A*24 | QYDQILIEI | 19 | 5 | None |
| Pol | B*07:02 | FPQGKAREL(F) | 26 (24) | 12 (8) | None |
| Pol | A*33:01 | YLS(A)WVPAHK | 6 (4) | 17 (25) | None |
| Vif | B*15 | ISKKAKRWFY | 18 | 6 | None |
| Vpr | A*02:06 | WTLELLEEL | 4 | 25 | None |
| Vpr | B*27:05 | SRIGITRQR | 5 | 20 | None |
| Vpr | B*51:01 | FPRVWLHGL | 10 | 10 | None |
| Tat | A*32:01 | CCFHCQVCF | 6 | 33 | None |
| Tat | B*58:02 | CCFHCQVCF | 4 | 25 | None |
| Tat | C*16:01 | CCFHCQVCF | 4 | 25 | None |
| Tat | C*06 | CCFHCQVCF | 7 | 14 | None |
| Tat | A*03 | GLGISYGRK | 30 | 17 | None |
| Env | A*01:01 | GPGPGRAFY | 10 | 30 | None |
| Env | A*01:01 | SFEPIPI(S)HY | 16 (16) | 19 (6) | A*2933, A*290234 |
| Nef | C*07:02 | QVPLRPMTY(F)K | 1 (20) | 100 (25) | A*0335, A*030136, A*110137 |
| Nef | C*02:02 | KRQDILDLW | 2 | 50 | None |
| Nef | C*04:01 | TRYPLTFGW | 18 | 39 | A*3315 |
| Nef | B*14:02 | TRYPLTFGW | 8 | 38 | A*3315 |
| Nef | B*18:01 | KEVLVWKF | 17 | 35 | None |
| Nef | C*03:03 | QVPLRPMTFK | 4 | 25 | None |
| Nef | B*42:01 | YPLTFGWCF | 4 | 25 | B*5338 |
| Nef | B*08:01 | AFHHMAREL | 14 | 21 | None |
| Nef | A*31:01 | DPEKEVLVWK | 5 | 20 | None |
| Nef | B*14 | KRQDILDLW | 6 | 17 | None |
| Nef | B*40:01 | MEDPEKEVL | 6 | 17 | None |
| Nef | B*08:01 | GALDLSHFL | 10 | 10 | None |
| Nef | B*44 | KRRDILDLW | 23 | 9 | None |
| Nef | C*04:01 | GAFDLSHFL | 31 | 6 | None |
| Nef | B*44 | GYFPDWQNY | 17 | 6 | None |
These epitopes were either tested in less than five individuals or the proportion of responders was less than 40%. Epitopes and variants were counted as a single possible novel epitope in this table. Data on variants of these epitopes with the same HLA-restriction are shown in brackets. Identical peptide sequences with different HLA-restrictions are listed separately.
Taking all responses detected against novel epitopes, known epitopes and minor variants of known epitopes, and presuming that the ex vivo peptide presentation was mediated by the predicted HLA allele, ELISpot testing in this cohort confirmed 190 (58%) of 327 HLA associations that had a predicted epitope and were ultimately able to be directly tested in our study, given the relevant HLA types available. There were 137 HLA-HIV associations for which we could not show minimal support for marking a primary site of T cell selection based on our immunological studies involving HLA directed ELISpot screens of 290 individuals (Figure 5). As previously mentioned, there were 507 HLA allele-specific polymorphisms identified in the original genetic analysis which did not have any known or putative epitope predicted in proximity to the association. Those HLA associations for which we could not assign any known epitope, any predicted novel epitope nor find at least one positive IFNγ response in the proteomic region spanning the association may therefore be considered more likely to represent secondary/compensatory amino acid co-variation or false positive associations, and less likely to indicate a primary site of T cell escape.
Figure 5. Summary of results of testing in IFNγ ELISpot assays.
Epitope-specific responses and associations with Epipred scores, autologous viral sequences viral loads and CD4 counts
In the subset of epitopes tested in at least five individuals, higher proportions of individuals responded to known epitopes compared with putative epitopes (median proportion of responders: A list = 33% and B list = 19% vs putative = 7%; p = 0.0003 and p = 0.003 respectively; Mann-Whitney test; Figure 6A). The magnitude of IFNγ responses for known epitopes was also higher than the magnitude of responses for putative epitopes (median epitope-specific responses: A-list = 420 SFUs/106 PBMCs and B-list = 200 SFUs/106 PBMCs vs putative = 70 SFUs/106 PBMCs; p = 0.003 and p = 0.02 respectively, taking all non-zero responses into account; Mann-Whitney test; Figure 6B). Amongst putative epitopes, we did not detect a statistically significant correlation between Epipred scores and either the proportion of positive responders (Spearman's r = 0.06; p = 0.5) or the magnitude of IFNγ responses (Spearman's r = 0.05; p = 0.6).
Figure 6. Proportions of responders and median magnitude of IFNγ responses for putative and known epitopes tested in five or more individuals.
The plots indicate sample medians (heavy white lines), interquartile ranges (limits of black boxes) and value ranges (limits of whiskers). Median (IQR) of number of individuals tested per epitope are: putative epitopes = 10 (6-16), A list epitopes = 10 (6-16) and B list epitopes = 10.5 (7-13); Median (IQR) of numbers of responses per epitope are: putative epitopes = 2 (1-5), A list epitopes = 5 (2-9) and B list epitopes = 4 (2-7). The proportions of responding individuals with epitope specific responses ≥ 100 SFUs/106 PBMCs (A) and median magnitudes of all non-zero IFNγ responses (B) elicited by putative epitopes were significantly lower in comparison to known epitopes with HLA-HIV associations (Mann-Whitney test).
At the individual level, baseline viral load and CD4 counts did not predict response (p > 0.1), but the probability of responding was significantly higher for non-adapted epitope sequences that matched the autologous viral sequences (p < 0.0001; generalized linear mixed effect models).
IFNγ responses to HLA- adapted epitopes
For the majority of epitopes, peptides with the non-adapted (susceptible/wildtype) sequence and the adapted (resistant/escaped) sequence were synthesized and tested to confirm HLA restricted immune reactivity to the non-adapted epitope, and loss or reduction of reactivity due to the specified HLA associated epitope variation. For 76 non-adapted epitopes tested in parallel with the paired adapted epitope, the HLA associated amino acid substitution occurred within the epitope, and for 32 of these; complete loss of an IFNγ response to the adapted epitope was seen in all cases. In the remainder, the HLA-adapted version of the epitope still elicited IFNγ responses ≥100 SFUs/106 PBMCs.
HLA associated polymorphisms occurred outside 66 epitopes tested in our study, representing potential sites of epitope processing escape. IFNγ responses were elicited by 30 of these epitopes (median magnitude = 1090 SFUs/106 PBMCs; interquartile range = 420 to 2690 SFUs/106 PBMCs). In addition, in 26 cases, mutations occurring within one putative or known epitope resulted in predictions of new possible epitopes (Epipred scores ≥0.4) adjacent to or partially overlapping the original epitope and associated with the same allele, suggesting that some “neo-epitopes” may remain available for HLA and T cell engagement despite being in an “HLA adapted” state. For example, an HLA-A*24:02 driven change from tyrosine (Y) to phenylalanine (F) at codon 135 in Nef RF10 (RYPLTFGWCF) (39) was still associated with an Epipred prediction (score = 0.61) of HLA-A*24:02 mediated recognition of FF9 (FPLTFGWCF). Both epitopes were tested in six individuals with carriage of HLA-A*24:02, with five individuals responding to the non-adapted epitope (median = 1580 SFUs/106 PBMCs; range = 360 to 4560 SFUs/106 PBMCs) and IFNγ responses elicited by the adapted epitope in three individuals (median = 440 SFUs/106 PBMCs; range = 200 to 520 SFUs/106 PBMCs).
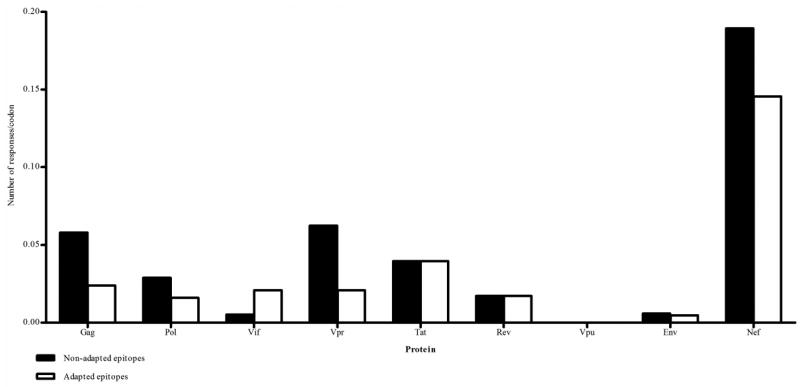
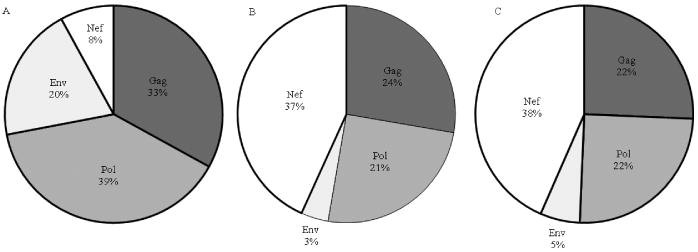
A substantial number of adapted epitopes elicited IFNγ responses ≥100 SFUs/106 PBMCs (n = 74), including 11 in which the mean magnitude of the response was two-fold higher for the adapted epitope relative to the non-adapted epitope in each individual tested. This appeared to be a general phenomenon, with examples in all proteins except Vpu, but was very prominent in Nef (Figure 7). There were some extremely complex patterns of new epitope-creation resulting from HLA associated changes in Nef as described above for the HLA-A*24:02-restricted epitope, RF10. This was particularly evident in the central region of Nef, where a 77 stretch of amino acids (positions 71 to 148) contained 21 partially overlapping epitopes created by polymorphism, which elicited IFNγ responses in our study cohort. Given the possible ramifications of this for vaccine induced immunity, we compared the proportion of CD8 T cell epitopes that would be in Nef compared to Gag, Pol and Env (as common vaccine antigens) if all HLA-specific variations and predicted epitopes were taken into account, versus the numbers of epitopes in these proteins in a single subtype B strain sequence (Figure 8A, B). This indicated an inflation of Nef epitopes and contraction of Pol, Gag and particularly Env epitopes associated with diversity coverage at the population level. This was further replicated when comparing proportions of epitopes which induce IFNγ responses, with Nef accounting for the greatest proportion of epitope-specific IFNγ responses relative to the other proteins in individuals in this study (Figure 8C).
Figure 7. Comparison of responses to non-adapted (■) and adapted (□) epitopes.
Number of positive IFNγ responses to adapted and non-adapted epitopes across the HIV-1 proteome. Numbers are adjusted for varying protein length by dividing by number of codons.
Figure 8. Marked inflation of Nef epitopes and Nef-specific IFNγ responses associated with HIV diversity.
The relative proportions of protein-specific epitopes in a single monovalent subtype B HIV-1 strain (A) compared with the proportions if all possible variants are included (B). C shows the proportion of protein-specific IFNγ responses derived from this study.
In order to determine whether responses to HLA-adapted epitopes could reflect general cross-reactivity phenomena, as opposed to de novo responses to the adapted epitope specifically, we sought to determine whether responses to adapted epitopes were more likely when there was a response to the non-adapted epitope despite a lack of match with the autologous viral epitope and therefore more cross-reactive response. As noted above, the probability of responding in general was significantly higher for non-adapted epitope sequences that matched the autologous viral sequences (p < 0.0001; generalized linear mixed effect model) and among those with demonstrated responses against a non-adapted epitope, those with match between the autologous sequence and non-adapted epitope sequence exhibited higher response rates to the adapted epitope (mean, adjusted for protein = 25%) compared with those where the individual's autologous viral sequence matched only the adapted epitope (14%; p = 0.05) or neither non-adapted or adapted epitope (14%; p = 0.02).
Discussion
To our knowledge this is the first large scale “reverse genomics” study in which the results of a genetic analysis were used to directly inform the selection and subsequent testing of particular viral antigens. Overall, we were able to provide immunological support for 190 HLA associated polymorphisms in subtype B HIV-1 as being sites of direct T cell recognition in vivo based on ex vivo IFNγ responses in the appropriate HLA background. This was 58% of the HLA associations tested in the study, representing an increase from only 35% that could have been explained by well characterised published CD8 T cell epitopes alone, prior to any cellular testing. For nine high probability epitopes there was a sufficiently frequent HLA type to show that the most likely HLA restriction of the epitopic response in the cohort matched that of the prediction, and there was sufficient frequency of testing and responses in at least 40% of cases to give the best level of evidence for immunoreactivity. A further set of possible novel epitopes was defined with responses rates of less than 40% but immunoreactivity in at least one individual with the predicted restricting HLA allele. It is notable however, that even well characterised published epitopes which have been used as a standard to validate genetic associations and as reagents in immunological studies had a mean response rate of only 33%. We therefore applied a higher standard of evidence for immunogenicity to potential novel epitopes compared with that observed for known epitopes in this study. The fact that cellular responsiveness was correlated with sequence match of the testing antigen to autologous virus, as shown in other studies (40) further confirms that viral diversity does influence the specificity of cellular responses within the individual. These data in general provide experimental evidence of a direct biological basis for 190 strongly HLA associated subtype B HIV-1 polymorphisms proteome-wide as sites of HIV-1 adaptation to HLA restricted T cell responses and should serve to guide further epitope characterisation and viral escape studies.
HIV-1 Nef was associated with the greatest number of epitopes which elicited IFNγ over the whole cohort and within individuals. This intense immunogenicity is in keeping with the extreme levels of HLA allele-specific selection in Nef shown in several population-based genetics studies (14, 19, 41) and mirrors the distribution of well characterised epitopes defined by cellular studies. As the majority of putative epitopes were tested in parallel with their “HLA-adapted” pair, we were also able to determine if any functional consequences of polymorphism within epitopes were apparent in a screening ELISpot assay. Marked reductions in IFNγ responses associated with the polymorphisms were seen in a proportion of cases supporting a role for loss of TCR engagement or HLA-peptide binding in vivo in these examples. There were also instances in which the HLA-adapted or “escaped” version of the epitope elicited equivalent or higher magnitude responses than the non-adapted versions. In a screening ELISpot with excess peptide concentrations, it is possible that such reactivity patterns result from T cell cross-reactivity, though this appeared to be more likely to occur with Nef epitopes, compared with other proteins and it is not clear why TCR clonotypes specific for Nef epitopes should be inherently more cross-reactive than other TCRs. Furthermore, we did not find that responses which appeared more inherently cross-reactive, as indicated by lack of match with autologous viral sequences were more likely to respond to the adapted epitopes. The general determinants of T cell recognition of viral variants have been explored in other studies (42, 43). It is important to emphasise we have tested specific epitope pairs based on population-signals of adaptation. In all these specific instances of positive responses to HLA-adapted epitopes, there was strong statistical evidence of the adapted residue being enriched in vivo in the selecting HLA-context in the original HLA associations analysis, suggesting that in the cellular studies here, either the true differences in peptide avidity were not apparent at excess peptide concentrations and would diverge with serial peptide dilutions, or alternatively that inducing immune responses to adapted variants provides some selective advantage to HIV-1 in vivo.
The formation of neo-epitopes as a result of T cell escape has been described in longitudinal studies (44) but our data suggest this could be reasonably common phenomena. We have described cases of HLA selection leading to high avidity, neo-epitope-specific responses in chronic progressive HIV infection (45) and have argued that this could represent a way for HIV mutations to promote “bad” immunodominance patterns in chronic infection and drive HIV evolution, not necessarily away from all immune recognition but to enhanced but ineffective recognition of a narrow range of epitopes. In this study, there were several extremely complex patterns of HLA associated polymorphisms in Nef leading to formation of new epitope targets for the same and new HLA alleles which were partially overlapping or distant from the original epitope. Given this combination of high variability with high density of reactive epitopes, including reactivity to many overlapping HLA adapted variants, it is not surprising that Nef epitopes as a proportion of all reactive epitopes are relatively inflated and the IFNγ responses to Nef dominates over all others when considered at a population level. If these Nef responses lead to a relative reduction in targeting of more structurally or functionally constrained proteins such as Gag or Pol in-vivo, where viral adaptations are more likely to incur fitness costs, then Nef-dominated immunity is conceivably more advantageous to the virus than the host. Since these immunodominance patterns characterise chronic infection where immune control has manifestly failed, recapitulating such immune hierarchies by a vaccine immunogen would seem empirically undesirable, particularly for therapeutic vaccines which could serve to boost this inflation. It is not known whether broad poly-specific vaccine-induced responses prior to viral exposure could block, not block or even enhance particular transmitting viral variants, though this data will emerge as more polyvalent strategies in preventative vaccines advance to clinical trials. Computational strategies which are based on conservation or are polyvalent but seek to minimise the inclusion of rare or unfavourable epitopes or are based on acute transmitted founder viruses may overcome this issue. This set of immunological data could be useful to help in scoring algorithms used to computationally optimize inclusion of important circulating acute variants and perhaps help in excluding particular variants that appear prone to interference or immunodominance phenomena in vivo.
Despite the large size of our study cohort, the extreme polymorphism of HLA molecules still limits the degree to which the HLA allele restriction of many responses could be defined analytically and limited stored cellular material on our study cohort subjects precluded further experimental studies. As we assigned a higher ranking to known or high probability HLA restrictions for those epitopes with overlapping HLA restrictions, our study is also inherently conservative, with a bias against assignment of novel epitope responses when there are limited numbers of individuals with that HLA. Furthermore, the use of an epitope prediction program trained on characteristics of known epitopes will inevitably tend to predict epitopes more similar to known epitopes and therefore the 507 associations for which no proximal epitope was predicted cannot be absolutely excluded as sites of true immune selection, particularly given the low mean response rate of even known epitopes shown here. However the additional peptide synthesis, sample and assay requirement of assessing all possible epitopic regions and variants spanning all associations is prohibitive at a practical level.
The challenges of translating the findings of genetic HLA polymorphism association studies to the functional, cellular level are considerable and include the extreme polymorphism of HLA and HIV-1 as discussed above (which necessitates large sample sizes), availability of samples and subjects for immunological testing, limitations in amount and quality of cryopreserved sample material (particularly from pre-treatment time-points), the general heterogeneity of T cell responses between subjects and over time, limitations of ex vivo based assays and single biomarkers such as IFNγ, and the false discovery rate of associations arising from any genetic associations study. Nevertheless, we were able to expand the base of immunological support for a number of subtype B HIV-1 polymorphisms being sites of immune selection. Apart from providing positive evidence for immune reactivity, the absence of any reactivity for some peptides can also be useful in studies of secondary or compensatory mutational networks. Indeed this study would suggest that only the minority of HLA-HIV polymorphisms (given a q-value cut-off of 0.2, with adjustment for viral phylogeny) can be explained by primary escape or co-targeting of multiple epitopes and many others are more likely secondary mutations affecting structurally or functionally interdependent residues. Mapping the mutational networks or genetic haplotypes in HIV-1 which determine viral fitness under diverse host environments will reveal more about the importance of specific residues in HIV replication and pathogenesis. The information provided here on mutations which are highly HLA allele specific but not within or near epitopes could help those modelling or studying such co-variation networks for both vaccine research and identifying novel ligands for antiviral drugs. More HLA association population-based studies will continue to be done in new and genetically diverse populations (46) and in larger populations, and the output from several studies have already been used as presumptive sites of viral escape in a number of secondary analyses. However, there is little value in generating vast numbers of hypotheses across these studies unless they are systemically tested and validated at the functional level where possible. The results of such testing can then be used to refine mapping of primary viral escape and compensatory pathways, iterate and validate analytical approaches to genetic studies, and understand the links between HIV polymorphism, adaptation and immunogenicity.
Supplementary Material
Acknowledgments
The authors would like to thank the study teams, study sites and participants in the US Adult ACTG A5142 (NCT00050895) and A5128 (NCT00031408) protocols as well as colleagues at the Centre for Clinical Immunology and Biomedical Statistics, Institute of Immunology and Infectious Diseases. We would also like to thank Donald Cooper, Shay Leary, Micheal Corkery and Daniel Piccoli for computing work associated with large scale high throughout ELISpot testing.
Abbreviations used in this paper
- SFUs
Spot forming units
Footnotes
This project was supported by grant number RO1 AI060460 from the National Institute of Allergy and Infectious Diseases (NIAID). The ACTG is supported by grant number AI-68636 and the Vanderbilt DNA Resources Core by grant number RR024975. The ACTG Clinical Trials Sites that collected DNA were supported by NIH grants AI64086, AI68636, AI68634, AI069471, AI27661, AI069439, AI25859, AI069477, AI069513, AI069452, AI27673, AI069419, AI069474, AI69411, AI69423, AI69494, AI069484, AI069472, AI069501, AI69467, AI069450, AI32782, AI69465, AI069424, AI38858, AI069447, AI069495, AI069502, AI069556, AI069432, AI46370, AI069532, AI046376, AI34853, and AI069434. The content of this study is the responsibility of the authors and does not necessarily represent the official views of NIAID or the National Institutes of Health (US). The project was additionally supported by the Australian National Health and Medical Research Council (program grant #384702) and the Bill and Melinda Gates Foundation (grant #31844).
References
- 1.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 2.Barouch DH, Korber B. HIV-1 vaccine development after STEP. Annu Rev Med. 2010;61:153–167. doi: 10.1146/annurev.med.042508.093728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33:542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 5.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson AC, Iversen AK, Chapman JM, de Oliviera T, Spotts G, McMichael AJ, Davenport MP, Hecht FM, Nixon DF. Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS One. 2007;2:e225. doi: 10.1371/journal.pone.0000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yerly D, Heckerman D, Allen T, Suscovich TJ, Jojic N, Kadie C, Pichler WJ, Cerny A, Brander C. Design, expression, and processing of epitomized hepatitis C virus-encoded CTL epitopes. J Immunol. 2008;181:6361–6370. doi: 10.4049/jimmunol.181.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch DH, O'Brien KL, Simmons NL, King SL, Abbink P, Maxfield LF, Sun YH, La Porte A, Riggs AM, Lynch DM, Clark SL, Backus K, Perry JR, Seaman MS, Carville A, Mansfield KG, Szinger JJ, Fischer W, Muldoon M, Korber B. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med. 2010;16:319–323. doi: 10.1038/nm.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 10.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci U S A. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 14.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, Eichbaum Q, Block BL, Baker B, Kadie C, Markowitz M, Jessen H, Kelleher AD, Rosenberg E, Kaldor J, Yuki Y, Carrington M, Allen TM, Mallal S, Altfeld M, Heckerman D, Walker BD. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, Kadie C, Mullins JI, Walker BD, Harrigan PR, Goulder PJ, Heckerman D. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, Matthews P, Payne R, Rolland M, Raugi DN, Maust BS, Learn GH, Nickle DC, Coovadia H, Ndung'u T, Frahm N, Brander C, Walker BD, Goulder PJ, Bhattacharya T, Heckerman DE, Korber BT, Mullins JI. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YE, Li B, Carlson JM, Streeck H, Gladden AD, Goodman R, Schneidewind A, Power KA, Toth I, Frahm N, Alter G, Brander C, Carrington M, Walker BD, Altfeld M, Heckerman D, Allen TM. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–1855. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John M, Heckerman D, James I, Park LP, Carlson JM, Chopra A, Gaudieri S, Nolan D, Haas DW, Riddler SA, Haubrich R, Mallal S. Adaptive interactions between HLA and HIV-1: highly divergent selection imposed by HLA class I molecules with common supertype motifs. J Immunol. 2010;184:4368–4377. doi: 10.4049/jimmunol.0903745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C, O'Connor G, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ, Buchbinder S, Kirk GD, Martin MP, Telenti A, Deeks SG, Walker BD, Goldstein D, McVicar DW, Moffett A, Carrington M. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, Garren KW, George T, Rooney JF, Brizz B, Lalloo UG, Murphy RL, Swindells S, Havlir D, Mellors JW. Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358:2095–2106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas DW, Wilkinson GR, Kuritzkes DR, Richman DD, Nicotera J, Mahon LF, Sutcliffe C, Siminski S, Andersen J, Coughlin K, Clayton EW, Haines J, Marshak A, Saag M, Lawrence J, Gustavson J, Anne Bennett J, Christensen R, Matula MA, Wood AJ. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 23.Heckerman D, Kadie C, Listgarten J. Leveraging information across HLA alleles/supertypes improves epitope prediction. J Comput Biol. 2007;14:736–746. doi: 10.1089/cmb.2007.R013. [DOI] [PubMed] [Google Scholar]
- 24.Almeida CA, Roberts SG, Laird R, McKinnon E, Ahmed I, Pfafferott K, Turley J, Keane NM, Lucas A, Rushton B, Chopra A, Mallal S, John M. Automation of the ELISpot assay for high-throughput detection of antigen-specific T-cell responses. J Immunol Methods. 2009;344:1–5. doi: 10.1016/j.jim.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SG, Joosten SA, Verscheure V, Pathan AA, McShane H, Ottenhoff TH, Dockrell HM, Mascart F. Identification of major factors influencing ELISpot-based monitoring of cellular responses to antigens from Mycobacterium tuberculosis. PLoS One. 2009;4:e7972. doi: 10.1371/journal.pone.0007972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Addo MM, Altfeld M, Rathod A, Yu M, Yu XG, Goulder PJ, Rosenberg ES, Walker BD. HIV-1 Vpu represents a minor target for cytotoxic T lymphocytes in HIV-1-infection. AIDS. 2002;16:1071–1073. doi: 10.1097/00002030-200205030-00015. [DOI] [PubMed] [Google Scholar]
- 28.Schellens IM, Kesmir C, Miedema F, van Baarle D, Borghans JA. An unanticipated lack of consensus cytotoxic T lymphocyte epitopes in HIV-1 databases: the contribution of prediction programs. AIDS. 2008;22:33–37. doi: 10.1097/QAD.0b013e3282f15622. [DOI] [PubMed] [Google Scholar]
- 29.Llano A, Frahm N, Brander C. How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection? In: Korber B, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2009. Los Alamos National Laboratory; Los Alamos, New Mexico: 2009. p. 3. [Google Scholar]
- 30.Goulder PJ, Tang Y, Pelton SI, Walker BD. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J Virol. 2000;74:5291–5299. doi: 10.1128/jvi.74.11.5291-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brander C, Walker B. HIV Molecular Immunology Database 1995. Los Alamos National Laboratory; Los Alamos, New Mexico: 1995. The HLA-Class I restricted CTL response in HIV-1 infection: identification of optimal epitopes; pp. IV--1–IV--9. [Google Scholar]
- 32.Brander C, Walker B. HIV Molecular Immunology Database 1997. Los Alamos National Laboratory; Los Alamos, New Mexico: 1997. Systematic identification of optimal HIV-1 CTL epitopes; pp. IV--1–IV--11. [Google Scholar]
- 33.Altfeld M, Rosenberg ES, Shankarappa R, Mukherjee JS, Hecht FM, Eldridge RL, Addo MM, Poon SH, Phillips MN, Robbins GK, Sax PE, Boswell S, Kahn JO, Brander C, Goulder PJ, Levy JA, Mullins JI, Walker BD. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J Exp Med. 2001;193:169–180. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culmann B, Gomard E, Kieny MP, Guy B, Dreyfus F, Saimot AG, Sereni D, Sicard D, Levy JP. Six epitopes reacting with human cytotoxic CD8+ T cells in the central region of the HIV-1 NEF protein. J Immunol. 1991;146:1560–1565. [PubMed] [Google Scholar]
- 36.Koenig S, Fuerst TR, Wood LV, Woods RM, Suzich JA, Jones GM, de la Cruz VF, Davey RT, Jr, Venkatesan S, Moss B, et al. Mapping the fine specificity of a cytolytic T cell response to HIV-1 nef protein. J Immunol. 1990;145:127–135. [PubMed] [Google Scholar]
- 37.Li L, Bouvier M. Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue. J Immunol. 2004;172:6175–6184. doi: 10.4049/jimmunol.172.10.6175. [DOI] [PubMed] [Google Scholar]
- 38.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 39.Tanuma J, Fujiwara M, Teruya K, Matsuoka S, Yamanaka H, Gatanaga H, Tachikawa N, Kikuchi Y, Takiguchi M, Oka S. HLA-A*2402-restricted HIV-1-specific cytotoxic T lymphocytes and escape mutation after ART with structured treatment interruptions. Microbes Infect. 2008;10:689–698. doi: 10.1016/j.micinf.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 40.Altfeld M, Addo MM, Shankarappa R, Lee PK, Allen TM, Yu XG, Rathod A, Harlow J, O'Sullivan K, Johnston MN, Goulder PJ, Mullins JI, Rosenberg ES, Brander C, Korber B, Walker BD. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77:7330–7340. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malhotra U, Nolin J, Horton H, Li F, Corey L, Mullins JI, McElrath MJ. Functional properties and epitope characteristics of T-cells recognizing natural HIV-1 variants. Vaccine. 2009;27:6678–6687. doi: 10.1016/j.vaccine.2009.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoof I, Perez CL, Buggert M, Gustafsson RK, Nielsen M, Lund O, Karlsson AC. Interdisciplinary analysis of HIV-specific CD8+ T cell responses against variant epitopes reveals restricted TCR promiscuity. J Immunol. 2010;184:5383–5391. doi: 10.4049/jimmunol.0903516. [DOI] [PubMed] [Google Scholar]
- 44.Allen TM, Yu XG, Kalife ET, Reyor LL, Lichterfeld M, John M, Cheng M, Allgaier RL, Mui S, Frahm N, Alter G, Brown NV, Johnston MN, Rosenberg ES, Mallal SA, Brander C, Walker BD, Altfeld M. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J Virol. 2005;79:12952–12960. doi: 10.1128/JVI.79.20.12952-12960.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keane NM, Roberts SG, Almeida C-A, Krishnan T, Chopra A, Demaine E, Laird R, Tschochner M, Carlson JM, Mallal S, et al. High-avidity, high-IFNγ-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol Cell Biol. 2011 doi: 10.1038/icb.2011.34. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avila-Rios S, Ormsby CE, Carlson JM, Valenzuela-Ponce H, Blanco-Heredia J, Garrido-Rodriguez D, Garcia-Morales C, Heckerman D, Brumme ZL, Mallal S, John M, Espinosa E, Reyes-Teran G. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology. 2009;6:72. doi: 10.1186/1742-4690-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.