Abstract
Uncoupling protein 3 (UCP3) is a member of the mitochondrial solute carrier superfamily that is enriched in skeletal muscle and controls mitochondrial reactive oxygen species (ROS) production, but the mechanisms underlying this function are unclear.
Aims
The goal of this work focused on the identification of mechanisms underlying UCP3 functions.
Results
Here we report that the N-terminal, intermembrane space (IMS)-localized hydrophilic domain of mouse UCP3 interacts with the N-terminal mitochondrial targeting signal of thioredoxin 2 (Trx2), a mitochondrial thiol reductase. Cellular immunoprecipitation and in vitro pull-down assays show that the UCP3–Trx2 complex forms directly, and that the Trx2 N-terminus is both necessary and sufficient to confer UCP3 binding. Mutation studies show that neither a catalytically inactivated Trx2 mutant, nor a mutant Trx2 bearing the N-terminal targeting sequence of cytochrome c oxidase (COXMTS-Trx2) bind UCP3. Biochemical analyses using permeabilized mitochondria, and live cell experiments using bimolecular fluorescence complementation show that the UCP3–Trx2 complex forms specifically in the IMS. Finally, studies in C2C12 myocytes stably overexpressing UCP3 (2.5-fold) and subjected to Trx2 knockdown show that Trx2 is required for the UCP3-dependent mitigation of complex III-driven mitochondrial ROS generation. UCP3 expression was increased in mice fed a high fat diet, leading to increased localization of Trx2 to the IMS. UCP3 overexpression also increased expression of the glucose transporter GLUT4 in a Trx2-dependent fashion.
Innovation
This is the first report of a mitochondrial protein–protein interaction with UCP3 and the first demonstration that UCP3 binds directly, and in cells and tissues with mitochondrial thioredoxin 2.
Conclusion
These studies identify a novel UCP3–Trx2 complex, a novel submitochondrial localization of Trx2, and a mechanism underlying UCP3-regulated mitochondrial ROS production. Antioxid. Redox Signal. 15, 2645–2661.
Introduction
Mitochondria generate the vast majority of cellular reactive oxygen species (ROS), but the complex mechanisms that govern ROS production are not fully understood. ROS generation mainly involves the slippage of electrons (derived from substrate oxidation) from their ubisemiquinone binding sites in complexes I and III, leading to the nonenzymatic reduction of molecular oxygen, to superoxide at topologically distinct sites in the mitochondrial matrix (complex I) and intermembrane space (complex III) (2, 32). Once formed, superoxide is subsequently converted by superoxide dismutases to the more lipid soluble and more reactive form hydrogen peroxide (H2O2). H2O2 can be further reduced to highly destructive hydroxyl radical species nonenzymatically in the presence of heavy metals (32). Under normal conditions, basal levels or inducible “bursts” of ROS generation are implicated in a variety of physiological cell signaling functions related to cell growth, differentiation, and survival (2). On the other hand, mitochondrial dysfunction induced by or in association with a variety of pathologic conditions including hypoxia, cancer (21, 23), aging (30), hyperglycemia (diabetes) (6), and chemical mitochondrial respiratory inhibition (49) can lead to a pathological increase in ROS generation and acceleration of age-related disease.
Uncoupling proteins (UCPs) comprise a subfamily of mitochondrial inner membrane anion carrier proteins that function prominently in the regulation of membrane proton conductance (1). UCP-dependent proton leak increases respiration and substrate oxidation that is uncoupled from ADP phosphorylation, and in brown fat and muscle is associated with thermogenesis (7). The prototypical UCP1 protein is largely, if not exclusively, expressed in brown fat and plays a critical role in mammalian thermoregulatory physiology in rodents, hibernators, and newborns; UCP1 is an indispensable mediator of heat production in response to cold and perhaps overfeeding (28). The close UCP1 homolog UCP3 was first identified in 1997 and is predominantly expressed in skeletal muscle, heart, and brown adipose tissue (5). UCP3 shares ∼72 percent amino acid sequence homology with UCP1, which suggests that it likely has at least some overlapping biochemical properties and functions with UCP1 (25). Despite intensive investigation, however, the physiological functions and underlying mechanisms of UCP3 action are still largely unknown. UCPs are regarded as the major, if not most important, regulators of de novo mitochondrial ROS generation, in part because of the well established relationship between the mitochondrial membrane potential and ROS production (25).
A growing body of evidence implicates UCP3 in the inhibition of mitochondrial oxidative damage, either by facilitating the export of unmetabolizable or oxidized mitochondrial fatty acids or by decreasing the generation of mitochondrial ROS (16, 43). Indeed, mitochondrial ROS production and irreversible protein oxidation (carbonylation) in response to various oxidative stress conditions have been demonstrated to be decreased in UCP3 overexpressing L6 and C2C12 muscle cells, respectively (3, 29). In animal studies, relative to wild-type mice, those lacking UCP3 had increased levels of oxidative damage markers and decreased activity of aconitase, a tricarboxylic acid enzyme with a defined superoxide-inhibited iron–sulfur cluster (51). Recent studies have demonstrated that UCP3 actively lowers the rate of ROS production in isolated energized skeletal muscle mitochondria in the absence of exogenous activators (48). However, the role of UCP3 in the regulation of mitochondrial membrane potential in muscle under physiological conditions is controversial, and the precise mechanism(s) by which UCP3 controls mitochondrial oxidant generation is unclear. Moreover, the paucity of information regarding possible post-translational modifications and protein–protein binding partners for the UCP family of proteins in general has impeded our understanding of their biochemical functions and mechanisms of action.
Thioredoxins are thiol reductase enzyme components of a major pathway mediating protein thiol reduction and ROS scavenging in cells (11). Thioredoxin 2 (Trx2) is a relatively oxidant resistant, mitochondrially-localized member of the thioredoxin family that is essential for survival in mice (12, 46). Trx2 has been demonstrated to block TNF-α-induced mitochondrial ROS generation in muscle (17), and heterozygous Trx2-deficient mice show impaired mitochondrial function and increased oxidative stress (37). Trx2 has a classical N-terminally-cleaved, 59 amino acid mitochondrial targeting signal (MTS) that directs its localization to the mitochondrial matrix (12). In the present study, we report that UCP3 binds with nonprocessed Trx2 in the mitochondrial intermembrane space (IMS) when overexpressed and at endogenous concentrations in vivo, and directly in vitro. This interaction with UCP3 directs Trx2 processing, submitochondrial localization, and antioxidative functions within the IMS compartment. The interaction surface between UCP3 and Trx2 was defined mutationally and consists of the N-terminal amino acids (1–16) of UCP3 and the N-terminal MTS of Trx2. Finally, the data show that the attenuation of mitochondrial H2O2 generation mediated by UCP3 is lost after Trx2 knockdown. These data illuminate a novel mechanism for the control of mitochondrial ROS generation involving a complex formed between Trx2 and UCP3 in the mitochondrial IMS.
Results
UCP3 associates with nonprocessed Trx2, but not processed Trx2 in mitochondria
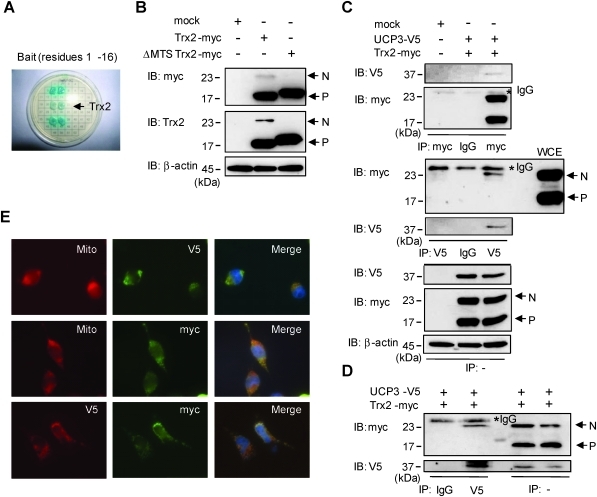
It has been reported that UCP3 protects cells against oxidative damage by attenuating the mitochondrial production of reactive oxygen species, but the mechanisms underlying UCP3-dependent control of ROS generation are unclear (16, 48). To identify novel UCP3 regulatory mechanisms, yeast two-hybrid screens were performed using a heart cDNA library with bait DNA coding sequences corresponding to the seven hydrophilic domains of the mouse UCP3 protein (subcloned into the pGBKT7 yeast expression vector, Clontech). We identified full-length Trx2 as a candidate binding partner with the N-terminal UCP3 hydrophilic domain consisting of amino acids 1–16 (Fig. 1A). In the first described characterization, Damdimopoulos and colleagues observed that in vitro translated rat Trx2 was approximately 20 kDa, and observed that rat mitochondrial lysates contained a processed, ∼14 kDa isoform (12). Similarly, after overexpression of full-length mouse Trx2-myc in transfected HeLa cells, both anti-myc (C terminal tag) and anti-Trx2 antibodies recognized both processed and nonprocessed isoforms (∼17 and ∼23 kDa, respectively) in whole cell lysates (Fig. 1B, middle lane). Furthermore, a Trx2 mutant lacking the putative 59 amino acid targeting signal (ΔMTS Trx2-myc) was also recognized by anti-myc and anti-Trx2 antibodies and migrated closely to the processed form of native Trx2 (Fig. 1B, right lane). Cell lysates from empty vector transfectants were used as negative controls for antibody specificity (mock, Fig. 1B, left lane). To confirm the yeast two-hybrid results, we performed co-immunoprecipitation (IP) assays in HeLa and C2C12 cells. Unlike in mock transfectants, in cells co-expressing UCP3-V5 and Trx2-myc, anti-myc antibody (Trx2-myc), but not IgG control, co-immunoprecipitated UCP3-V5 (IB: anti-V5) (Fig. 1C, upper panel). Interestingly, when the reverse IP was performed with anti-UCP3-V5 followed by immunoblotting for captured Trx2-myc, UCP3 was found to interact specifically with non-processed (N) Trx2 but not with the processed (P) form (Fig. 1C, panel 3). Expression of the transfected proteins and equal protein loading was confirmed by immunoblotting for V5, myc, and β-actin in whole cell lysates from the indicated transfectants (Fig. 1C, lower three panels). The UCP3-Trx2 complex could also be detected in co-transfected C2C12 myoblasts (Fig. 1D). We also performed immunostaining in HeLa cells co-transfected with UCP3 and Trx2. As expected, both UCP3-V5 and Trx2-myc each co-localized (Merge) in a punctate, perinuclear fashion with the mitochondrial dye Mitotracker red (Mito) (Fig. 1E, upper and middle panels). Moreover, co-immunostaining for UCP3 (V5) and Trx2 (myc) shows that the expressed proteins interact in an overlapping mitochondrial pattern (Fig. 1E, lower panels).
FIG. 1.
Interaction of UCP3 and thioredoxin 2 (Trx2). (A) Trx2 was identified as a UCP3 interacting partner by yeast two-hybrid analysis. The coding sequences for the hydrophilic amino acid residues 1–16 of the UCP3 gene were cloned into the pGBKT7 vector and used as bait. With a heart cDNA library used as prey, three potential UCP3 binding candidates were identified by yeast growth, with the arrow representing the Trx2-positive colony. (B) HeLa cells were transfected with empty vector (mock), myc-tagged Trx2 (Trx2-myc), and myc-tagged Trx2 with a truncated MTS (ΔMTS Trx2-myc). Cell extracts (20 μg/lane) were subjected to SDS-PAGE, followed by immunoblotting (IB) with anti-myc, anti-Trx2, and anti-β-actin antibodies. N, nonprocessed Trx2; P, processed Trx2. (C and D) HeLa (C) and C2C12 (D) cell extracts (100 μg) transfected with Mock, Trx2-myc, and V5-tagged UCP3 (UCP3-V5) were immunoprecipitated (IP) with anti-IgG, anti-myc, or anti-V5 antibodies and analyzed by immunoblotting to detect V5 (UCP3) and myc (Trx2). β-Actin was used as an internal standard. WCE, whole cell extracts. *indicates the nonspecific IgG band on the immunoblot. (E) HeLa cells transfected with UCP3-V5 (top) or Trx2-myc (middle) were immunostained with V5 (green) or myc (green) antibodies and co-stained with the mitochondrial indicator MitoTracker Red (red), as indicated. HeLa cells cotransfected with UCP3-V5 and Trx2-myc (bottom) were co-immunostained with V5 (red) and myc (green) antibodies. The overlay shows that UCP3 co-localized with Trx2. IB, IP, and microscopy experiments are representative of at least three independent studies. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
The Trx2 active site cysteines are dispensable for UCP3 binding
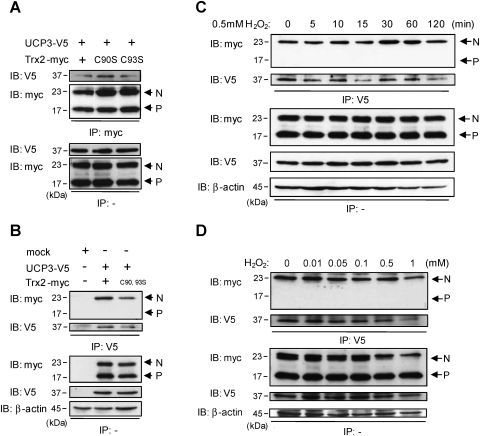
Trx2 has been shown to play a crucial role in the scavenging of ROS in mitochondria and in regulation of the mitochondrial apoptosis signaling pathway (47). Trx2 canonically mediates the reduction of target protein thiols through the conserved thioredoxin active site motif Trp-Cys-Gly-Pro-Cys, which binds and reduces oxidized cysteine residues in proteins such as peroxiredoxin-3 and ASK1 (53, 54). Full-length UCP3 contains seven cysteine residues; one in the IMS region, five in the inner membrane domains, and one on the matrix side (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/ars), suggesting the possibility that it could interact with the active site of Trx2. To determine whether the interaction between UCP3 and Trx2 requires the catalytic cysteine residues of Trx2, we performed immunoprecipitation experiments in cells expressing either single or double active site cysteine residue Trx2 mutants (C90S–Trx2, C93S–Trx2 and C90, 93S–Trx2) along with wild-type UCP3 or empty vector (mock). Unlike with cells transfected with empty vector, wild-type, C90S, and C93S Trx2 mutants immunoprecipitated UCP3-V5 (Fig. 2A, IP: myc, IB: V5), and similarly, full-length UCP3-V5 specifically immunoprecipitated only the nonprocessed versions (N) of the wild-type and C90S/C93S double mutant Trx2-myc (Fig. 2B, IP: V5, IB: myc). Consistent with the idea that the Trx2 active site is not involved in UCP3 binding, neither treatment of cells with 0.5 mM H2O2 over a 2-hour time course, nor varying doses of H2O2 influenced the interaction between UCP3 and nonprocessed Trx2 (Figs. 2C and 2D, IP: V5, IB:myc). These results suggest that the Trx2:UCP3 interaction is atypical and may not be mediated by the redox-sensitive Trx2 active site.
FIG. 2.
Effects of Trx2 active site mutations and oxidative stress on the interaction of UCP3 and Trx2. (A and B) HeLa cells were transfected with mock, UCP3-V5, Trx2-myc, C90STrx2-myc, C93STrx2-myc, or the double active site mutant (C90, 93S Trx2-myc) as indicated. Cells extracts (100 μg) were immunoprecipitated (IP) with anti-myc and anti-V5 antibodies and analyzed by immunoblotting (IB) with anti-V5 and anti-myc antibodies, respectively. (C and D) Cells transfected with UCP3-V5 and Trx2-myc were treated with hydrogen peroxide (H2O2) (0.5 mM) for 0–120 min (C) or 0.1 mM H2O2 for 120 min (D) and cells were immnoprecipitated (IP) with anti-V5 antibodies and analyzed by IB with anti-myc antibodies. Equal protein loading was determined by immunoblotting for β-actin. N, nonprocessed Trx2; P, processed Trx2. IB, IP experiments are representative of at least three independent studies.
The Trx2-UCP3 interaction is mediated by the N-termini
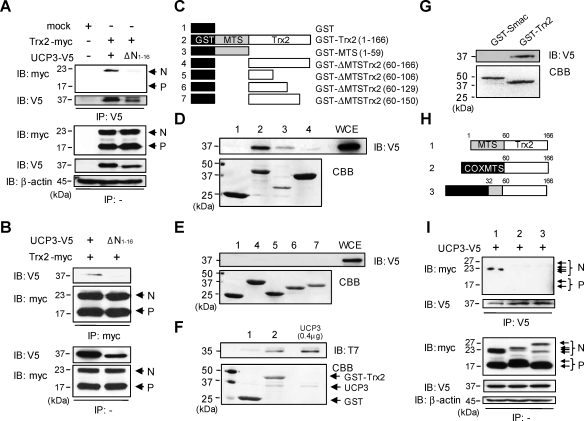
As shown in Figure 1, the N-terminal 16 amino acids of UCP3 were predicted to mediate complex formation with Trx2. To test the role of the UCP3 N-terminus in Trx2 binding, we constructed an N-terminal truncation mutant of UCP3 (UCP3 ΔN1–16) that exhibits appropriate mitochondrial localization (Supplementary Fig. S2A). Unlike full-length UCP3, the UCP3 ΔN1–16 truncation mutant lost the ability to bind nonprocessed Trx2 (Fig. 3A, IP:V5, IB:myc; Fig. 3B, IP:myc, IB:V5). These results confirm yeast two hybrid data and indicate that the UCP3 N-terminus is required for Trx2 binding.
FIG. 3.
Mapping the interacting domains of UCP3 and Trx2 in cells and in vitro. (A and B) The N terminal residues of UCP3 required for binding to Trx2 were defined using mutagenesis and immunoprecipitations. HeLa cells were transfected with full-length UCP3 and truncated mutants (residues1–16, UCP3 ΔN1–16). Cells extracts (100 μg) were immunoprecipitated (IP) with anti-myc antibodies and analyzed by immunoblotting (IB) with anti-V5 antibodies. (C) Diagram for expression constructs of (1) GST, (2) GST-Trx2 (residues 1–166), (3) GST-MTS alone (Trx2 residues 1–59), (4) GST-MTS truncation mutant (ΔMTS-Trx2, residues 60–166), (5) GST-ΔMTSTrx2 (residues 60–106), (6) GST-ΔMTSTrx2 (residues 60–129) and (7) GST-ΔMTSTrx2 (residues 60–150). (D and E) The residues of Trx2 required for direct binding to UCP3 were determined with GST pulldown assays. GST-fusion proteins were purified and stained by Coomassie brilliant blue (CBB) as indicated. In HeLa cell extracts expressing UCP3-V5, the ability of the truncated Trx2 fusion proteins to bind to UCP3 was analyzed by immunoblotting against V5. WCE, whole cell extracts. (F) Direct protein interactions were determined by incubating purified recombinant T7-tagged UCP3 (1 μg) with GST-Trx2 or GST. The samples from the GST pulldown assay were subjected to SDS-PAGE and immunoblotting against T7. Purified UCP3 (0.4 μg) protein was used as a positive control. (G) The specificity of the interaction between UCP3 and the Trx2-MTS was determined using GST-Smac, another mitochondrial intermembrane space protein containing a mitochondrial targeting signaling sequence. GST-fusion proteins were purified and stained by CBB. The ability of the proteins to bind to UCP3 in HeLa cell extracts expressing UCP3-V5 was analyzed by immunoblotting against V5. (H) Diagram for expression constructs of (1) Trx2 (residues 1–166), (2) COXMTS–Trx2Δ1–59 (residues 60–166), and (3) COXMTS–Trx2Δ1–31 (residues 32–166). COXMTS derived from the subunit VIII of human cytochrome c oxidase. (I) Cell extracts (100 μg) all transfected with UCP3-V5 plus (1) Trx2 (residues 1–166), (2) COXMTS–Trx2Δ1–59 (residues 60–166) or (3) COXMTS–Trx2Δ1–31 (residues 32–166) were immunoprecipitated (IP) with anti-V5 antibodies and analyzed by IB with anti-myc antibodies. N, nonprocessed Trx2; P, processed Trx2. IB, IP, and pulldown experiments are representative of at least three independent studies.
Since UCP3 was shown to bind specifically to nonprocessed Trx2, we constructed, bacterially expressed, and purified a variety of N- and C-terminal serial truncation mutants of Trx2 in frame with N-terminal glutathione S transferase (GST) using glutathione beads (constructs are shown in Fig. 3C). Importantly, bacteria lack the ability to process mammalian MTS-containing proteins. We tested the ability of GST, wild-type Trx2, and its mutant derivatives to bind UCP3-V5 from transfected whole cell extracts (WCE) using GST pull-down assays. As revealed by UCP3 immunoblotting, unlike GST alone (construct 1) both full-length Trx2 (GST–MTS–Trx2, construct 2) and the Trx2 N-terminus alone (GST–MTS, construct 3) bound to full-length UCP3 (Fig. 3D, upper panel, constructs 1–4, IB: V5); however, a Trx2 mutant lacking the Trx2MTS amino acids 1–59 (MTS, construct 4) was unable to do so. Shown also is a positive control (WCE) for UCP3 immunoreactivity (Fig. 3D, WCE, IB:V5) and a Coomassie brilliant blue-stained gel (CBB) to confirm construct expression (Fig. 3D, lower panel, CBB). Similarly, none of the Trx2 mutants lacking the MTS (GST–ΔMTS–Trx2) bound UCP3 (Fig. 3E, upper panel, constructs 4, 5, 6, 7), despite similar expression levels (Fig. 3E, lower panel, CBB).
To test whether these interactions are direct, we performed in vitro binding reactions using purified recombinant GST, GST-Trx2, and T7-tagged UCP3 proteins (expression shown in Fig. 3F, CBB, lanes 1 and 2). As shown in Figure 3F, lane 2, recombinant GST-Trx2 (containing the MTS) bound efficiently to full length UCP3-T7 (IB: T7). In this assay, we also confirmed whether UCP3 binding is specific for the MTS of Trx2 (as opposed to all MTS) using GST-Smac, another mitochondrial intermembrane space protein that contains a mitochondrial targeting signal. As expected, UCP3 is unable to interact with Smac, unlike Trx2 (Fig. 3G). To further characterize the Trx2 MTS interaction with UCP3 observed in GST pull-down assays, we performed domain-swapping experiments where either the entire Trx2 MTS (59 amino acids) or only the first half (31 amino acids) of the MTS was replaced with tandem repeats of the MTS from subunit VIII of human cytochrome c oxidase (Fig. 3H, construct 2, COXMTS-Trx2Δ1–59; construct 3, COXMTS–Trx2Δ1–31). The COXMTS strongly targets proteins to the mitochondrion in mammalian cells (40). As expected, COXMTS-Trx2Δ1–59 and COXMTS–Trx2Δ1–31 correctly localized to mitochondria when expressed in mammalian cells (Supplementary Figs. S2B and S2C). However, unlike full-length wild-type Trx2, which interacted with UCP3 (Fig. 3I, construct 1, IP:V5, IB: myc), neither COXMTS-Trx2Δ1–59 nor COXMTS–Trx2Δ1–31 interacted with UCP3 in HeLa cells (Fig. 3I, constructs 2 and 3). Combined, these results indicate that MTS amino acids 1–31 of Trx2 are both necessary and sufficient for direct binding to UCP3.
Full-length but not processed Trx2 interacts with UCP3 at the IMS side of the inner mitochondrial membrane
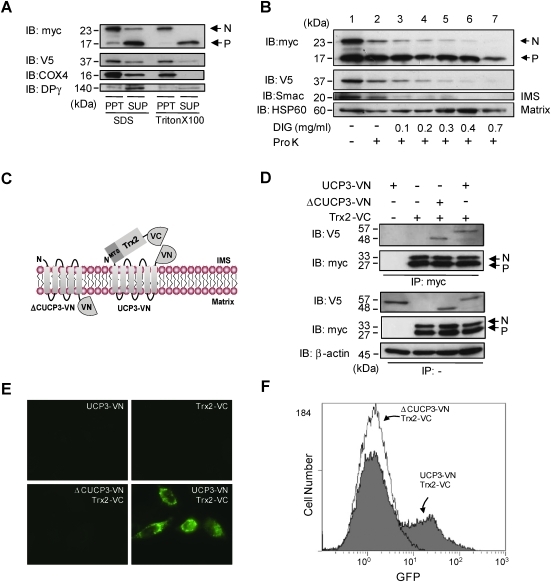
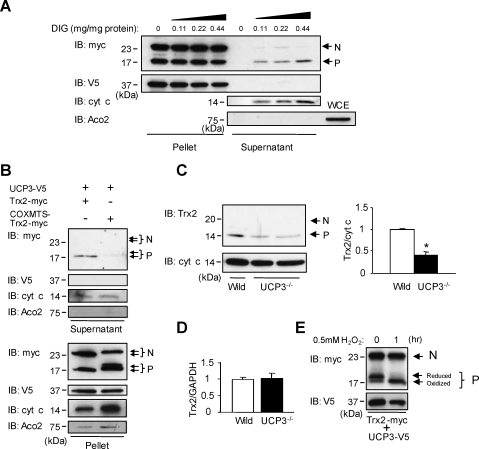
Based on the crystal structure of the adenine nucleotide carrier, the N-terminal hydrophilic sequences of UCP3 are strongly predicted to localize to the mitochondrial IMS (Supplementary Fig. S1). Thus, in addition to its demonstrated localization to the mitochondrial matrix, where it is known to bind thiol moieties (35), Trx2 should also be present in the IMS. To examine this notion, we used complementary biochemical and live cell approaches to define the submitochondrial localization of Trx2 and its interaction with UCP3. First, we examined Trx2 localization in the soluble and membrane fractions of detergent-treated (either SDS or Triton X-100), isolated mitochondria from HeLa cells co-expressing Trx2-myc and UCP3-V5. We found that processed Trx2-myc was localized predominantly in the supernatant fraction (SUP) containing the soluble matrix resident DNA polymerase γ (Fig. 4A, IB: myc, DPγ). In contrast, nonprocessed Trx2 was strongly and selectively localized in the membrane fraction (PPT), along with UCP3 and cytochrome c oxidase 4 (Fig. 4A, IB: Myc, V5, COX4). To further define the mitochondrial localization of Trx2, we performed a submitochondrial localization assay using varying concentrations of digitonin (DIG) in combination with protease K (Pro K) treatment. Immunoreactivity of nonprocessed Trx2 (myc) and UCP3 (V5) was more resistant to DIG when compared with the IMS resident Smac (Fig. 4B, lane 1–5), indicating that the UCP3:Trx2 complex binding surface localizes to the IMS. In contrast, the matrix resident HSP60 and processed Trx2 (V5) required increased DIG concentrations for proteolysis (Fig. 4B, lane 1–7). Next, we used bimolecular fluorescence complementation (BiFC) (22), and mutational techniques to directly visualize and define the mitochondrial localization of the UCP3–Trx2 complex in live cells. The BiFC system we chose uses a split-Venus fluorophore tag expression system in which Venus protein fluorescence can be reconstituted from individual split-Venus tags (N-terminal=VN, C-terminal=VC) when protein interactions occur (22). As illustrated in Figure 4C, for this set of studies we generated the following fusion proteins: full-length UCP3 with the N-terminus of Venus (residues 1–172) fused to the UCP3 C-terminus that extends into the IMS (UCP3-VN), C-terminally truncated UCP3 (residues 1–234) with the N-terminus of Venus extending into the matrix (ΔCUCP3-VN), and full-length Trx2 with the C-terminus of Venus (residues 155–238) fused to the C-terminus of Trx2. In this BiFC assay, ΔCUCP3 was also useful as a negative control because Trx2 interacts through the N-terminus of UCP3. Localization of Trx2-VC, UCP3-VN, and ΔCUCP3-VN in mitochondria is the same as that observed in Figure 4B (Supplementary Fig. S3A), indicating that the Venus protein does not disturb the localization of the tagged proteins. As expected, in cell lysates from co-transfected (but not single transfected) HeLa cells, both V5-tagged UCP3-VN and ΔCUCP3-VN co-immunoprecipitated with myc-tagged Trx2-VC (Fig. 4C, IP: myc, IB:V5). In live cells, expression of UCP3-VN or Trx2-VC alone yielded no fluorescence, nor did co-expression of ΔCUCP3-VN and Trx2-VC. However, strong mitochondrial fluorescence was obtained with co-transfection of UCP3-VN and Trx2-VC (Fig. 4E and Supplementary Fig. S3B). We used FACS analysis to quantify the BiFC fluorescence intensity in order to determine the efficiency of fluorescence complementation in cells. As shown in Figure 4F, the fluorescence intensity significantly increased in cells co-transfected with UCP3-VN and Trx2-VC compared to cells expressing ΔCUCP3-VN and Trx2-VC. Combined, these results strongly suggest that nonprocessed Trx2 binds to the N-terminus of UCP3 in the IMS.
FIG. 4.
Submitochondrial localization of the UCP3/Trx2 interaction. (A) Mitochondria isolated from HeLa cells transfected with UCP3 and Trx2 were suspended in RIPA buffer+SDS or lysis buffer+Triton X-100, respectively. Mitochondrial pellets and supernatants were subjected to SDS-PAGE followed by immunoblotting (IB) with anti-myc, anti-V5, anti-COX4, and anti-DNA polymerase γ (DPγ) antibodies. PPT, mitochondrial pellet after centrifugation; SDS, RIPA buffer; SUP, supernatant after centrifugation; Triton X-100; lysis buffer. (B) Mitochondria isolated from HeLa cells transfected with UCP3-V5 and Trx2-myc were subjected to mitochondrial sublocalization assay, as described in the Materials and Methods. Mitochondrial pellets and supernatants were resuspended in SDS-PAGE buffer, and then loaded onto an SDS-PAGE gel. Immunoblots show the presence of the intermembrane space (IMS) resident Smac and the matrix resident HSP60 in mitochondria that were untreated or treated with proteinase K (Pro K) (lanes 2–7) and increasing concentrations of digitonin (DIG) (lanes 3–7, 0.1 to 0.7 mg/ml DIG). (C) N (VN)- and C (VC)- terminal fragments of Venus fluorescent proteins were fused to the C-terminus of UCP3 (resides 1–308) (UCP3-VN) or ΔCUCP3 (residues1–234) (ΔCUCP3-VN), and the C-terminus of Trx2, respectively. The fragments of fluorescent proteins of belonging to UCP3-VN and ΔCUCP3-VN were localized to the mitochondrial IMS and matrix, respectively. (D) HeLa cells were transfected with UCP3-VN, Trx2-VC, ΔCUCP3-VN and Trx2-VC, and UCP3-VN and Trx2-VC. Cell extracts (100 μg) were immunoprecipitated (IP) with anti-myc antibodies and analyzed by IB to detect V5 (UCP3) and myc (Trx2). β-Actin was used as an internal standard. (E) Fluorescent images of HeLa cells transfected with UCP3-VN, Trx2-VC, ΔCUCP3-VN and Trx2-VC, and UCP3-VN and Trx2-VC as indicated in each panel. (F) HeLa cells transfected with ΔCUCP3-VN or UCP3-VN, and Trx2-VC were analyzed by flow cytometry. N, nonprocessed Trx2; P, processed Trx2. IB, IP, microscopy, and flow cytometry experiments are representative of at least three independent studies. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
UCP3 physiologically regulates the submitochondrial localization of Trx2
Proteins bearing an N-terminally cleaved MTS are processed by the mitochondrial intermediate peptidase (MIP) within the matrix, and the recently identified intermembrane peptidase (IMP) in the IMS (15). Binding of Trx2 to the N-terminal hydrophilic domain of UCP3 predicts that Trx2 could be localized and processed in the mitochondrial IMS as well as the matrix. As shown in Supplementary Figure S4, this notion is supported by the presence of contiguous consensus sequences for both IMP and MIP-mediated cleavage near the established processing site (∼amino acid position 60) of the pro-form of Trx2 (46). To begin to understand the impact of changes in UCP3 function on Trx2 mitochondrial localization, we generated mitoplasts from UCP3-V5 and Trx2-myc co-transfected HeLa cells using increasing concentrations of digitonin to selectively permeabilize the outer mitochondrial membrane and fractionate the matrix (Pellet) from the IMS (Supernatant). As shown in Figure 5A, the majority of Trx2 (IB:myc) and all of UCP3 immunoreactivity (IB:V5) were localized to the matrix. In addition, a pool of primarily processed Trx2 co-localized in the IMS with the IMS resident cytochrome c (IB:cyt c). In contrast, the matrix resident aconitase 2 (Aco2) was not released by digitonin treatment (IB:Aco2), but could be detected in whole cell extracts (Fig. 5A, bottom panel, WCE). Moreover, consistent with the domain-switching studies showing that COXMTS–Trx2 fails to interact with UCP3 (Fig. 3H), we failed to detect the processed form of COXMTX–Trx2 in the IMS fraction when co-expressed with UCP3-V5 (Fig. 5B, IB:myc).
FIG. 5.
UCP3-dependent regulation of Trx2 in the IMS. (A) Mitochondria isolated from HeLa cells transfected with UCP3 and Trx2 were subjected to digitonin (DIG) treatment at the concentrations shown. Mitochondrial pellets and supernatants were resuspended in SAS-PAGE buffer, and then loaded onto an SDS-PAGE gel, followed by immunoblotting (IB) with anti-myc, anti-V5, anti-cytochrome c (cyt c, IMS marker) and anti-aconitase 2 (Aco2, matrix marker) antibodies. (B) Mitochondria isolated from HeLa cells transfected with UCP3, Trx2 (residues 1–166) and COXMTS-Trx2Δ1–59 (residues 60–166) were treated with DIG. Supernatants after centrifugation were resuspended in SAS-PAGE buffer, and then loaded onto an SDS-PAGE, followed by IB with anti-myc, anti-V5, anti-cyt c, and anti-Aco2 antibodies. The upper and lower band of nonprocessed or processed Trx2 represents COXMTS-Trx2Δ1–59 and Trx2, respectively. (C) Mitochondria isolated from wild-type and UCP3-/- mice were treated with DIG. Supernatants after centrifugation were resuspended in SDS-PAGE buffer, and loaded onto an SDS-PAGE, followed by IB with anti-Trx2 and anti-cyt c antibodies. The ratio of Trx2 to cyt c was calculated by densitometric analysis. *p<0.05, compared with wild-type mice. (D) Total RNA extracted from gastrocnemius muscle in wild-type and UCP3-/- mice was subjected to quantitative RT-PCR. The intensity ratio of Trx2 to GAPDH was calculated. (E) HeLa cells were transfected with UCP3-V5 and Trx2-myc. AMS treatment was then used to identify the redox status of Trx2. The upper band represents the AMS-modified dithiol (reduced), active form of Trx2, while the disulfide form of Trx2 (oxidized) is detected as the lower band that lacks AMS labeling. H2O2 (0.5 mM) treatment was used as an oxidizing agent where indicated. N, nonprocessed Trx2; P, processed Trx2; WCE, whole cell extracts. IB and IP experiments are representative of at least three independent studies.
The physiological significance of these results is underscored by observations that the levels of processed Trx2 released from the IMS fraction in digitonin-treated UCP3-/- mouse muscle mitoplasts was significantly decreased relative to wild-type muscle (Fig. 5C), whereas there was no difference in Trx2 mRNA level in wild-type and UCP3-/- mice (Fig. 5D). This result combined with the lack of detection of transfected COXMTS–Trx2 in the IMS in UCP3-expressing cells argues strongly that the UCP3-dependent partitioning of Trx2 is mediated by the UCP3–Trx2 interaction, and not by the overaccumulation of transfected molecules or any electrogenic effect on import machinery. Finally, the UCP3-regulated processing and localization of Trx2 influences its function. Using a common alkylation assay with the thiol-specific reagent AMS, we found that IMS-localized (released from mitoplasts), processed Trx2 can undergo thiol alkylation with AMS in a manner blocked by H2O2. However, nonprocessed, IMS-localized Trx2 failed to bind AMS, implying that Trx2 acts as a ROS scavenger in the IMS only in its processed form (Fig. 5E).
Trx2 regulates UCP3-dependent suppression of mitochondrial ROS generation
Because both Trx2 and UCP3 regulate ROS production or scavenging, we sought to understand whether they function in a common pathway that regulates mitochondrial ROS generation using DHE, MitoSox, and Amplex Red dyes (48). C2C12 myoblast cell lines stably overexpressing UCP3 within the physiological range of induction (approximately 2.5-fold) were generated (Fig. 6A). UCP3-infected cells had comparable membrane potentials to mock-infected control cells, but not CCCP-treated cells (Fig. 6B). Mock and UCP3-infected cells showed similar levels of mitochondrial matrix superoxide (measured by MitoSOX fluorescence, data not shown). Addition of antimycin A with succinate drastically increased superoxide release from the mitochondria, as measured by DHE, however, this effect was significantly suppressed in UCP3-infected cells compared with Mock-infected controls (Fig. 6C). Consistent with these results, the rate of H2O2 production by isolated mitochondria from UCP3-infected C2C12 myoblast cells was significantly decreased compared to mock-infected control cells (Fig. 6D). In contrast, there were no significant differences between mock- and UCP3-infected cells in the rate of H2O2 production by mitochondria in the presence of pyruvate/malate plus antimycin A or rotenone (Fig. 6D).
FIG. 6.
Effects of UCP3 on mitochondrial superoxide and H2O2 production. (A) C2C12 myoblasts were stably infected with FG9-mock and -UCP3 lentivirus. Cell extracts (20 μg/lane) were subjected to SDS-PAGE followed by immunoblotting (IB) with anti-UCP3 and anti-β-actin antibodies. The ratio of UCP3 to β-actin was calculated by densitometric analysis. (B) Mock and UCP3-infected C2C12 myoblasts were assayed for membrane potential by flow cytometry using the fluorescent dye TMRM. (C) The rate of superoxide from mock- and UCP3-infected C2C12 myoblastic cells was assessed by dihydroethidium (DHE) fluorescence. Succinate and antimycin A were used as respiratory complex II substrate and complex III inhibitors, respectively. (D) The rate of H2O2 production from isolated mitochondria in mock- and UCP3-infected C2C12 myoblastic cells was assessed by Amplex Red fluorescence. Succinate and pyruvate/malate were used as respiratory substrates. Rotenone was used as a complex I inhibitor. For ROS production experiments, means±SD from three independent experiments performed in duplicate are shown. *p<0.05, compared with mock-infected cells. IB and flow cytometry experiments are representative of at least three independent studies.
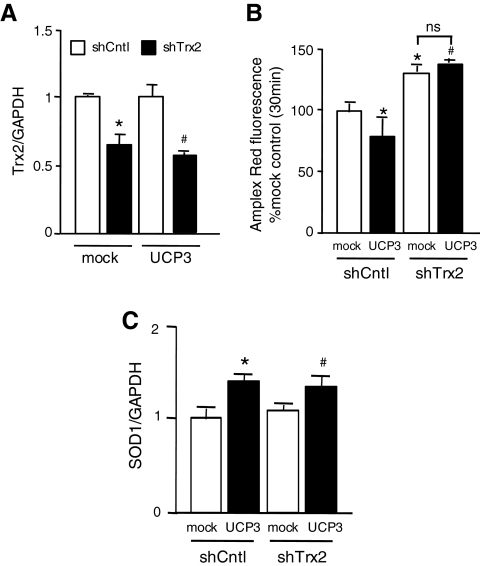
Since complete absence of Trx2 expression is embryonically lethal and cytotoxic (33), we used moderate Trx2 knockdown to determine whether the decrease in H2O2 production observed in UCP3-infected cells involved Trx2. As shown in Figure 7A, the amount of Trx2 mRNA in Trx2 shRNA-transfected cells stably expressing control or UCP3 lentiviral constructs was decreased approximately 40% compared with shRNA control cells. Importantly, the capacity for UCP3 to decrease the rate of succinate-driven H2O2 production from isolated mitochondria in control shRNA-transfected cells (Fig. 7B, shCntl) was abolished in cells after Trx2 knockdown (Fig. 7B, shTrx2). Moreover, Trx2 knockdown led to significant, similar increases in H2O2 production in both control and UCP3 overexpressing cells (Fig. 7B). Under these conditions, the amount of SOD1 mRNA in UCP3-infected cells significantly increased compared with mock-infected cells, while Trx2 knockdown did not affect SOD1 mRNA level (Fig. 7C). Combined, these results imply that Trx2 plays a mechanistic role in UCP3-dependent mitochondrial ROS suppression.
Fig. 7.
Effects of Trx2 knockdown on UCP3-mediated abrogation of mitochondrial H2O2 production. (A) Mock and UCP3-infected C2C12 myoblasts were transfected with scrambled and Trx2 shRNA using TransIT-LT1 (Mirus Bio) transfection reagent. Total RNA was extracted and subjected to quantitative RT-PCR. The intensity ratio of Trx2 to GAPDH was calculated. Data are mean±SD (n=3). *p<0.05, #p<0.05, compared with mock- and UCP3-infected control cells, respectively. (B) The rate of H2O2 production from isolated mitochondria in scrambled and Trx2 shRNA-transfected cells overexpressing mock DNA and UCP3 was measured using Amplex Red fluorescence 30 min after respiratory energization of mitochondria with succinate. (C) Total RNA from cells was extracted and subjected to quantitative RT-PCR. The intensity ratio of superoxide dismutase 1 (SOD1) to GAPDH was calculated. Data are mean±SD (n=3). *p<0.05, #p<0.05, compared with mock- and UCP3-infected control cells, respectively. Shown are the means±SD from three independent experiments, each one performed in duplicate. *p<0.05, #p<0.05, compared with mock- and UCP3-infected control cells, respectively. Quantitative RT-PCR production experiments are representative of at least three independent studies.
The UCP3–Trx2 complex is increased in response to high fat diet and regulates members of the insulin signaling pathway
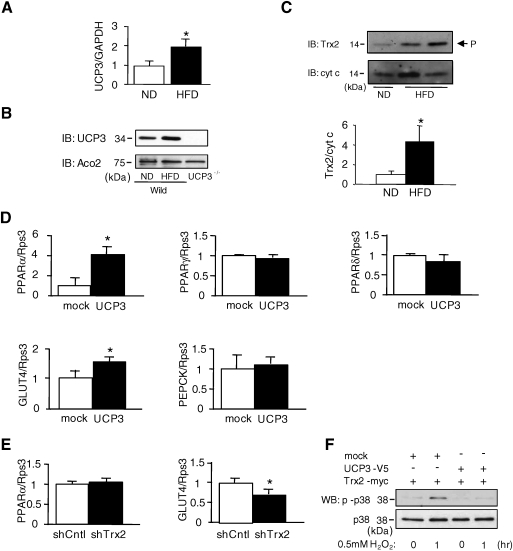
Finally, we examined the role of the UCP3–Trx2 complex in pathological conditions. Because it has been reported that increased levels of oxidative stress are correlated with the development of obesity-related insulin resistance and type 2 diabetes (34), we reasoned that the UCP3–Trx2 complex may be involved in protection from the excess ROS produced under these conditions. UCP3 mRNA and protein expression in skeletal muscle was significantly increased in mice fed a high fat diet (HFD, ∼45% calories from fat) compared with normal diet (ND)-fed mice (Figs. 8A and 8B). Accordingly, the amount of processed Trx2 released into the IMS fraction in the HFD-fed mice was significantly increased relative to ND-fed mice (Fig. 8C). This implies that the UCP3–Trx2 complex may be upregulated as a mechanism of cytoprotection in pathological conditions that result in elevated cellular ROS.
FIG. 8.
Effect of pathological conditions on the UCP3–Trx2 complex, and implications for the insulin signaling pathway. (A) Total RNA extracted from muscle of mice fed high fat diet (HFD) and normal diet (ND) for 5 weeks was subjected to quantitative RT-PCR. The intensity ratio of UCP3 to GAPDH was calculated. Data are mean±SD (n=6). *p<0.05, compared with muscle of mice fed ND. (B) Mitochondrial extracts (40 μg/lane) isolated from muscle of mice fed HFD and ND for 5 weeks were subjected to SDS-PAGE, followed by immunoblotting (IB) with anti-UCP3 and anti-aconitase 2 (Aco2) antibodies. Mitochondria extracted from UCP3-/- mice were used as a negative control. (C) Mitochondria isolated from muscle of mice fed HFD and ND for 5 weeks were treated with DIG and probed with anti-Trx2 and anti-cyt c antibodies. The ratio of Trx2 to cyt c was calculated by densitometric analysis. *p<0.05, compared with ND. (D) Total RNA from cells was extracted and subjected to quantitative RT-PCR. The intensity ratio of PPARα, PPARγ, PPARδ, GLUT4, and PEPCK to ribosomal protein S3 (Rps3), a house keeping gene, was calculated. Data are mean±SD (n=3). *p<0.05, compared with mock-infected control cells. (E) Expression level of PPARα and GLUT4 transcripts in scramble and Trx2 shRNA transfected UCP3 overexpressing cells was assessed by quantitative RT-PCR. The intensity ratio of PPARα, and GLUT4 to Rps3 was calculated. Data are mean±SD (n=3). *p<0.05, compared with scramble RNA transfected UCP3 overexpressing control cells. (F) Cells transfected with empty vector (mock), UCP3-V5, and Trx2-myc were treated with hydrogen peroxide (H2O2) (0.5 mM), and suspended in lysis buffer (50 mM Tris-HCl buffer, pH 7.5, 150 mM NaCl, 1% Triton X-100, protease inhibitors, phosphatase inhibitors containing 0.5 mM Na3VO4 and 20 mM Na4P2O7). Cell extracts (20 μg/lane) were subjected to SDS-PAGE, followed by immunoblotting (IB) with anti-phospho-p38 and anti-p38 antibodies.
Since it is known that UCP3 expression levels influence insulin sensitivity, fatty acid oxidation and related signaling pathways (44), we also investigated whether the UCP3–Trx2 complex was associated with changes in glucose homeostasis and lipid metabolism. As shown in Figure 8D, the amounts of PPARα and GLUT4 mRNA levels in UCP3-infected cells were significantly higher than those in mock-infected cells, while there were no significant differences in PPARγ, PPARδ, and PEPCK gene expression between mock- and UCP3 infected cells. Interestingly, Trx2 knockdown only affected GLUT4 expression (Fig. 8E), suggesting that the UCP3–Trx2 complex may regulate a GLUT4-related signaling pathway. In fact, H2O2 treatment induced phosphorylation of p38 in mock transfected cells, whereas UCP3 transfection significantly decreased H2O2-mediated phosphorylation of p38 (Fig. 8F).
Discussion
The results of this study provide the first direct biological and biochemical evidence that UCP3 interacts with and controls the partitioning of Trx2 in the mitochondrial IMS via the N-terminus of UCP3 and the N-terminus of nonprocessed Trx2. To our knowledge, this is also the first demonstration to show that Trx2 can localize to the IMS, and among the few reports of Trx binding mechanisms not mediated by the Trx active site cysteines. Additionally, these observations illuminate a novel protein–protein interaction functionally involved in the UCP3-dependent control of mitochondrial ROS generation. Together, these results add to an increasingly complex understanding of the regulation of uncoupling protein function, especially as it relates to mitochondrial redox control mechanisms.
The mitochondrial respiratory chain generates roughly 90% of the overall cellular burden of superoxide and its reactive derivatives, such as hydrogen peroxide and hydroxyl radicals. Topologically, mitochondrial complex I generates superoxide on the matrix side of the inner membrane, and complex III releases ROS on the IMS side and ultimately into the cytoplasm (9, 31). Thus, the IMS is an important region for the regulation of redox crosstalk between the mitochondrial respiratory chain and extramitochondrial compartments. Although antioxidant systems were identified long ago in multiple subcellular compartments, only very recently have redox and disulfide-relay control mechanisms been demonstrated to exist in the IMS. Indeed, very recent reports indicate that the IMS contains a large number of small (<30 kDa) disulfide bond-containing proteins (8, 18, 24). Interestingly, the Erv1–Mia40 component disulfide relay system transfers disulfides to these IMS proteins. During disulfide formation and exchange, this system utilizes the electron transport chain as a terminal electron acceptor and is tightly coupled to both mitochondrial protein import and antioxidant defense (4). The antioxidant Cu-Zn superoxide dismutase has recently been confirmed to localize to the IMS along with glutathione (24), and thioredoxin reductase-1, which reduces and re-activates thioredoxins (20). Similarly, the eubacterial thioredoxins DsbC and DsbG localize to the bacterial periplasm, the phylogenetic origin of the eukaryotic IMS, where they mediate thiol reduction (13). We found that both overexpressed and endogenous Trx2 is localized in a UCP3-binding dependent manner in the IMS of isolated mitochondria ex vivo, and that the IMS interaction of Trx2 and UCP3 could be visualized in live cells in vivo (Figs. 5A and 5C).
The capacity for Trx2 to dynamically localize in both the matrix and the IMS compartments can be inferred by the presence of protease cleavage sites for both the matrix polypeptidase (MIP) and the IMS peptidase (IMP). Mouse Trx2 is known to be cleaved by MIP between Leu-59 and Thr-60 (46). Trx2 also has several sites that can potentially be processed by IMP, which localizes to the outer face of the mitochondrial inner membrane and cleaves the pro-form of proteins at an asparagine residue, position −1 (38). Moreover, mitochondrial thioredoxin reductase also contains an IMP cleavage site near the predicted processing site for cleavage of the N-terminal targeting sequence (not shown). We detected processed Trx2 in the IMS that was similar in size to that of processed Trx2 in the mitochondrial matrix fraction, and noted that the cleavage sites on Trx2 are distinctly different between IMP and MIP. Many mitochondrial protein precursors are processed in two sequential steps by mitochondrial processing peptidases (MPP) and MIP (15). These precursors are characterized by the motif: R-X↓(F/L/I)-X-X-(T/S/G)-X-X-X-X↓. MPP first cleaves the motif two peptide bonds after the arginine residue, yielding a processing intermediate with a typical N-terminal octapeptide that is sequentially cleaved by MIP, yielding the mature-size protein. Trx2 also contains a (V/I)-X-X-T-X-X-X-X↓motif that can be recognized by MIP between Val-64 and Gln-65 (Supplementary Fig. S4). In fact, when we generated a truncation mutant based on the previously reported cleavage site (yielding a processed intermediate containing residues 60–166 plus methionine), it migrated by SDS-PAGE slightly slower than the major processed version of Trx2 in mitochondria (Figs. 1B, 3H, and 5B). These results suggest that the predicted cleavage site may be slightly upstream of the actual cleavage site, which interestingly has overlapping cleavage site homology for both MIP and IMP proteases. Further studies are necessary to define the processing mechanisms of Trx2.
A growing body of evidence shows that UCP3 decreases mitochondrial ROS production, protects from oxidative sequelae (e.g., lipid peroxidation), and exports metabolizable and/or oxidized fatty acid peroxides across the inner membrane to protect the matrix from oxidative stress. However, the mechanism of UCP3-dependent redox control is hotly debated, and the role of the membrane potential in this process is unclear. Toime and Brand demonstrated that UCP3 decreases ROS production through a membrane potential-independent mechanism in isolated mitochondria (48). Similarly, we found that a 2.5-fold increase in UCP3 expression has no effect on membrane potential, but significantly attenuates ROS production in isolated mitochondria. Additionally, the capacity for UCP3 to blunt mitochondrial ROS generation disappears when levels of Trx2 are decreased. It has also been reported that Trx2 forms complexes with cytochrome c, a potent and direct superoxide and lipid peroxide reductant, but no information was provided regarding the submitochondrial distribution of this interaction (36, 47). Interestingly, the bulk of the nontransmembrane component of cytochrome c extends into the IMS, and only a very small portion extends into the matrix. Thus, it is tempting to speculate that UCP3 may simultaneously regulate the detoxification and export of fatty acid peroxides and complex III-generated superoxide through the putative Trx2-cytochrome c complex.
Complex III is a major site of mitochondrial ROS generation (9, 31) and is comprised of two ubisemiquinone redox centers: the Q0 center is oriented toward the IMS; and the Q1 center is localized in the inner membrane and faces the mitochondrial matrix. Antimycin A, a strong antagonist of the Q1 center, strongly activates superoxide anion production and release into the IMS from the Q0 center (26, 50). Since Trx2 interacts with UCP3 on the IMS side of the inner membrane, we focused on ROS production through the Q0 center. Modestly increased UCP3 expression significantly attenuates ROS production in isolated mitochondria respiring on succinate (complex II substrate), but not pyruvate/malate (complex I substrate) (Fig. 6D). Furthermore, UCP3 attenuates generation of ROS by antimycin A-inhibited complex III, which releases superoxide to both sides of the mitochondrial inner membrane, whereas there is no difference in rotenone-inhibited complex I, which directs ROS into the mitochondrial matrix (Figs. 6C and 6D). These results are consistent with a previous report that UCP3 actively lowers the rate of ROS production in isolated, energized skeletal muscle mitochondria, in absence of exogenous activators such as rotenone (48). The significantly decreased ROS production mediated by UCP3 is abolished by moderate Trx2 knock down. Our data are in agreement with previous work showing a significant increase in ROS production in the presence of respiratory substrates for complex II in Trx2 heterozygous knockout mice (37). Although Trx2 gene deletion in mice is embryonically lethal (33), conditional Trx2 knockdown in DT40 cells leads to increased ROS production (47, 52). Combined, the studies suggest that Trx2 acts as an oxidant scavenger in the IMS.
Oxidative stress may contribute to obesity-induced insulin resistance and type 2 diabetes. In this study, a high fat diet for 5 weeks led to upregulation of UCP3 protein levels and increased release of Trx2 in the IMS. These findings suggest that increased UCP3 may regulate the processing of Trx2, and the resultant increase of processed Trx2 in the IMS may mitigate oxidative stress induced during pathological conditions such as obesity. It has been noted that oxidative stress markedly decreases insulin-stimulated glucose metabolism, and is associated with decreased GLUT4-mRNA and protein expression. Furthermore, overexpression of UCP3 in the skeletal muscle of transgenic mice results in increased energy expenditure, resistance to diet-induced obesity, and increased glucose tolerance and insulin sensitivity (10). Oxidative stress also results in the phosphorylation and activation of p38 MAPK, which may play a role in the oxidant stress-induced impairment of insulin-stimulated glucose transport activity (14). In accordance with these findings, we found that increased UCP3 expression improved glucose metabolism by increasing GLUT4, and that this effect was abrogated by Trx2 knock down. At the same time, UCP3 expression also inhibited oxidative stress-induced phosphorylation of p38. Thus, our results suggest that the UCP3–Trx2 complex may play a role in the regulation of increased ROS production in the IMS seen under pathological conditions such as obesity, thereby leading to a significant inhibition of the oxidative stress-mediated impairment of insulin signaling.
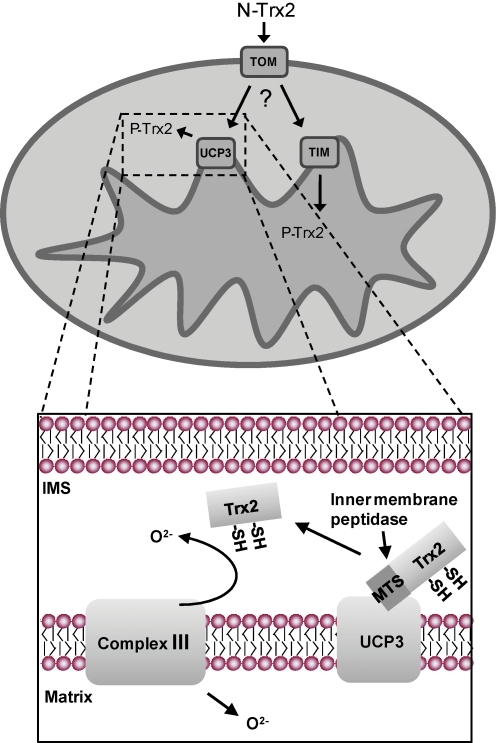
In summary, we present the first evidence of a novel, redox-regulatory UCP3–Trx2 complex in the mammalian IMS of skeletal muscle, and illuminate a mechanistic role for Trx2 in the UCP3-dependent attenuation of mitochondrial ROS production (Fig. 9). The data support a hypothetical model wherein factors that influence UCP3 expression and activity (e.g., fatty acids, lipid peroxides) may regulate the relative matrix versus IMS pools of Trx2. Obesity and fasting are associated with increased skeletal muscle UCP3 expression, and overnutrition and refeeding can induce supraphysiological mitochondrial ROS production. Thus, UCP3 may act to localize Trx2 to the IMS under these conditions as a protective measure against excess or pathologic ROS effects, such as signaling or damage, in cells undergoing nutritional stress. Future work is necessary to explore the physiological functions of the UCP3–Trx2 complex, and the mechanisms for its regulation.
FIG. 9.
Model of Trx2-mediated redox regulation by UCP3 in the mitochondrial intermembrane space (IMS). The hydrophilic sequences of the N-terminal IMS facing region of UCP3 are necessary for binding to Trx2 and interact with the MTS of nonprocessed Trx2 in the IMS. Trx2 bound to UCP3 can then be processed in the IMS by the intermembrane peptidase (IMP), thereby activating the IMS pool of Trx2. ROS released from complex III into the IMS can then be detoxified through a Trx2-regulated pathway prior to escape into the cytoplasm. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Materials and Methods
Yeast two-hybrid
The hydrophilic sequence in the UCP3 N-terminus (nucleotides encoding amino acids 1–16) was PCR-amplified from a full-length mouse UCP3 cDNA clone. This fragment was fused to the GAL4 DNA binding domain in the pGBKT7 vector, and used as bait in a two-hybrid screen from a heart cDNA library. From this screen, we identified a cDNA encoding thioredoxin 2 that was characterized by sequencing using the BLAST algorithm.
Plasmid DNA Constructs
Mouse UCP3 and Trx2 cDNA was isolated from a mouse skeletal muscle cDNA library by PCR. The isolated UCP3 and Trx2 PCR products were subcloned into mammalian expression plasmid vectors pcDNA3.1-V5 and pCNDA6.1-myc (Invitrogen, Carlsbad, CA), respectively. Point mutations were made in the template of pCDNA6.1-Trx2-myc using the QuikChange site directed mutagenesis technique of Stratagene Cloning Systems (Stratagene, La Jolla, CA). The C90S and C93S mutants, as well as the C90S and C93S double mutant, were generated by PCR. The mutational primers were as follows: C90S Trx2, forward: 5′-ATGCACAGTGG AGTGGCCCCT-3′, reverse: 5′-AGGGGCCACTCCACTGTG CAT-3′; C93S Trx2, forward: 5′-GTGTGGCCCCAGCAAGATCC-3′, reverse: 5′-GGATCTTGCTGGGGCCACAC-3′. The N-terminal deletion of UCP3 (residues 17–308) was constructed using the modified PCR technique of Stratagene Cloning Systems. The following oligonucleotide primers were used for amplification: 5′- TTCCTGGGGGCCGGCACTG-3′ and 5′-CATGGTGGATCCGAGCTCGGTAC-3′ for UCP3. The Trx2 mutants that replaced the MTS of Trx2 with that of human cytochrome c oxidase, subunit VIII (COXMTS) were subcloned from the pHyper-dMito vector (Evrogen, Moscow, Russia) and ligated in frame into the pCDNA6.1-Trx2-myc. For the construction of the expression plasmids for GST-tagged Trx2 (GST–Trx2) and T7 tagged-UCP3, the DNA fragments encoding mouse Trx2 and UCP3 were subcloned from pCDNA6.1-Trx2 and pCNDA3.1-UCP3 vectors and ligated in frame into the pGEX6P-1 and pET23 vector, respectively. The truncated forms of GST-Trx2 were constructed by modified PCR. The mouse Smac sequence was subcloned by PCR and ligated in frame into the pGEX6P-1 vector. UCP3 was cloned into the FG9 lentiviral vector (kindly provided by Dr. Casey W. Wright, University of Texas-Austin, TX). For the generation of shRNAs, oligonucleotides to Trx2 (5′-GATCCCCCC AGTTGTTGTGGACTTTCATTTCAAGAGAATGAAAGTCC ACAACAACTGGTTTTTGGAAA -3′; 5′-AGCTTTTCCAA AAACCAGTTGTTGTGGACTTTCATTCTCTTGAAATGAA AGTCCACAACAACTGGGGG-3′) or a scrambled control (sense: 5′-GATCCCCACCGTCGATTTCACCCGGGTTCAAGAGACCCGGGTGAAATCGACGGTTTTTGGAAA-3′; antisense: 5′-AGCTTTTCCAAAAACCGTCGATTTCACCCGG GTCTCTTGAACCCGGGTGAAATCGACGGTGGG-3′) were annealed to generate sticky ends (underlined) and immediately cloned into the BglII–HindIII sites of pSuper (OligoEngine, Seattle, WA). The boxed sequence corresponds to the 9-nucleotide hairpin that is generated after transcription.
Cell culture
HeLa and C2C12-Mock and -UCP3 cells (an isolated clone of C2C12 cells stably infected with empty vector (Mock) or the UCP3 gene) were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% 100X PenG-Streptomycin (Invitrogen). Cells were incubated at 37°C with 5% CO2. Maintenance of C2C12-Mock and -UCP3 cells was performed by treating with hygromycin (300 μg/ml). HeLa and C2C12-Mock and -UCP3 cells were transiently transfected using Lipofectamine (Invitrogen) and TransIT-LT1 (Mirus Bio, Madison WI) transfection regents, respectively. Cells were allowed to incubate for 24 h prior to experiments.
Diet-induced obesity studies
Six-week-old wild-type FVB/N and UCP3-knockout (UCP3-/-) FVB/N mice (n=3) were placed on either a normal diet (5.3% calories from fat) or a high-fat diet (45% calories from fat) for 5 weeks, following which mice were sacrificed by cervical dislocation. Skeletal muscle (gastrocnemius) was isolated and used for subsequent analyses, as described below.
Isolation of mitochondria
Mitochondria from HeLa, C2C12-Mock and -UCP3 cells, and skeletal muscle from wild-type C57BL6/j, FVB/N, UCP3-/- C57BL6/j, and UCP3-/-FVB/N mice were prepared according to previously established protocols (45). Briefly, tissues or cells were immediately harvested and minced in ice-cold CP-1 buffer (100 mM KCl, 50 mM Tris-HCl, 2 mM EGTA, pH 7.4). After grinding in a glass homogenizer, supernatants were applied to a 40 μM cell strainer. Mitochondria were then separated by differential centrifugation and lysed with either RIPA+SDS (50 mM Tris-HCl, pH 8.0, containing 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, and protease inhibitors) or lysis buffer+Triton X-100 (50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl, 1% Triton X-100, and protease inhibitors).
Submitochondrial localization assay
Mitochondria were isolated from HeLa cells as described above. Water-soluble digitonin (MP Biomedical, Cleveland, OH) was used for permeabilization. Following 5 μg/ml proteinase K treatment, 100 μM phenylmethylsulfonyl fluoride (PMSF) was added to stop the enzyme's activity. Anti-Smac (Cell Signaling, Danvers, MA) and anti-HSP60 (Enzo Life Sciences, Plymouth Meeting, PA) primary antibodies were used as markers of the mitochondrial intermembrane space and matrix, respectively.
Isolation of mitochondrial IMS contents
The mitochondrial outer membrane was selectively disrupted by treating mitochondria (40 mg/ml) with digitonin (0.11 mg/mg of protein) (20). The IMS contents were then removed from mitoplasts by centrifugation at 10,000 g for 15 min at 4°C.
GST pull-down assay
Purified GST-tagged proteins were prepared as previously described (39). Five micrograms of each purified GST fusion protein or GST protein alone was added to 100 μg of cell extracts from HeLa cells and incubated in GST lysis buffer (20 mM Tris-HCl, pH 7.4, 0.5 mM DTT, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, plus protease inhibitors) for 3 h at 4°C. After addition of 25 μl of glutathione-Sepharose 4B, the binding was allowed to continue for an additional 16 h at 4°C. The bound beads were washed four times with the same lysis buffer and analyzed by SDS-PAGE.
Immunoblotting
Cells were prepared in 50 mM Tris-HCl buffer, pH 7.5, containing 150 mM NaCl, 1% Triton X-100, 200 μM PMSF, 1 μg/ml Leupeptin, 1 μg/ml Pepstatin A and 100 U/ml Aprotinin, and samples were homogenized using a sonicator. The Pierce BCA Assay (Pierce Biotechnology, Rockford, IL) was used to quantify proteins. Protein samples were combined with 4X sample buffer (250 mM Tris-HCl, 8% SDS, 40% glycerol, 8% betamercaptoethanol, 0.02% bromophenol blue) and separated on a 12% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and probed with primary antibody according to the manufacturer's instructions; anti-V5, anti-UCP3, anti-aconitase 2, anti-β-actin (Abcam, Cambridge, MA), anti-cytochrome c (BD Biosciences, Franklin Lakes, NJ), anti-myc, anti-phospho-p38, anti-p38 (Cell Signaling), anti-COX4 (Clontech, Mountain View, CA), anti-DNA polymerase γ (Neo Markers, Fremont, CA), anti-T7 (Novagen, Gibbstown, NJ), and anti-thioredoxin 2 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies were used. Secondary antibodies were donkey anti-rabbit 1:5000 and sheep anti-mouse 1:5000 (Amersham Biosciences, Piscataway, NJ). Membranes were developed using Pierce Super Signal West Pico chemiluminescent substrate (Pierce Biotechnology).
Immunoprecipitation
Immunoprecipitation (IP) samples were prepared using 100 μg protein and adjusted to 300 μl with lysis buffer. They were incubated with either 2 μg of primary antibody or IgG (for controls) at 4°C for 4 h with rotation. Thirty microliters of Protein G Sepharose beads (Amersham Biosciences) were added to each IP sample and rotated at 4°C for 16 h. Samples were washed six times in wash buffer, after which samples were prepared for SDS-PAGE as described above.
Redox Western blotting for Trx2
The redox state of Trx2 was determined by analysis of cell extracts pre-incubated with AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid) as previously described (27). Briefly, cells were precipitated with 10% ice-cold trichloroacetic acid for 30 min at 4°C. Samples were centrifuged at 12,000 g for 30 min, resuspended in 100% acetone and incubated at 4°C for 30 min. Following centrifugation at 12,000 g for 10 min, the acetone was removed and protein pellets were dissolved in 20 mM Tris/HCl, pH 8.0, containing 15 mM AMS and incubated at room temperature (25°C) for 3 h. Trx2 redox forms were separated by SDS-PAGE in the presence of nonreducing loading buffer.
Quantitative RT-PCR
Total RNA was extracted from C2C12 cells and mouse skeletal muscle with an acid guanidinium thiocyanate-phenol-chloroform mixture (TRIZOL; Invitrogen). Quantitative RT-PCR was performed with the primers and SYBR Green dye by using a real-time PCR system (CFX96™ Real-Time PCR Detection System; Bio-Rad, Hercules, CA), as described previously (19). The following primers were used for amplification: 5′-TTCATGCACAGTGGTGTGG-3′ and 5′-GGCAC AGCTGACACCTCATA-3′ for mouse Trx2; 5′-GGAGTCTC ACCTGTTTACTGACAACT-3′ and 5′-GCACAGAAGCCA GCTCCAA-3′ for mouse UCP3; 5′-GGGTTCCACGTCCATC AGTA-3′ and 5′-CTTTCCAGCAGTCACATTGC-3′ for mouse SOD1; 5′-TCACAGACACCCTCTCTCCA-3′ and 5′-TTCGACACTCGATGTTCAGG-3′ for mouse PPARα; 5′-CCC ATCGAGGACATCCAA-3′ and 5′-CACGTGCTCTGTGACG ATCT-3′ for mouse PPARγ; 5′-GCAGCCTCTTCCTCAAT GAC-3′ and 5′-GGAGACTTCGCAAGAACTCG-3′ for mouse PPARδ; 5′-GCTCTCCTGCAGCTGATTCT-3′ and 5′-CAGC TCAGCTAGTGCGTCAG-3′ for mouse GLUT4; 5′-GGTT CCGGAAGGACAAAGAT-3′ and 5′-CCTTAGGGATGTAG CCGATG-3′ for mouse PEPCK; 5′-ATCAGAGAGTTGAC CGCAGTTG-3′ and 5′-AATGAACCGAAGCACACCA TAG-3′ for mouse ribosomal protein S3 (Rps3); 5′-AGAA CATCATCCCTGCATCC-3′ and 5′-GGTCCTCAGTGTAGC CCAAG-3′ for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Immunofluorescence staining
Cells were cultured on coverslips and loaded with 50 nM MitoTracker® Red CMXROS (Invitrogen) for 30 min at 37°C in the dark, then fixed in 2% paraformaldehyde for 10 min at 25°C, and permeabilized with 0.2% Triton X-100 for 10 min at 4°C. The cells were incubated with anti-V5 and anti-myc antibodies, followed by secondary anti-rabbit Alexa 568, anti-rabbit Alexa 488, or anti-mouse Alexa 488 (Invitrogen). All fluorescent images were acquired with a Nikon Eclipse Ti-S microscope, using a 100X Nikon Plan Apo VC Oil objective with a numerical aperture of 1.40. Images were taken with a Photometrics Coolsnap EZ camera and processed using NIS Elements BR 3.0 software.
BiFC analysis
The sequences encoding amino acid residues 1–172 and 155–238 of Venus were subcloned from the pCE-BiFC-VN173 and pCE-BiFC-VC155 vectors (Addgene, Cambridge, MA), and ligated in frame into the pCNDA3.1-UCP3-V5 (resides 1–308) or pCNDA3.1-ΔCUCP3-V5 (residues 1–234), and pCDNA6.1-Trx2-myc (Trx2–VC) vectors, respectively. The fluorescence signals were analyzed using a Nikon Eclipse Ti-S fluorescent microscope or by flow cytometry.
Assessment of mitochondrial membrane potential and superoxide levels
C2C12-Mock and -UCP3 cells were harvested by trypsin digestion, washed with PBS, and incubated with HBSS containing 25 nM tetramethylrhodamine methyl ester (TMRM) for 15 min (assessment of mitochondrial membrane potential), or 5 μM MitoSox for 10 min (superoxide detection in the mitochondrial matrix) at 37°C in the dark. TMRM and MitoSox fluorescence was analyzed by flow cytometry (Beckman-Coulter, Fullerton, CA). Dihydroethidium (DHE) was used as a second method for detection of superoxide. C2C12-Mock and -UCP3 cells were plated in 96-well plates and incubated with HBSS containing 5 μM DHE. Fluorescence was recorded by a microplate reader (VICTOR™ 3V; Pelkin Elmer, Waltham, MA) with 490 nm excitation and 595 nm emission wavelengths.
Detection of H2O2 production
The rate of H2O2 production by isolated mitochondria was measured using the oxidation of the fluorogenic indicator Amplex Red, as previously described (41). Fluorescence was recorded using a microplate reader with 531 nm excitation and 595 nm emission wavelengths. Standard curves obtained by adding known amounts of H2O2 to the assay medium in the presence of reactants were linear up to 2 μM. Mitochondria from C2C12 cells were suspended in a buffer containing 5 mM MOPS (pH 7.4), 70 mM sucrose, and 220 mM mannitol, and mitochondrial protein concentration was determined using a BCA protein assay (Pierce Biotechnology, Rockford, IL). Mitochondria (5 μg mitochondrial protein/well) were incubated in a reaction mixture containing 50 μM Amplex Red, 0.2 U/ml horseradish peroxidase (HRP), 30 U/ml superoxide dismutase (SOD), 10 μM antimycin A or 2.4 μM rotenone, and pyruvate (2.5 mM)/malate (2.5 mM) or succinate (10 mM) as substrate at room temperature for 30 min, protected from light. SOD was added to convert all superoxide into H2O2. The rate of H2O2 production was linear with respect to mg of mitochondrial protein.
Statistics
Analysis of variance comparisons for immunoblotting, quantitative RT-PCR, and detection of H2O2 production were analyzed by single factor ANOVA followed by a Dunnet's post hoc test, with a p<0.05 set a priori as statistically significant.
Supplementary Material
Abbreviations Used
- AMS
4-acetamido-4′maleimidylstilbene-2,2′-disulfonic acid
- BiFC
bimolecular fluorescence complementation
- CBB
Coomassie brilliant blue
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- COXMTS
cytochrome c oxidase subunit VIII mitochondrial targeting signal
- DHE
dihydroethidium
- DIG
digitonin
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione S transferase
- HFD
high fat diet
- HRP
horseradish peroxidase
- IMP
inner membrane peptidase
- IMS
intermembrane space
- MIP
mitochondrial intermediate peptidase
- MOPS
3-(N-morpholino)propanesulfonic acid
- MPP
mitochondrial processing peptidase
- MTS
mitochondrial targeting signal
- ND
normal diet
- NP40
nonidet-P40
- PAGE
polyacrylamide gel electrophoresis
- PMSF
phenylmethylsulfonyl fluoride
- RIPA
radio immunoprecipitation assay
- ROS
reactive oxygen species
- SDS
sodium dodecyl sulfate
- SOD
superoxide dismutase
- TMRM
tetramethylrhodamine-methyl-ester
- Trx2
thioredoxin 2
- UCP3
uncoupling protein 3
Acknowledgments
This work was supported in part by Grant 1R01DK089224 (EMM) from the National Institutes of Health and a JSPS Postdoctoral Fellowships for Research Abroad (KH). We thank Dr. Casey W. Wright for providing the FG9 lentiviral vector.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Azzu V. Brand MD. The on-off switches of the mitochondrial uncoupling proteins. Trends Biochem Sci. 2010;35:298–307. doi: 10.1016/j.tibs.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban RS. Nemoto S. Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Barreiro E. Garcia-Martínez C. Mas S. Ametller E. Gea J. Argilés JM. Busquets S. López-Soriano FJ. UCP3 overexpression neutralizes oxidative stress rather than nitrosative stress in mouse myotubes. FEBS Lett. 2009;583:350–356. doi: 10.1016/j.febslet.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 4.Bihlmaier K. Mesecke N. Terziyska N. Bien M. Hell K. Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boss O. Samec S. Paoloni-Giacobino A. Rossier C. Dulloo A. Seydoux J. Muzzin P. Giacobino JP. Uncoupling protein-3: A new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408:39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 6.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 7.Cannon B. Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 8.Chacinska A. Koehler CM. Milenkovic D. Lithgow T. Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q. Vazquez EJ. Moghaddas S. Hoppel CL. Lesnefsky EJ. Production of reactive oxygen species by mitochondria: Central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 10.Choi CS. Fillmore JJ. Kim JK. Liu ZX. Kim S. Collier EF. Kulkarni A. Distefano A. Hwang YJ. Kahn M. Chen Y. Yu C. Moore IK. Reznick RM. Higashimori T. Shulman GI. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J Clin Invest. 2007;117:1995–2003. doi: 10.1172/JCI13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collet JF. Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 12.Damdimopoulos AE. Miranda-Vizuete A. Pelto-Huikko M. Gustafsson JA. Spyrou G. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J Biol Chem. 2002;277:33249–33257. doi: 10.1074/jbc.M203036200. [DOI] [PubMed] [Google Scholar]
- 13.Depuydt M. Leonard SE. Vertommen D. Denoncin K. Morsomme P. Wahni K. Messens J. Carroll KS. Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 14.Diamond-Stanic MK. Marchionne EM. Teachey MK. Durazo DE. Kim JS. Henriksen EJ. Critical role of the transient activation of p38 MAPK in the etiology of skeletal muscle insulin resistance induced by low-level in vitro oxidant stress. Biochem Biophys Res Commun. 2011;405:439–444. doi: 10.1016/j.bbrc.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gakh O. Cavadini P. Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- 16.Goglia F. Skulachev VP. A function for novel uncoupling proteins: Antioxidant defense of mitochondrial matrix by translocating fatty acid peroxides from the inner to the outer membrane leaflet. FASEB J. 2003;17:1585–1591. doi: 10.1096/fj.03-0159hyp. [DOI] [PubMed] [Google Scholar]
- 17.Hansen JM. Zhang H. Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann JM. Köhl R. Catch me if you can! Oxidative protein trapping in the intermembrane space of mitochondria. J Cell Biol. 2007;176:559–563. doi: 10.1083/jcb.200611060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirasaka K. Kohno S. Goto J. Furochi H. Mawatari K. Harada N. Hosaka T. Nakaya Y. Ishidoh K. Obata T. Ebina Y. Gu H. Takeda S. Kishi K. Nikawa T. Deficiency of Cbl-b gene enhances infiltration and activation of macrophages in adipose tissue and causes peripheral insulin resistance in mice. Diabetes. 2007;56:2511–2522. doi: 10.2337/db06-1768. [DOI] [PubMed] [Google Scholar]
- 20.Iñarrea P. Moini H. Han D. Rettori D. Aguiló I. Alava MA. Iturralde M. Cadenas E. Mitochondrial respiratory chain and thioredoxin reductase regulate intermembrane Cu, Zn-superoxide dismutase activity: Implications for mitochondrial energy metabolism and apoptosis. Biochem J. 2007;405:173–179. doi: 10.1042/BJ20061809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irani K. Xia Y. Zweier JL. Sollott SJ. Der CJ. Fearon ER. Sundaresan M. Finkel T. Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 22.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimova T. Chandel NS. Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ. 2008;15:660–666. doi: 10.1038/sj.cdd.4402307. [DOI] [PubMed] [Google Scholar]
- 24.Koehler CM. Beverly KN. Leverich EP. Redox pathways of the mitochondrion. Antioxid Redox Signal. 2006;8:813–822. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- 25.Krauss S. Zhang CY. Lowell BB. The mitochondrial uncoupling-protein homologues. Nat Rev Mol Cell Biol. 2005;6:248–261. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 26.Ksenzenko M. Konstantinov AA. Khomutov GB. Tikhonov AN. Ruuge EK. Effect of electron transfer inhibitors on superoxide generation in the cytochrome bc1 site of the mitochondrial respiratory chain. FEBS Lett. 1983;155:19–24. doi: 10.1016/0014-5793(83)80200-2. [DOI] [PubMed] [Google Scholar]
- 27.Lim PL. Liu J. Go ML. Boelsterli UA. The mitochondrial superoxide/thioredoxin-2/Ask1 signaling pathway is critically involved in troglitazone-induced cell injury to human hepatocytes. Toxicol Sci. 2008;101:341–349. doi: 10.1093/toxsci/kfm273. [DOI] [PubMed] [Google Scholar]
- 28.Liu X. Rossmeisl M. McClaine J. Riachi M. Harper ME. Kozak LP. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. J Clin Invest. 2003;111:399–407. doi: 10.1172/JCI15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLellan JD. Gerrits MF. Gowing A. Smith PJ. Wheeler MB. Harper ME. Physiological increases in uncoupling protein 3 augment fatty acid oxidation and decrease reactive oxygen species production without uncoupling respiration in muscle cells. Diabetes. 2005;54:2343–2350. doi: 10.2337/diabetes.54.8.2343. [DOI] [PubMed] [Google Scholar]
- 30.Moghaddas S. Hoppel CL. Lesnefsky EJ. Aging defect at the QO site of complex III augments oxyradical production in rat heart interfibrillar mitochondria. Arch Biochem Biophys. 2003;414:59–66. doi: 10.1016/s0003-9861(03)00166-8. [DOI] [PubMed] [Google Scholar]
- 31.Muller FL. Liu Y. Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonn L. Williams RR. Erickson RP. Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nourooz-Zadeh J. Rahimi A. Tajaddini-Sarmadi J. Tritschler H. Rosen P. Halliwell B. Betteridge DJ. Relationships between plasma measures of oxidative stress and metabolic control in NIDDM. Diabetologia. 1997;40:647–653. doi: 10.1007/s001250050729. [DOI] [PubMed] [Google Scholar]
- 35.Patenaude A. Ven Murthy MR. Mirault ME. Mitochondrial thioredoxin system: Effects of TrxR2 overexpression on redox balance, cell growth, and apoptosis. J Biol Chem. 2004;279:27302–27314. doi: 10.1074/jbc.M402496200. [DOI] [PubMed] [Google Scholar]
- 36.Pereverzev MO. Vygodina TV. Konstantinov AA. Skulachev VP. Cytochrome c, an ideal antioxidant. Biochem Soc Trans. 2003;31:1312–1315. doi: 10.1042/bst0311312. [DOI] [PubMed] [Google Scholar]
- 37.Pérez VI. Lew CM. Cortez LA. Webb CR. Rodriguez M. Liu Y. Qi W. Li Y. Chaudhuri A. Van Remmen H. Richardson A. Ikeno Y. Thioredoxin 2 haploinsufficiency in mice results in impaired mitochondrial function and increased oxidative stress. Free Radic Biol Med. 2008;44:882–892. doi: 10.1016/j.freeradbiomed.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 38.Pratje E. Guiard B. One nuclear gene controls the removal of transient pre-sequences from two yeast proteins: One encoded by the nuclear the other by the mitochondrial genome. EMBO J. 1986;5:1313–1317. doi: 10.1002/j.1460-2075.1986.tb04361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Psarra AM. Hermann S. Panayotou G. Spyrou G. Interaction of mitochondrial thioredoxin with glucocorticoid receptor and NF-kappaB modulates glucocorticoid receptor and NF-kappaB signalling in HEK-293 cells. Biochem J. 2009;422:521–531. doi: 10.1042/BJ20090107. [DOI] [PubMed] [Google Scholar]
- 40.Rizzuto R. Brini M. Pizzo P. Murgia M. Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 41.Rohrbach S. Gruenler S. Teschner M. Holtz J. The thioredoxin system in aging muscle: Key role of mitochondrial thioredoxin reductase in the protective effects of caloric restriction? Am J Physiol Regul Integr Comp Physiol. 2006;291:R927–R935. doi: 10.1152/ajpregu.00890.2005. [DOI] [PubMed] [Google Scholar]
- 42.Schleiff E. McBride H. The central matrix loop drives import of uncoupling protein 1 into mitochondria. J Cell Sci. 2000;113:2267–2272. doi: 10.1242/jcs.113.12.2267. [DOI] [PubMed] [Google Scholar]
- 43.Schrauwen P. Saris WH. Hesselink MK. An alternative function for human uncoupling protein 3: Protection of mitochondria against accumulation of nonesterified fatty acids inside the mitochondrial matrix. FASEB J. 2001;15:2497–2502. doi: 10.1096/fj.01-0400hyp. [DOI] [PubMed] [Google Scholar]
- 44.Senese R. Valli V. Moreno M. Lombardi A. Busiello RA. Cioffi F. Silvestri E. Goglia F. Lanni A. de Lange P. Uncoupling protein 3 expression levels influence insulin sensitivity, fatty acid oxidation, and related signaling pathways. Pflugers Arch. 2011;461:153–164. doi: 10.1007/s00424-010-0892-3. [DOI] [PubMed] [Google Scholar]
- 45.Sprague JE. Yang X. Sommers J. Gilman TL. Mills EM. Roles of norepinephrine, free fatty acids, thyroid status, and skeletal muscle uncoupling protein 3 expression in sympathomimetic-induced thermogenesis. J Pharmacol Exp Ther. 2007;320:274–280. doi: 10.1124/jpet.106.107755. [DOI] [PubMed] [Google Scholar]
- 46.Spyrou G. Enmark E. Miranda-Vizuete A. Gustafsson J. Cloning and expression of a novel mammalian thioredoxin. J Biol Chem. 1997;272:2936–2941. doi: 10.1074/jbc.272.5.2936. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka T. Hosoi F. Yamaguchi-Iwai Y. Nakamura H. Masutani H. Ueda S. Nishiyama A. Takeda S. Wada H. Spyrou G. Yodoi J. Thioredoxin-2 (TRX2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 2002;21:1695–1703. doi: 10.1093/emboj/21.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toime LJ. Brand MD. Uncoupling protein-3 lowers reactive oxygen species production in isolated mitochondria. Free Radic Biol Med. 2010;49:606–611. doi: 10.1016/j.freeradbiomed.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turrens JF. Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turrens JF. Alexandre A. Lehninger AL. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 51.Vidal-Puig AJ. Grujic D. Zhang CY. Hagen T. Boss O. Ido Y. Szczepanik A. Wade J. Mootha V. Cortright R. Muoio DM. Lowell BB. Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem. 2000;275:16258–16266. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]
- 52.Wang D. Masutani H. Oka S. Tanaka T. Yamaguchi-Iwai Y. Nakamura H. Yodoi J. Control of mitochondrial outer membrane permeabilization and Bcl-xL levels by thioredoxin 2 in DT40 cells. J Biol Chem. 2006;281:7384–7391. doi: 10.1074/jbc.M509876200. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H. Go YM. Jones DP. Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch Biochem Biophys. 2007;465:119–126. doi: 10.1016/j.abb.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Zhang R. Al-Lamki R. Bai L. Streb JW. Miano JM. Bradley J. Min W. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res. 2004;94:1483–1491. doi: 10.1161/01.RES.0000130525.37646.a7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.