MicroRNAs are important regulators of gene expression, yet the functional outputs of most microRNA-target interactions remain elusive. Here we introduce the Drosophila microRNA sponge (miR-SP) as a powerful transgenic technology to dissect the function of every microRNA with precise spatiotemporal resolution. We demonstrate that miR-SPs can be used to characterize tissue-specific microRNA loss-of-function phenotypes, define the spatial regulation of their effectors, and uncover interactions between microRNAs and other genes. identify an essential role of the conserved microRNA miR-8, in neuromuscular junction (NMJ) formation. Tissue-specific silencing reveals that postsynaptic activity of miR-8 is important for normal NMJ morphogenesis. Given that miR-SPs rely on a bipartite modular expression system, they could be used to elucidate the endogenous function of microRNAs in any species where conditional expression can be achieved.
MicroRNAs are post-transcriptional regulators of gene expression with functions linked to development, physiology and disease1. These ~22-nucleotide non-coding RNAs bind to specific target sequences frequently located within the 3'UTR of protein-coding genes, and suppress expression by means of mRNA decay or translational inhibition2. Hundreds of microRNAs exist in organisms from plants to humans, yet the biological function of relatively few microRNA-target interactions have been thoroughly characterized in vivo1. As the diversity of microRNA identity and expression patterns unfolds3, biologists face an increasing challenge to discern microRNA contributions in highly dynamic or complex biological processes.
While genomic techniques facilitated rapid identification of microRNAs4, uncovering their functions has been limited by available technologies for in vivo analysis. For instance, a body of evidence concerning microRNA functions has been inferred from misexpression studies1. In Drosophila, overexpression of miR-7, miR-iab-4-5p and miR-315, has implicated these microRNAs in important signaling pathways, such as Notch, Ubx and wingless respectively1, 5. Because every microRNA is predicted to interact with many mRNAs6, misexpression may lead to silencing of genes that normally escape regulation due to spatial, temporal or quantitative restrictions, thus generating neomorphic phenotypes1. For this reason, most microRNA gain-of-function experiments require validation with loss-of-function data.
The tools currently used for microRNA loss-of-function analysis present several caveats. Classical genetic knockouts are time-consuming, unable to simultaneously silence “seed” sequence-redundant microRNAs, and offer only limited flexibility for spatial and temporal analysis. Chemically modified synthetic oligonucleotides7 have the ability to silence both individual and families of microRNAs but have narrow applicability for developmental or physiological studies. More recently, modified antisense oligonucleotides, originally termed microRNA “sponges”8, expressed from plasmids or lentiviral vectors offered a promising advance towards dissecting microRNA function, albeit still restricted to cell culture systems or transplanted tissues8, 9. A major challenge for both chemically modified oligonucleotide inhibitors and microRNA “sponges” has been targeted and effective in vivo delivery. While high doses of these reagents can be attained, these technologies are limited in their spatial or temporal resolution in living organisms. The most versatile methods for robust spatial and/or temporal-specific transgene expression are modular expression systems based on exogenous DNA recombinases or transcription factors such as Cre and Gal4, respectively10, 11. Here, we generated transgenic miR-SPs by placing modified microRNA complementary oligonucleotides downstream of repetitive UAS sequences. Our results demonstrate that combining these constructs with specific Gal4 drivers renders sufficient expression of the microRNA silencer to achieve spatiotemporal microRNA inhibition in intact animals.
RESULTS
MiR-SP technology – design and proof of principle
To generate miR-SP elements, ten repetitive sequences complementary to a microRNA with mismatches at positions 9–12 (ref. 8) for enhanced stability, were introduced into the 3'UTR of EGFP or mCherry in a pUAST expression vector (Fig. 1a–b). Transgenic animals were generated carrying one or two copies of the miR-SP cassette.
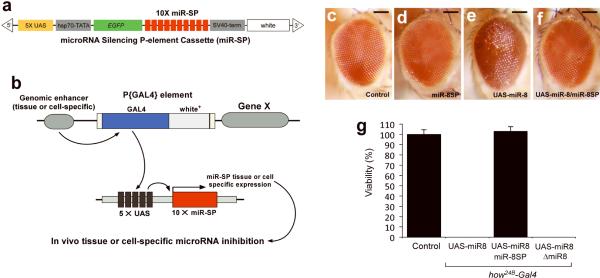
FIGURE 1. Design and functionality of transgenic miR-SP elements.
a, Ten microRNA binding sites (red) containing a mismatches at positions 9–12 were inserted downstream of EGFP (green) in a UAS-containing P-element vector. b, The resulting transgenic animals can be crossed to specific Gal4 lines. c–f, Expression of UAS-miR-8 results in a rough eye phenotype that can be rescued by co-expression of miR-8SP. Adult eye morphology in Control GMR-Gal4/+ (c), GMR-Gal4/miR-8SP;miR-8SP/+ (d), GMRGal4/+;UAS-miR-8/+ (e), GMR-Gal4/miR-8SP;UAS-miR-8/miR-8SP (f). Scale bars are 100 μm. g, Histogram illustrating percent viability in miR-8 null mutants and modified miR-8 genetic backgrounds (n > 50 for all genotypes; Error bars: ± SEM).
To assess the specificity and efficiency of the miR-SP elements, we analyzed a neomorphic phenotype in the compound eye. MiR-8 loss of function either by a null mutant (not shown) or by expressing a miR-SP construct (miR-8SP) using the eye-specific driver GMR-Gal4 (Fig. 1d) does not affect adult eye morphogenesis. However, overexpression of miR-8 (UAS-miR-8) results in a rough eye phenotype (Fig. 1e) relative to GMR-Gal4 controls (Fig. 1c). In contrast, animals co-expressing UAS-miR-8 and miR-8SP displayed wild-type morphology indicating complete suppression of this neomorphic phenotype as a consequence of miR-8 silencing (Fig. 1f). To demonstrate that miR-SPs function independent of tissue context, we analyzed their activity in muscles using the mesodermal how24B-Gal4 driver. Overexpression of miR-8 in muscle results in fully penetrant embryonic lethality (Fig. 1g), even when expressed in a miR-8 null background. However, co-expression of miR-8SP and miR-8 under the same how24B-Gal4 driver restores viability to wild-type levels (Fig. 1g). These results demonstrate that miR-SP elements can inhibit microRNA activity in vivo with high efficiency, even when expressed under the same Gal4 driver.
MiR-SPs effectively inhibit endogenous microRNA activity
To ask if miR-SPs can interfere with endogenous microRNA activity we first employed a known microRNA-target regulatory network. The Drosophila neuronal-specific transcription factor Nerfin-1 has been shown to be under stringent microRNA-mediated post-transcriptional regulation12, 13. Analysis of a transgenic sensor construct consisting of nerfin-1 3'UTR fused to GFP suggested that microRNA activity spatially restricts Nerfin-1 expression12. The miR-9 family plays an important role in endogenous nerfin-1 regulation13. Indeed, the 3'UTR of nerfin-1 contains 4 target sites for miR-9 of which two are highly conserved in Drosophila species12, 13. Moreover the GFP nerfin-1 3'UTR sensor was found to be silenced in larval wing imaginal discs12 where miR-9a is expressed14. Thus, it was likely that miR-9a regulates sensor expression in this context, providing a means to determine if miR-SPs can inhibit endogenous microRNA function. To test this, we expressed the GFP nerfin-1 3'UTR sensor throughout the wing imaginal disc and targeted miR-9aSP in a subset of cells along the anterior-posterior compartment boundary using the ptc-Gal4 driver. We expressed miR-9aSP or GFP nerfin-1 3'UTR alone as controls. Analysis of third instar larval discs revealed a specific increase in GFP signal in the ptc-Gal4 expressing cells but not in the control samples (Fig. 2a and data not shown). Thus miR-SP elements can be employed for targeted inhibition of endogenous microRNA activity. In addition, given that microRNA inhibition often leads to upregulation of target expression, this suggests that the miR-SP technology can be used to identify microRNA targets with precise spatial resolution.
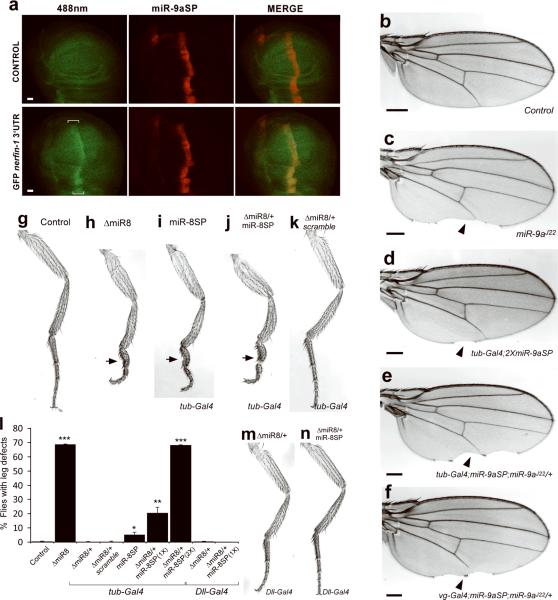
FIGURE 2. Effective tissue-specific silencing of endogenous microRNA activity by miR-SP.
a, Wing imaginal discs from ptcGal4/miR-9aSP;miR-9aE39/+ (CONTROL) and tubulinEGFP nerfin-1 3'UTR, ptcGal4/miR-9aSP;miR-9aE39/+ (GFP nerfin-1 3'UTR,) imaged for GFP or mCherry. The cells along the anterior-posterior compartment boundary display increased GFP expression (brackets). Scale bars are 20 μm. b–f, MiR-9a silencing results in specific posterior wing margin defects. Cuticle preparations of adult wings from wild type (b), miR-9aJ22/miR-9aJ22 (c), miR-9aSP/+; miR-9aSP/tubulin-Gal4 (d), miR-9aSP/+; miR-9aJ22/tubulin-Gal4 (e), miR-9aSP/vg-Gal4; miR-9aJ22/+ (f). Scale bars are 100 μm. g–k, Inhibition of miR-8 activity by genomic knockdown or miR-SP silencing results in a deformed third leg pair. Cuticle preparations of adult third legs from wild type (g), ΔmiR-8/ΔmiR-8 (h), miR-8SP/+; tubulin-Gal4/miR-8SP (i), miR-8SP/ΔmiR-8; tubulin-Gal4/miR-8SP (j), ΔmiR-8/+; tubulin-Gal4/Scramble-SP (k). l, Percentage of adult flies displaying malformed third leg phenotype in control and miR-8 modified genetic backgrounds (Student's t-test, *P=0.05; **P < 0.03; ***P < 10−4; Error bars: ± SEM). m, n, Cuticle preparation of adult third legs from Dll-Gal4/ΔmiR-8 (m) and Dll-Gal4/ΔmiR-8; miR-8SP/+ (n).
We next sought to establish the effectiveness of the miR-SP system to uncover bona fide loss of function phenotypes in intact animals. Of the few Drosophila microRNAs analyzed to date, only a small number have striking morphological phenotypes14–16. MiR-9a regulates sensory organ formation and controls wing morphogenesis in adults14. MiR-9a mutant flies display an increased number of sensory bristles on the anterior part of the wing and a notched posterior wing margin14. Similar to null mutant animals, expression of miR-9aSP using the ubiquitous tubulin-Gal4 driver displayed posterior wing margin defects in adult progeny (Fig. 2b–d). Moreover, we observed increased expressivity, reaching null mutant severity, by the addition of a heterozygous miR-9a mutant background (Fig. 2e). No defects were observed in miR-9a heterozygous mutant animals (not shown). This phenotype can be rescued by overexpression of miR-9a in the wing imaginal disc cells along the dorsalventral boundary using the vg-Gal4 driver14. The strength of the miR-SP technology lies in the ability to uncover discrete tissue-specific microRNA functions. To determine whether miR-9a activity is required ubiquitously to control wing morphogenesis or only in a subset of cells, we expressed miR9a-SP with vg-Gal4. The progeny displayed the same wing margin phenotype observed in miR-9a null mutants or miR-9aSP/tubulin-Gal4 animals, indicating that the function of miR-9a in wing morphogenesis is restricted to this subset of cells along the dorsal-ventral boundary (Fig. 2f). These results highlight the versatility of the miR-SP technology in attaining tissue-specific inhibition of microRNA function, required for the analysis of complex developmental processes.
To establish the broad applicability of the miR-SP technology, we explored another well characterized microRNA loss-of-function phenotype. Genomic loss of miR-8 causes a malformed third leg phenotype (Fig. 2g, h, l, ref. 16). Expression of miR-8SP using the ubiquitous tubulin-Gal4 driver results in the same, albeit less penetrant, third leg phenotype (Fig. 2i, l). We also found a dose-dependent elevation in penetrance that reaches null severity by increasing miR-SP copy number, or by addition of a heterozygous miR-8 mutant background (Fig. 2j, l). Thus, the miR-SP system provides the opportunity to generate allelic series, a tool essential for genetic interaction studies. Expression of a control miR-SP with scrambled sequence did not induce any phenotype even in a miR-8 heterozygous mutant background (Fig. 2k, l).
Despite a highly penetrant adult leg defect, early leg disc patterning and morphogenesis appear to be normal in miR-8 mutant flies, suggesting that the observed phenotype results from a late systemic defect, such as mechanical damage during late pupariation, rather than an early leg-specific effect16. Consequently, one would expect that localized inhibition of miR-8 in the developing leg should not render a miR-8 phenotype. In contrast to ubiquitous miR-SP expression (Fig. 2i, j, l) or a miR-8 null mutant (Fig. 2h, l), animals expressing miR-8SP with the leg-specific driver Dll-Gal4 were indistinguishable from control even in a miR-8 heterozygous mutant background (Fig. 2m, n, l).
To determine if miR-SP elements can be used for more sophisticated genetic interaction studies, we exploited a previously characterized microRNA-dependent signaling pathway. MiR-7 negatively regulates the expression of transcription factor Yan during photoreceptor differentiation15. Tissue-specific overexpression of YanACT, a transgenic Yan protein that undergoes reduced turnover, results in a reduced glassy eye phenotype (Supplementary Fig. 1 online, ref. 15) exacerbated by loss of miR-7 function15. Similar to miR-7 depletion15, expression of miR-7SP in a wild type background had no detectable effect on the overall adult eye morphology (Supplementary Fig. 1 online). However, co-expression of YanACT and miR-7SP using the eye-specific driver eyeless-Gal4 strongly enhanced the YanACT eye phenotype (Supplementary Fig. 1 online).
Intrinsic to the Gal4-UAS system and supported by our analysis of three independent microRNAs, this method provides a rapid and straightforward tool for studies in which tissue-specific inhibition may be essential for a comprehensive analysis of microRNA functions.
Defining tissue-specific microRNA site of action using miR-SP
Many microRNAs are expressed during nervous system development suggesting that they control a repertoire of neuronal processes across a diverse range of species17, 18. The Drosophila neuromuscular junction (NMJ) has emerged as a powerful system to study synaptic development, neurotransmission and cell biology19. In a forward genetic screen we identified the conserved Drosophila miR-8 as a candidate regulator of NMJ formation (CSL, CML and DVV, unpublished observations). We assessed third instar larval NMJ morphology in animals expressing miR-8SP under the ubiquitous tubulin-Gal4 driver. Analysis of the well-characterized muscle 6/7 NMJ (Fig. 3 a, b diagrams) revealed a striking phenotype in transgenic animals characterized by reduced size and complexity of the NMJ that could be quantified using three morphological parameters: (i) a decreased number of synaptic boutons, (ii) reduced expansion of the presynaptic terminal arbor, and (iii) fewer axonal branches (Fig. 3d, g). Ubiquitous expression of either two copies of miR-8SP alone, or one copy in a ΔmiR-8 heterozygous background, revealed a significant and dose-dependent decrease in synaptic bouton number (up to 20%, P<0,001; Fig. 3g left graph), NMJ expansion (up to 37%, P<0,001; Fig. 3g middle graph) and NMJ branches (up to 25%, P<0,001; Fig. 3g right graph). Analysis of miR-8 null mutants revealed comparable albeit slightly more robust NMJ morphological phenotypes than ubiquitous miR-8SP expression (Fig. 3e, f, g). These results demonstrate that miR-8 plays an important role in NMJ morphogenesis.
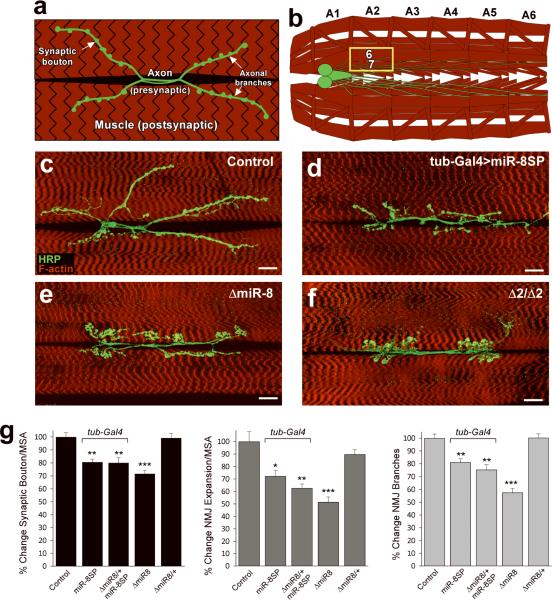
FIGURE 3. Discovery of novel microRNA functions using miR-SP elements.
a,b, Schematic of the Drosophila third instar larval NMJ (a) and ventral body wall musculature (b). Abdominal segments are labeled A1–A6; anterior is left. The presynaptic compartment and CNS are shown in green and postsynaptic compartment in red; muscles 6 and 7 are highlighted in (a) and yellow box in (b). c–f, Immunofluorescence of larval NMJs from the indicated genotypes. All panels show confocal images of NMJ at muscles 6/7 in segment A2 of wandering third instar larvae stained for HRP (green, Horseradish Peroxidase) and F-actin (red). Scale bars are 20 μm. g, Quantification of synaptic bouton number, NMJ expansion and NMJ branch number as percentage of isogenized wild-type control. (*P<0.005; **P<0.001; ***P <10−7; by two-tailed Student's t-test; Error bars: mean ± SEM; n=20 for all genotypes and parameters). For NMJ analysis, statistical significance was determined with respect to the corresponding wild type or ΔmiR-8 heterozygous mutant background. Synaptic bouton numbers and NMJ expansion were normalized to muscle surface area (MSA). In all panels, “>” stands for driver-induced transgene expression.
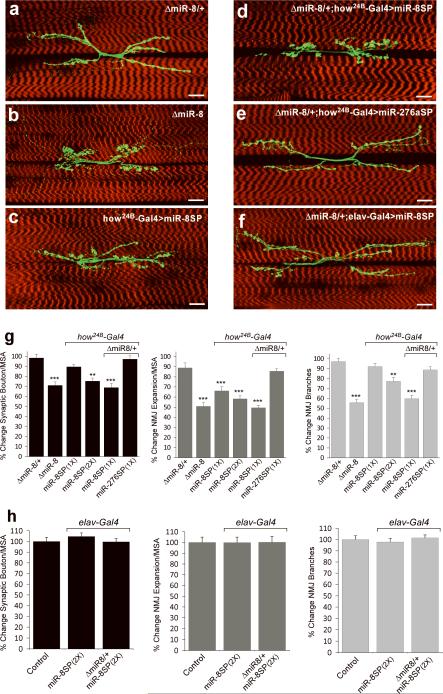
Signaling between pre- and postsynaptic membranes is essential to coordinate NMJ morphogenesis and function20. MiR-8 could potentially regulate NMJ formation by regulating genes on either side of the synapse. This issue can be addressed with miR-SP elements using tissue-specific Gal4 drivers. Expression of miR8-SP using the muscle-specific driver how24B-Gal4 resulted in the same NMJ defects observed with either the tubulin-Gal4 driver or with miR-8 null mutants (Fig. 4b, c, g). The penetrance of this NMJ phenotype increased with the miR-SP copy number indicating dose-dependence (Fig. 4d, g). In contrast, expression of miR-8SP using the strong pan-neural driver elav-Gal4 generated no detectable NMJ phenotype even when one copy of miR-8 was removed (Fig. 4f, h). To control for non-specific effects of miR-SP expression, we generated a miR-SP for an unrelated microRNA, Drosophila miR-276a (miR-276aSP). Expression of miR-276aSP under how24B-Gal4 was indistinguishable from controls even in a miR-8 heterozygous background (Fig. 4e, g). To verify that the NMJ phenotype is caused by a loss of miR-8 function, we asked if expression of transgenic miR-8 could suppress the effect of miR8-SP. Overexpression of miR-8 in muscle results in embryonic lethality, which is suppressed when co-expressed with miR-8SP (Fig. 1g), thus allowing NMJ analysis at late larval stages. Analysis of these larvae revealed rescue of the NMJ defects, confirming that this phenotype is induced by muscle-specific depletion of miR-8 (Supplementary Fig. 2 online). Thus, by virtue of tissue-specific transgenic microRNA inhibition, we find that muscle cells play an essential role in miR-8 mediated control over synaptic development.
FIGURE 4. MiR-SPs define microRNA activity with spatial specificity.
a–f, Immunofluorescence of third instar larval NMJs from the indicated genotypes. Scale bars are 20 μm. g, h, Quantification of synaptic bouton number, NMJ expansion and NMJ branch number as percentage of isogenized wild-type control in a tissue-specific miR-8-modified genetic background in the post- (g) or pre-synaptic (h) compartments. ΔmiR-8/+ samples were statistically indistinguishable from wild-type NMJs and used as control reference for NMJ analysis of Δmir-8 heterozygous backgrounds. Post-synaptic miR-8SP expression results in a dose- dependent NMJ phenotype (g) similar to ΔmiR-8 null animals. In contrast, larvae expressing miR-8SP pan-neurally appeared statistically identical to wild-type controls (h) (Error bars are ± SEM; **P<0.0001; ***P<10−6 by two-tailed Student's t-test; n>17 for all genotypes and parameters)
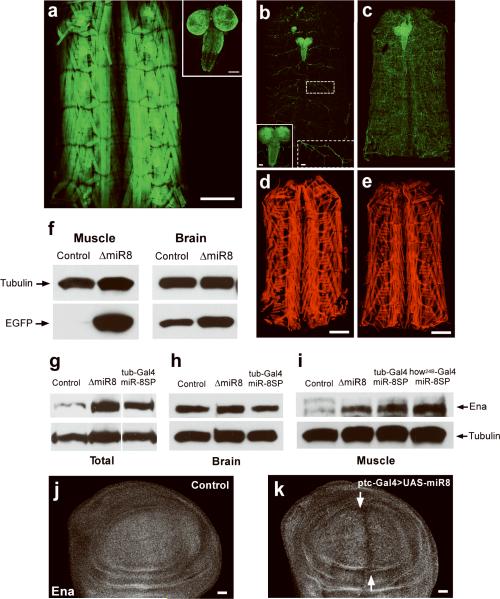
The prevalence of postsynaptic miR-8 activity during NMJ development could reflect restricted miR-8 expression or tissue-specific differences in miR-8 activity. Analysis of EGFP expression driven by a miR-8-Gal4 enhancer trap suggests that miR-8 is broadly expressed in muscle (Fig. 5a) and central nervous system (CNS) (Fig. 5a inset, ref. 16). However, immunofluorescence analysis of wild-type larvae ubiquitously expressing a miR-8-EGFP reporter construct21 showed robust miR-8 activity in the body wall musculature (Fig. 5b, d), and no obvious activity in the CNS (Fig. 5b solid inset) or motor neurons (Fig. 5b dashed inset). Reporter levels were significantly upregulated in the muscle of miR-8 null larvae, confirming that reporter silencing in wild-type body wall musculature was due to endogenous miR-8 activity (Fig. 5c and e). A quantitative analysis of total EGFP levels by Western blotting confirmed nearly complete reporter inhibition in the larval body wall musculature, and also revealed a subtle yet reproducible miR-8 activity in dissociated larval CNS extracts (Fig. 5f and Supplementary Fig. 5 online).
FIGURE 5. MiR-SP elements can uncover tissue-specific microRNA function.
a. EGFP under the control of a MiR-8 Gal4 driver is robustly expressed in the body wall musculature (a) and in the brain and ventral nerve cord (a, inset). b, c Confocal images of TubulinmiR-8-EGPF fluorescence in total dissected larvae (b, c), CNS (b solid inset) and motorneurons (b dashed inset) from wild-type controls compared to ΔmiR-8 mutants (c, e). Scale bars are 50 μm in (b) insets, and 500 μm in (d, e). f, Total miR-8-EGFP levels isolated from fresh muscle and brain extracts of control and ΔmiR-8 mutant animals, assessed by Western blotting using an anti-GFP antibody. g–i, Western blot analysis of Ena and tubulin expression in total dissected third instar larvae (g), CNS extracts (h) or muscle (i), in wild-type larvae (control), or larvae in which miR-8 was inhibited by ubiquitous expression of miR-8SP, or by homozygous miR-8 deletion (ΔmiR-8). Ten larvae were used to obtain the CNS homogenate in (h) and two larvae were used for muscle extracts in (i). j, k Third instar larval wing imaginal discs from wild type (j) and ptc-Gal4/UAS-miR-8 animals (k) immunostained with an Ena antibody. Cells expressing miR-8 along the anterior-posterior boundary, indicated by white arrows display reduced levels of Ena. Scale bars are 20 μm.
Identification of tissue-specific microRNA effectors using miR-SP
To determine if miR-SPs can be used to identify genes that are differentially regulated by microRNAs in a particular tissue, developmental stage or process we sought to uncover the effector(s) underlying miR-8-mediated regulation of NMJ morphogenesis. The 3'UTRs of over 300 genes are predicted to contain one or more miR-8 target sites (TargetScanFly release 5.0). Directly related to neuronal morphogenesis, the actin regulatory protein Enabled (Ena)22 contains one predicted miR-8 target site in the 3'UTR that is highly conserved across Drosophila species (Supplementary Fig. 3 online).
To determine if Ena is a downstream effector of miR-8 during Drosophila larval development, we analyzed Ena levels in miR-8 modified genetic backgrounds. Western blotting of total larval extracts revealed a marked increase in Ena protein levels in tubulin-Gal4:miR-8SP animals compared to control samples, (Fig. 5g and Supplementary Fig. 6 online). MiR-8 null mutants revealed a similar change in Ena levels (Fig. 5g). Given the postsynaptic function revealed by miR-8SP, key miR-8 effectors at the NMJ should also be regulated in muscle. Comparison of dissected muscle and CNS tissues from tubulin-Gal4;miR-8SP and miR-8 null mutant animals demonstrated a substantial increase of Ena levels in the muscle (Fig. 5i), but not in dissociated CNS extracts (Fig. 5h), when compared to controls. This provided an ideal platform to test the capacity of the miR-SP system to reveal tissue-specific microRNA-mediated gene regulation in the intact animal without the need of separating different tissues. Indeed, postsynaptic expression of miR8-SP with how24B-Gal4 resulted in an increase of Ena levels comparable to ubiquitous loss of miR-8 (Fig. 5i). To confirm that miR-8 can repress Ena expression in vivo we analyzed the effect of ectopic miR-8 expression (UAS-miR-8) in Drosophila wing imaginal discs using the ptc-Gal4 driver. Analysis of the wing pouch revealed decreased Ena protein levels in miR-8 expressing cells along the anterior-posterior compartment boundary (Fig. 5 j, k).
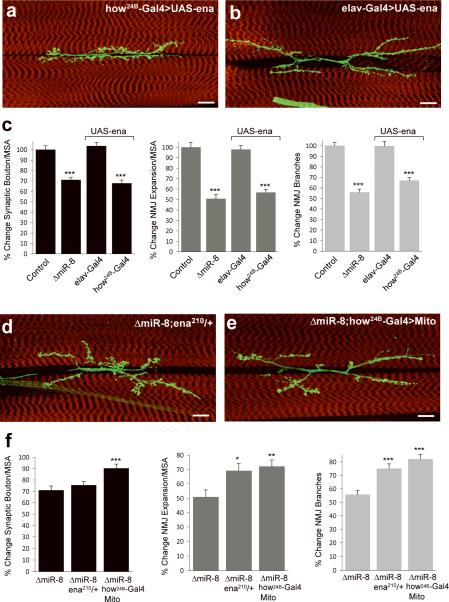
If Ena is a key effector underlying miR-8 function during NMJ development, then increased Ena levels should mimic the miR-8 null phenotype whereas reduced levels of Ena should rescue the miR-8 null induced defects in NMJ growth, in the same tissue-specific pattern. As predicted, postsynaptic Ena overexpression using how24BGal4 elicited the same NMJ phenotype as miR-8 null mutant animals (Fig. 6a, c). In contrast, the NMJs of animals overexpressing Ena under the pan-neural elav-Gal4 driver were indistinguishable from controls (Fig. 6b, c). Conversely, removing one copy of ena by a loss-of-function allele (ena210) resulted in a partial yet consistent rescue of miR-8 null NMJ defects despite the presence of one wild type ena allele (Fig. 6d, f). However, expression of UAS-FP4-mito, a construct that renders endogenous Ena protein inert by sequestering it to mitochondria and thus mimicking an Ena null mutant23, results in a robust rescue of all miR-8 null NMJ phenotypic parameters (Fig. 6e, f). Neither removal of one copy of ena nor postsynaptic overexpression of UAS-FP4-mito display any detectable NMJ phenotypes in a wild-type genetic background (Supplementary Fig. 4 online). Taken together, these results indicate that the miR-SP technology is an effective tool to dissect the tissue-specific logic of microRNA effector regulation.
FIGURE 6. Genetic dissection of Ena function confirms miR-8-mediated postsynaptic regulation of NMJ morphogenesis.
Analysis of NMJs at muscles 6/7 in larvae over-expressing UAS-ena under the muscle-specific how24B-Gal4 driver (a, c). The NMJs of larvae expressing UAS-ena pan-neurally under the control of the elav-Gal4 driver appear indistinguishable from wild-type controls (b, c). Reducing Ena levels by a heterozygous loss-of-function allele (d) or suppressing Ena activity postsynaptically by expressing UAS-FP4-mito (e) under the muscle-specific driver how24B-Gal4. Scale bars are 20 μm. Quantitatively, the synaptic bouton phenotype was rescued by 15 and 63%, NMJ expansion by 37 and 58%, and the reduction in branch number by 43 and 67% in the ena210 heterozygous and how24B-Gal4:UAS-FP4-mito backgrounds, respectively(f). (Error bars are ± SEM; for (c) ***P<10−6 relative to control; for (f) *P<0.005; **P<0.001; ***P<0.0001 relative to ΔmiR8 homozygous animals by two-tailed Student's t-test; n>17 for all genotypes and parameters).
DISCUSSION
Our transgenic miR-SP technology utilizes the modular Gal4-UAS system11 to achieve consistent inhibition of microRNA function from the entire living organism to a single tissue, organ or cell type. Thus, Drosophila miR-SP is the first versatile technology that allows transgenic microRNA silencing with precise spatial resolution at any stage of organismal life. miR-SPs can be used to rapidly identify microRNA phenotypes in multiple contexts, thus providing an efficient alternative to synthetic chemically modified oligonucleotide inhibitor approaches or classical genetic knockouts, both of which still present significant limitations9. In addition, as demonstrated by our studies in eye, wing, leg, muscle and CNS, miR-SPs will allow researchers to define the full repertoire of microRNA functions across a complex spectrum of developmental, physiological and behavioral processes. Moreover, because the levels of transgene expression can be precisely controlled, miR-SPs are also powerful tools to investigate genetic interactions between microRNAs and their functional partners in vivo. Given the extensive collection of different Gal4 driver strains, and the ease of generating complete miR-SP libraries, this technology will allow biologists to rapidly explore the entire landscape of microRNA functions in living organisms.
We uncovered the cellular and molecular logic with which miR-8 acts to promote presynaptic growth by limiting postsynaptic expression of Ena. Despite expression on both sides of the synapse, tissue-specific depletion shows that postsynaptic miR-8 activity is required for NMJ development. Presynaptic expression of miR-8SP showed no effects, suggesting either that presynaptic miR-8 depletion is incomplete, or that miR-8 is intrinsically less active in larval motoneurons than in muscle cells. Although an in vivo measure of presynaptic miR-8 depletion is needed to rule out the former, two observations support the latter hypothesis for the effector Ena: (i) decrease of miR-8 expression using null mutations does not alter Ena levels in the CNS, and (iii) elevation of Ena levels in neurons does not emulate miR-8 NMJ defects. These findings imply that cell type-specific mechanisms influence miR-8 regulation of ena.
The analysis using ptc-Gal4 for miR-9a and how24B-Gal4 for miR-8 illustrates that the miR-SP technology can be used to genetically dissect the regulation of microRNA target genes or effectors. Combined with quantitative genome and proteome-wide expression analysis, miR-SPs can yield essential information regarding in vivo genes that are differentially regulated in a particular tissue, developmental stage or process.
Given that modular and conditional expression systems have been developed in other model organisms10, 24, the utility of the miR-SP technology is not restricted to flies. In addition to a recent modification of the Cre-loxP system to achieve tissue-specific gene expression in mice10, recent studies reported the use of Gal4-UAS bipartite modular expression in zebrafish24. The extensive collections of tissue-specific Cre mice25, and the recent generation of zebrafish Gal-4 libraries24, will provide a platform for miR-SP-mediated tissue-specific inhibition of microRNA function in these higher organisms.
ONLINE METHODS
Drosophila Genetics
All stocks were maintained and crossed at 25°C according to standard procedures. Stocks were obtained from the Bloomington Stock Center unless otherwise specified. The following loss-of-function alleles and transgenic lines were used: GMR-YanAct (ref. 15, 26) was a gift from R.W. Carthew, UAS-FP4-mitoEGFP (ref. 23) was a gift from M. Peifer, ena210 and UAS-ena (ref. 27) were gifts from F. M. Hoffmann, miR-9aJ22 and miR-9aE39 (ref. 14) was a gift from F.B. Gao; miR-8-GFP Sensor (ref. 21) was a gift from H. Ruohola-Baker, EP-atro, and P{EPgy2}GugEY14339. miR-8Δ2 (ref. 16) was a gift from S. Cohen; an independent miR-8 null allele ΔmiR-8 was generated as part of a separate study in our laboratory (C.S.L., C.M.L. and D.VV. unpublished observations), tubulinEGFP nerfin-1 3'UTR reporter12 was a gift from J. Brennecke. For the analysis of miR-9a activity in wing imaginal discs, the tubulinEGFP nerfin-1 3'UTR reporter was recombined with ptc-Gal4 on the second autosomal chromosome. The following Gal4 drivers were used to drive ubiquitous, eye, leg, wing disc, pan-neural and mesodermal expression: tubulin-Gal4, GMR-Gal4 and eyeless-Gal4, Dll-Gal4, ptc-Gal4, elav-Gal4 and how24B-Gal4 respectively. The miR-8-Gal4 line was obtained from the Drosophila Genetic Resource Center (DGRC) and used to drive expression of UAS-CD8GFP.
To generate the allelic combination of ena heterozygous/ΔmiR-8 homozygous genetic background, the ena210 allele was recombined with ΔmiR-8 allele on the second autosomal chromosome and the miR-8 NMJ phenotypes were assessed as described above. To test the effect of postsynaptic Ena inhibition on ΔmiR-8 induced NMJ phenotype, UAS-FP4-mito was expressed using the how24B-Gal4 driver, in a ΔmiR-8 homozygous mutant background. The specificity of the UAS-FP4-mito has been previously described23.
Molecular Biology
For the sequences of all DNA primers and miR-SP constructs used in this study see Supplementary tables. Each miR-SP construct was generated by introducing ten repetitive microRNA complementary sequences separated by a variable four-nucleotide linker into the 3'UTR of EGFP. Each sequence was designed with a mismatches at positions 9–12 as previously described8, and the entire cassette was cloned into the pUAST-EGFP vector according to company protocols (Bio Basic Inc). Transgenic animals carrying one or two copies of the miR-SP cassette were obtained following P element transposon-mediated genomic integration (BestGene Inc).
To generate the miR-8 overexpression construct (UAS-miR-8) ~450 bp of genomic DNA including the miR-8 hairpin were amplified by PCR and cloned into the pUAST vector28.
Immunohistochemistry and Quantitation of NMJ Morphology
Wandering third instars were dissected in Ca2+-free saline and fixed in 4% paraformaldehyde for 30 min, except for Ena immunostaining which required a fixation in a mix of 37% formaldehyde and 10–15% methanol for 3 min. Fixed larvae were washed 3 times in PBS, 0.1% Triton-X100 (PBT) and blocked for 1 h in 5% NGS in PBT. The following primary antibodies were used for immunohistochemistry: anti-HRP 1:1000 (Jackson ImmunoResearch), anti-Ena 1G6 (1:4) (a gift from F.M. Hoffmann) and FITC-conjugated anti-GFP 1:1500 (Abcam). F-actin was visualized using Alexa Fluor® 633 phalloidin 1:400 (Invitrogen). Secondary antibodies Alexa Fluor-488 and -568 (Invitrogen) were used at a 1:400 dilution. RP3 and MN6/7b terminals of muscle 6 and 7 in the abdominal segment A2 of wandering third instar larvae were used for the quantification of all morphological parameters. This analysis was carried out using a Zeiss Axioplan2 microscope and a Hamamatsu ORCA wide-field digital camera. Three parameters were used to assess NMJ morphology: (i) Synaptic boutons were counted as previously described29. (ii) NMJ expansion was quantified using the Elements (Versions 2.3–3.0) Nikon Imaging Software by tracing a polygon along the axonal arbor where each vertex consists of a NMJ terminal branch point. Pixel number was then converted to μm, and the area enclosed by the traced figure was measured. Both bouton number and NMJ expansion were normalized to muscle surface area. (iii) Branch count was defined as the number of branch tips per NMJ. To avoid potential biased interpretation, all experiments were blinded to genotype.
Confocal and Epifluorescence Microscopy
Confocal microscopy was performed using a Zeiss LSM 510 META upright microscope. Max-intensity projections were obtained using the Zeiss LSM Software package. For the miR-8GFPsensor experiments, control and ΔmiR-8 samples were dissected and fixed simultaneously under identical conditions. Prior to acquisition, laser parameters were adjusted to obtain non-saturating conditions. Confocal images of the entire third instar body wall musculature and CNS were taken at 5× magnification using identical confocal acquisition parameters (laser intensity, gain and pinhole settings). 2×2 grids were captured and automatically rendered by the Zeiss LSM Multi-Time application. Epifluorecent images of miR-8-Gal4 driving UAS-CD8GFP were taken with a Zeiss Stemi SV6 microscope and 1XHRD 100-NIK digital camera (Diagnostic Instruments Inc.) and processed using OpenLab 5.0.1 software.
Bioinformatics
The TargetScan algorithm30 was used to identify ena as a conserved miR-8 target. Sequence alignment of miR-8 and the ena 3'UTR was obtained using the Bibiserv RNAhybrid algorithm31 (http://bibiserv.techfak.unibielefeld.de/rnahybrid/submission.html)
Biochemistry
To obtain total brain and body wall musculature homogenates, wandering third instar larvae were filleted in dissection buffer (PBS, 1 mM EGTA, 1× Complete mini protease inhibitor cocktail (Roche)), transferred to 25 μl of lysis buffer (PBS, 0.05% Tween, 1 mM EGTA, 1× Complete mini protease inhibitor cocktail), homogenized and boiled for 10 min in 1× SDS Sample buffer (Sigma). To dissociate the CNS from the muscle tissue, wandering third instar larvae were filleted in dissection buffer and the CNS was carefully removed from the body wall musculature. 10 brains and 2 muscle samples were homogenized in lysis buffer as described above. Samples were loaded into 10% SDS-PAGE gels (Lonza), and analyzed by immunoblotting according to standard protocols. The following primary antibodies were used for Western blotting: mouse anti-Ena 5G2 (DSHB) 1:20, rabbit anti-tubulin (Abcam) 1:50,000 and mouse anti-GFP (Living Colors Monoclonal Antibody (JL-8; Clontech) 1:50,000. HRP-conjugated anti-rabbit and anti-mouse secondary antibodies (Jackson ImmunoResearch) were used at a 1:10,000 dilution.
Supplementary Material
Acknowledgements
We thank Drs. Michael Greenberg, Norbert Perrimon, Thomas Schwarz, Mel Feany and John Flanagan for critical feedback on the manuscript and Ms. Margaret Ebert for advice on designing the miR-SP constructs and for comments on the manuscript. We thank Ms. Vasanthi Sridhar and the Harvard NeuroDiscovery Center Optical Imaging program for technical assistance. We are grateful to Drs. R.W. Carthew, F.B. Gao, M. Peifer, F. M. Hoffmann, H. Ruohola-Baker, J. Brennecke and S. Cohen for providing critical reagents and strains. We thank the Developmental Studies Hybridoma Bank (DSHB) at the University of Iowa, Department of Biological Sciences for antibodies. C.M.L, C.S.L and D.VV. were supported by a grant from the National Institutes of Health (NS40043). T.A.F. was supported in part by a fellowship from the John Douglas French Alzheimer's Foundation.
Footnotes
The authors do not have any competing financial interests
Author contributions TAF conceived and designed the miR-SP technology. CML and TAF designed and performed the experiments described in the study, analyzed the data and interpreted the results. CSL generated the miR-8 null mutant and independent of this study performed preliminary NMJ analysis and proposed Ena as a candidate miR-8 effector. As principal investigator of the lab DVV supervised the study and provided material and salary support for CML. CSL and TAF. The text and figures were drafted by TAF, and were then edited by TAF, DVV and CML.
Note: Supplementary Information is available on the Nature Methods website
References
- 1.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 3.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci U S A. 2005;102:18017–22. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genet. 2008;4:e22. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silver SJ, Hagen JW, Okamura K, Perrimon N, Lai EC. Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc Natl Acad Sci U S A. 2007;104:18151–6. doi: 10.1073/pnas.0706673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–85. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 10.Stern P, et al. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci U S A. 2008;105:13895–900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott DA, Brand AH. The GAL4 system : a versatile system for the expression of genes. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- 12.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 13.Kuzin A, Kundu M, Brody T, Odenwald WF. The Drosophila nerfin-1 mRNA requires multiple microRNAs to regulate its spatial and temporal translation dynamics in the developing nervous system. Dev Biol. 2007;310:35–43. doi: 10.1016/j.ydbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–77. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–45. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 17.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 18.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–61. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Keshishian H, Kim YS. Orchestrating development and function: retrograde BMP signaling in the Drosophila nervous system. Trends Neurosci. 2004;27:143–7. doi: 10.1016/j.tins.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Shcherbata HR, et al. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell. 2007;1:698–709. doi: 10.1016/j.stem.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drees F, Gertler FB. Ena/VASP: proteins at the tip of the nervous system. Curr Opin Neurobiol. 2008;18:53–9. doi: 10.1016/j.conb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gates J, et al. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–39. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- 24.Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50:391–9. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 25.Sauer B, Henderson N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989;17:147–61. doi: 10.1093/nar/17.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 27.Comer AR, Ahern-Djamali SM, Juang JL, Jackson PD, Hoffmann FM. Phosphorylation of Enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein-protein interactions of Enabled. Mol Cell Biol. 1998;18:152–60. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 29.Johnson KG, et al. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006;49:517–31. doi: 10.1016/j.neuron.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.