Abstract
A West Nile virus (WNV) isolate from Mexico (TM171-03) and BIRD1153, a unique genotype from Texas, have exhibited reduced murine neuroinvasive phenotypes. To determine if murine neuroinvasive capacity equates to avian virulence potential, American crow (Corvus brachyrhynchos) and house sparrows (Passer domesticus) were experimentally inoculated with representative murine neuroinvasive/non-neuroinvasive strains. In both avian species, a plaque variant from Mexico that was E-glycosylation competent produced higher viremias than an E-glycosylation–incompetent variant, indicating the potential importance of E-glycosylation for avian replication. The murine non-neuroinvasive BIRD1153 strain was significantly attenuated in American crows but not house sparrows when compared with the murine neuroinvasive Texas strain. Despite the loss of murine neuroinvasive properties of nonglycosylated variants from Mexico, our data indicate avian replication potential of these strains and that unique WNV virulence characteristics exist between murine and avian models. The implications of reduced avian replication of variants from Mexico for restricted WNV transmission in Latin America is discussed.
Introduction
West Nile virus (WNV), which was introduced into North America in 1999,1 is a member of the family Flaviviridae and the genus Flavivirus.2 Before the identification of mortality in commercial geese and migratory storks in Israel,3 WNV had not been associated with mortality in avian species. Subsequent WNV outbreaks in New York City in 19994 and elsewhere in North America have been characterized by extensive avian death, with corvids such as the American crow (Corvus brachyrhynchos) being the most susceptible.5 Studies using experimentally infected birds have identified that the strain introduced into North America in 1999 exhibited an increased avian virulence phenotype for American crows6 compared with WNV strains from Australia and Kenya.7 This difference in virulence was associated with a single mutation within the viral helicase protein identified to be under positive selection,8 indicating that high replication rates of WNV could be evolutionarily beneficial in certain avian species. Additional studies have also indicated enhanced viremia and virulence in house sparrows (Passer domesticus) inoculated with a North American WNV strain compared with a Kunjin virus from Australia.9
Despite the wide-scale association of WNV transmission in the United States and Canada with avian virulence, few bird deaths have been reported in Mexico.10,11 Because many viral isolations of WNV come from dead bird specimens in the United States and Canada, this lack of avian mortality in Mexico could help to explain the lack of viral isolates from this region. This fact, coupled with the isolation of a WNV strain from Mexico with reduced murine neuroinvasive properties,12 also could indicate that strains circulating in Mexico have reduced virulence properties within avian hosts. This isolate from Mexico appears to be representative of an intermediary clade between the genotype introduced to North America (East Coast genotype) and the genotype (WN02) that has subsequently predominated in North America since 2002.13
Glycosylation status of the virion envelope glycoprotein (E) of WNV strains has been associated with decreased neuroinvasive properties in the mouse model,14–16 reduced replication in tissue culture17 and reduced replication in chicks (Gallus domesticus).18 Altered virion stability at low pH14 and intracellular maturation17 of nonglycosylated viral particles have been associated with decreased circulating viremias and a subsequent reduced neuroinvasive phenotype in a murine model.19 The first WNV isolate from Mexico, TM171-03, was identified to contain a mixture of large and small plaque phenotype viruses,12 where the small plaque variants were identified to be defective in E protein glycosylation and were not neuroinvasive in a murine model after intraperitoneal inoculation.12
A unique WN02 genotype from southeastern Texas identified in 2003, BIRD1153, also has been associated with reduced neuroinvasiveness in murine hosts, temperature sensitivity, and a small plaque phenotype.20 This genotype has not been subsequently isolated in nature after it was initially identified in 2003. In this study, the avian virulence of representative 2003 mouse neuroinvasive (BIRD1461, TM171-03-pp5 and NY99; WN02, intermediary and East Coast genotypes, respectively) and non-neuroinvasive (TM171-03-pp1 and BIRD1153; intermediary and unique 2003 Texas genotypes, respectively) strains of WNV from Texas and Mexico were characterized. The concordance of murine12,15,20 and avian virulence and the potential for natural murine attenuated WNV strains to circulate within avian hosts are discussed.
Materials and Methods
Detection of pre-existing flaviviral antibodies.
Wild American crows were collected by cannon nets in Bellvue, Colorado, and house sparrows were collected by mist nets in Bakersfield, California. To confirm that American crows and house sparrows had not been exposed to WNV or St. Louis encephalitis virus (SLEV), birds were bled before inoculation and serum was tested by plaque reduction neutralization assays (PRNTs) with WNV or SLEV as described.6 A 90% reduction in plaque-forming units (PFU) (PRNT90), compared with the serum-negative control, was used as the determinant of neutralization. Detection of any neutralizing activity (titers ≥ 1:10) in serum of any bird to either SLEV or WNV precluded use for experimental inoculation.
Virus inoculation.
Viral stocks were diluted to 1,500 PFU/0.1 mL in minimal essential media (MEM) containing 20% fetal bovine serum. One hundred microliters of diluted stocks were inoculated subcutaneously on the breast region of eight American crows or six house sparrows in four infection groups and one group of five house sparrows with the NY99 virus. American crows and house sparrows were inoculated with WNV strains/variants BIRD1461, BIRD1153, TM171-03-pp5, or TM171-03-pp1 (Table 1). Serum collected on 5 days post-inoculation (dpi) from one American crow (123) inoculated with the BIRD1153 virus in which an increased viremia of 9.9 log10 PFU/mL had developed (see Results) was diluted to 1,500 PFU/0.1 mL and used to inoculate an additional six American crows. All American crows and house sparrows were examined for signs of disease twice a day for 14 dpi, and bled once a day through 6 or 8 dpi to measure viremia for house sparrows and American crows, respectively. Blood samples from American crows and house sparrows were collected from the jugular or brachial vein by using a 27- or 28-gauge needle; 0.2 mL or 0.1 mL of blood was added to 0.9 mL or 0.45 mL of MEM supplemented with 20% fetal bovine serum to obtain approximately a 10−1 serum dilution for American crows and house sparrows, respectively. Coagulation was allowed to take place at room temperature for 30 minutes, after which samples were centrifuged at 3,500 × g for 10 minutes to pellet clotted cells. Supernatants from these samples were frozen at –80°C until samples were titrated for infectious virus.
Table 1.
Strains used for West Nile virus virulence testing in American crows and house sparrows*
| Virus strain | Glycosylation at E154–156 | Phenotype | Source | Passage history† | Location, year of isolation | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|
| BIRD1461 | +(N-Y-S) | NIV (+), LP, ts (−) | Blue jay 1461 | V2 | Texas, 2003 | AY712947 | 20 |
| BIRD1153 | +(N-Y-S) | NIV (−), SP, ts (+) | Mourning dove 1153 | V2 | Texas, 2003 | AY712945 | 20 |
| TM171-03-pp1 | −(N-Y-P) | NIV (−), SP, ts (−) | Raven, small plaque | V2 | Mexico, 2003 | AY660002 | 12 |
| TM171-03-pp5 | +(N-Y-S) | NIV (+), LP, ts (−) | Raven, large plaque | V2 | Mexico, 2003 | AY660002 | 12 |
NIV (±); = neuroinvasive, non-neuroinvasive in mice; LP = large plaque on Vero cells; ts = temperature sensitive; SP = small plaque on Vero cells.
Viruses were propagated in Vero (V) cells. Numbers after passage source represent the number of viral passages.
Assaying for infectious virus/plaque size evaluation.
Daily blood samples were assayed by plaque formation in monolayers of Vero cells. In addition, inocula for all viruses were back-titrated by plaque assay to confirm the uniformity of the doses administered. Briefly, serial 10-fold dilutions of serum were inoculated in Vero cells that were overlaid as described.6 Infectivity was enumerated at 72 hours and multiplied by the dilution factor to determine viral titers per milliliter of serum. An initial 1:10 dilution of serum and use of 200 μL of the lowest dilution resulted in a limit of viral PFU detection of 1.7 log10 PFU/mL of serum. At 72 hours, monolayer cultures were fixed with a 20% methanol, 0.5% crystal violet solution, and plaque size was grouped as large (> 2.5 mm), intermediate (2.5–1.5 mm), and small (< 1.5 mm) diameter size groups.
Sequence analyses of inoculum strains and viruses after passage in American crows.
To assess the potential for genetic reversion to a glycosylation-competent motif, the E156–159 nucleotide region was sequenced for the nonglycosylated TM171-03-pp1 virus after isolation at 4 dpi from serum of American crows. Viral RNA was extracted from 4 dpi serum of individual American crows. Additionally, RNA from virus isolated from the brains of American crows that died following infection with the TM171-03-pp1 variant virus was similarly processed. Reverse transcription was performed by using the primer WNc2506 (5′-GCTCTTGCCGGCTGATGTCTATG-3′). Generation of an amplicon encoding the N-linked glycosylation sequence from the resulting cDNA was performed by using primers WN1346 (5′-TGCCTGCTCTACCAAGGCAATAGG-3′) and WNc1797 (5′-ATGACCCGACGTCAACTTGACAGT-3′). Primer WN1346 (5′-TGCCTGCTCTACCAAGGCAATAGG-3′) was used to sequence the glycosylation site, and primers WNc1797, WN898 (5′-GGCAGCCGTCATTGGTTGGATGC-3′), WN7574 (5′-CAACAACTGCCATCGGACTC-3′), WNc10,116 (5′-TCGCACCACCAGCCACCATTGTCG-3′), WNc10,993 (5′-CTCGCACCACCAGCCACCATTGTCG-3′), and WN10,552 (5′-ACCGGAAGTTGAGTAGACGGTGCTG-3′) were used to sequence the genetic differences between the BIRD1153 and NY99 strains from two American crows that died following infection with this viral strain. Sequencing of the complete genomes of the unpassaged TM171-03-pp1 and pp5 plaque variants were performed as described.12
Determination of cross-protection.
Blood (0.6 mL) was drawn at 14 dpi from American crows to determine titers by using a two-way beta PRNT. Briefly, two-fold dilutions of bird serum samples were incubated at 56°C for 30 minutes and mixed with 100 PFU of NY99 (strain 382-99) virus as described.6 Plaques were counted and neutralization was reported as a 90% reduction in plaque formation compared with the serum-negative control. Crows that survived through 14 dpi were subsequently challenged with 1,500 PFU of strain 382-99. Challenged crows were bled daily through 7 dpi. Serum samples from the seven daily bleedings were diluted 1:10 in MEM diluent, centrifuged, and stored at –80°C. Samples were thawed and titrated in Vero cells as described above.
Statistical analyses.
Analyses of variance was used to compare mean peak viremia, duration of viremia, day of viremia onset, and day of death among viruses for experimentally infected American crows. Means were compared between viral strains/variants by using Tukey's honestly significant difference adjustment for multiple comparisons. Clinical outcome (survival versus death) comparisons of viremia magnitudes were made by using the Student t-test with Welch's modification for unequal variances. Similarly, Student t-tests were used to compare BIRD1153 initial versus re-infection daily viremia magnitudes and peak viremias for American crows inoculated with the BIRD1153 strain. Mortality and morbidity proportions were compared by using Fisher's exact test. Viremias of house sparrows were compared among viruses by using two-way analysis of variance, blocked by dpi. Because of the relatively low sample sizes (n = 6 or 8) in this study, a threshold for significance of α = 0.10 was applied to increase the power of our statistical tests.
Results
None of the pre-infection serum from American crows showed the presence of flaviviral-reactive antibody by PRNT90, whereas 20% of house sparrows had neutralizing antibody to WNV and were excluded. This seroprevalence was consistent with previous findings in California.21
Mortality rates for American crows.
Mortality rates ranged from 100% (BIRD1461) to 25% (BIRD1153) for the Texas WNV strains. The WNV variants TM171-03-pp5 and TM171-03-pp1 from Mexico had mortality rates of 63% and 50%, respectively (Table 2 and Figure 1A). When compared by Fisher's exact test, a significantly higher proportion of American crows inoculated with the BIRD1461 strain died because of infection than with the BIRD1153 and gly(−) variant from Mexico (P < 0.007 and P < 0.08, respectively), but not the gly(+) variant from Mexico (P = 0.2). The E gly(−) variant from Mexico was significantly slower in eliciting a mortality response than the BIRD1461, BIRD1153, NY99 virus, and the gly(+) variant from Mexico (P < 0.05). The nonglycosylated variant from Mexico induced a significantly reduced mortality rate (P = 0.077) than the BIRD1461 strain. The mortality rate of the glycosylated variant from Mexico did not differ significantly (P = 0.2) from that of the highly virulent Texas strain (BIRD1461) (Table 2 and Figure 1A).
Table 2.
Viremia and mortality profile of American crows inoculated with West Nile viruses
| Virus | Mean ± SD peak viremias* | Mean ± SD day of maximum viremia | % Mortality† | Mean ± SD day of death‡§ |
|---|---|---|---|---|
| BIRD1461 | 10.0 ± 0.4 | 4.4 ± 0.5 | 100 (8) | 4.9 ± 0.2 (8) |
| BIRD1153 | 6.3 ± 1.7 | 4.1 ± 0.7 | 25 (8) | 6.3 ± 0.4 (2) |
| TM171-03-pp1 | 4.4 ± 1.3 | 4.4 ± 0.7 | 50 (8) | 7.5 ± 0.6 (4) |
| TM171-03-pp5 | 6.9 ± 3.4 | 4.2 ± 0.4 | 63 (8) | 5.9 ± 0.2 (5) |
| P991 (NY99) | 9.4 ± 0.8 | 4 | 100 (8)¶ | 5.5 ± 0.5 (8)¶ |
Numbers represent log10 plaque-forming units/mL of serum titers determined by plaque assay on Vero cells.
Numbers in parentheses represent numbers of American crows inoculated.
Mean day of death is represented in days post-inoculation.
Numbers represent the total number of decedent American crows used for the calculation of mean day of death.
Data from Kinney and others.7
Figure 1.
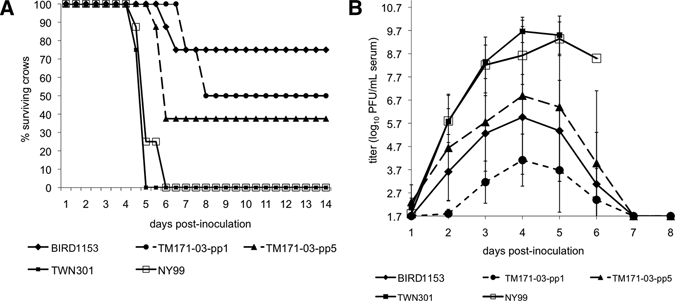
A, Percentage survivorship of eight American crows exposed to 1,500 plaque-forming units (PFU) of representative West Nile viruses (WNVs). American crows were checked twice a day for signs of disease. B, Mean (±SD) viremia response of eight American crows exposed to 1,500 PFU of representative WNVs. Viremias were determined by plaque assay on Vero cells with a detection limit of 1.7 log10 PFU/mL of serum. Only positive error bars and offset data points are presented for clarity.
Viremia profiles for American crows.
The mean ± SD peak viremia for American crows infected with the BIRD1461 WNV strain from Texas was 10.0 ± 0.4 log10 PFU/mL of serum and occurred on 4.4 ± 0.5 dpi (Table 2 and Figure 1B). Viremia peak, onset, and day of peak viremia were not distinguishable between the BIRD1461 and NY996 viruses (P > 0.55). Onset of viremia was detected in all crows inoculated with the BIRD1461 strain at 24 hours post-infection. In contrast, the poorly murine neuroinvasive strain BIRD1153 produced a mean peak viremia in American crows of only 6.3 ± 1.7 log10 PFU/mL of serum that occurred on 4.1 ± 0.7 dpi (Table 2 and Figure 1B). Magnitude of viremia in crows was significantly different (P < 0.01) between the murine neuroinvasive and non-neuroinvasive strains on dpi 2–4. The approximate 5,000-fold difference in peak viremia was statistically significant (P < 0.0001), but the timing of the peak viremia was not significant (P > 0.6).
Mean viremia was significantly higher and day of onset of viremia was significantly earlier for the TM171-03-pp5 E gly(+) strain than the TM171-03-pp1 E gly(−) variant. However, the day of peak viremias for the TM171-03-pp1 and TM171-03-pp5 variants was indistinguishable and occurred on 4.4 ± 0.7 dpi and 4.2 ± 0.4 dpi, respectively (Table 2 and Figure 1B).
The nonglycosylated, TM171-03-pp1 WNV variant from Mexico produced a significantly lower mean peak viremia (4.4 ± 1.3 log10 PFU/mL of serum) than the Texas murine neuroinvasive strain BIRD1461 (10.0 ± 0.4 log10 PFU/mL of serum) (P < 0.0001) and the Texas BIRD1153 strain on 2 dpi (P = 0.1) and 3 dpi (P < 0.07). However, when compared with the nonglycosylated variant, the glycosylation competent [E gly(+); TM171-03-pp5] variant from Mexico had a significantly higher (P = 0.000002) mean peak viremia of 6.9 ± 3.4 log10 PFU/mL of serum (Table 2 and Figure 1B) that was indistinguishable (P > 0.36) from that of the BIRD1461 strain.
Survival partitioning in American crows.
Considerable variability of viremia response was identified within the BIRD1153 and TM171-03-pp5 E gly(+) American crow infection groups. When viremia data were analyzed on the basis of partitioning of clinical outcome of infection (Figure 2), significant differences were identified between crows that survived or died because of infection for the TM171-03-pp5 (Figure 2A) and BIRD1153 viruses (Figure 2C). A mean peak viremia of 8.4 ± 1.0 log10 PFU/mL was identified for American crows that died because of infection (euthanized) with the BIRD1153 strain, whereas the surviving crows had mean peak viremia titers of only 5.5 ± 0.3 log10 PFU/mL of serum. When daily viremias were compared, only 5 dpi values were significantly different between survivors (4.3 log10 PFU/mL of serum) and decedents (8.5 log10 PFU/mL of serum) (P = 0.077) (Figure 2C). Despite these marginally significant differences in the magnitude of viremia, statistical analyses failed to identity more days in which viremias were significantly different because of the small number of decedent American crows within this infection group (n = 2).
Figure 2.
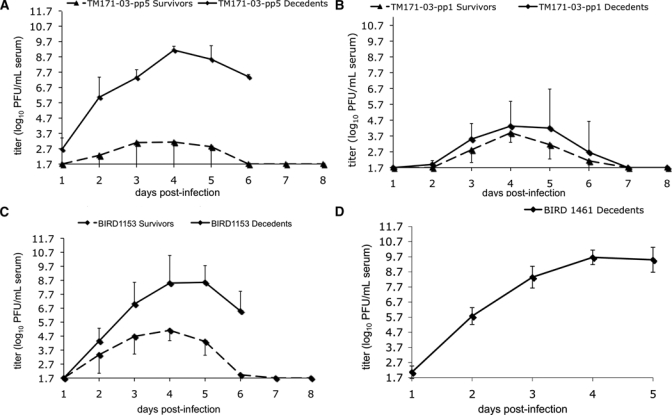
Mean (±SD) viremia profile comparisons for American crows inoculated with A, TM171-03-pp1, B, TM171-03-pp5; C, BIRD1153; and D, BIRD1461 West Nile viruses. Triangles indicate survivors and boxes indicate decedents. Bars represent SDs from the means. Either positive or negative error bars are presented for clarity. PFU = plaque-forming units.
When viremia data for the TM171-03-pp5 infection group were assessed on the basis of outcome, mean peak viremias in the group of American crows that died because of infection were 1,500-fold higher than those for crows who survived infection, and mean daily viremia peak differences were up to 6 log10 PFU/mL of serum higher for birds that died than for birds that survived on 4 dpi (Figure 2A). Daily viremia values on 1–6 dpi were significantly different between survivors and decedents for this gly(+) strain from Mexico (P < 0.09). Birds that died because of infection in this group had an onset of viremia at 1 dpi and mean peak titers that reached 9.1 ± 0.3 log10 PFU/mL of serum compared with a mean peak titer of 3.1 ± 2.4 log10 PFU/mL for crows that survived. Variability within the peak daily viremia values of this group was considerable; two of the three American crows that survived infection with the TM171-03-pp5 variant did not have a viremia on any of the eight days sampled after infection (< 1.7 PFU/mL of serum). Interestingly, no significant differences in peak viremia, mean daily peak titers, or viremia onset were identified between the American crows that survived or died because of infection with the TM171-03-pp1 variant.
Inoculation of an additional six American crows with virus derived from an American crow (AMCR 123) in which an extremely high serum viremia (9.9 log10 PFU/mL) had developed after infection with the BIRD1153 strain resulted in an approximately four-fold higher peak viremia (6.8 log10 PFU/mL of serum versus 6.3 log10 PFU/mL of serum) that occurred one day earlier (3 dpi versus 4 dpi for the mean of all American crows originally inoculated with the BIRD1153 strain) (Figure 3). However, this mean peak viremia was not significantly different (P = 0.5). Interestingly, the mean viremia value derived from six secondarily inoculated American crows was significantly lower (P < 0.0007) (mean 6.6 ± 0.7 log10 PFU/mL of serum, range = 6.1–8.4 log10 PFU/mL) than the original American crow 123 that peaked at 9.9 log10 PFU/mL of serum of the passaged virus (Figure 3). Comparison of the daily mean viremia titers of BIRD1153 American crow passage 1 with the non-passaged BIRD1153 single American crow viremia profile indicated significant differences on 3–5 dpi (P < 0.003). In addition, significant differences in the daily viremia means of the BIRD1153-AMCR-P1 profile compared with the non-passaged BIRD1153 profile were identified on 2 dpi (4.6 log10 PFU/mL of sera versus 3.6 log10 PFU/mL of serum, P < 0.08) and on 3 dpi (6.6 log10 PFU/mL of serum versus 5.3 log10 PFU/mL of serum; P < 0.07) (Figure 3).
Figure 3.
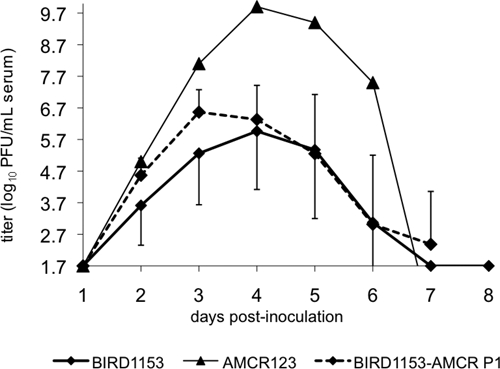
Viremia response of American crows infected with the BIRD1153 West Nile virus strain (n = 8; positive error bars representing SDs from the mean are presented). The single profile of American crow 123 infected with BIRD1153 and the secondary infection of American crows (n = 6; negative error bars representing SDs from the mean are presented) inoculated with virus derived from 4 day post-inoculation American crow 123. PFU = plaque-forming units.
Sequencing of variants and American crow–passaged viruses.
Full genome nucleotide sequencing of the parental TM171-03 isolate from Mexico identified four amino acid differences when compared with the NY99 strain at premembrane 141 (prM141) (Ile in NY99 to Thr in TM171-03), envelope 156 (E156) (Ser to Pro), NS4B-245 (Ile to Val) and NS5-898 (Thr to Ile).12 Sequence analyses of the TM171-03-pp5 and TM171-03-pp1 variants confirmed the presence of these mutations in both variants, with the exception of the E156 being identified to be a Ser in -pp5 and Pro in -pp1. Additional polymorphisms were identified between the plaque variants at 5′ untranslated region (UTR) position 85 (G to A), NS5-224 (Arg to Gly), and 3′UTR position 10989 (A/G mixture to G) (Table 3). RNA was extracted from 4 dpi serum of American crow 123 in whom an increased viremia developed after inoculation of the BIRD1153 strain (peak viremia of 9.9 log10 PFU/mL of serum) to determine if reversion had occurred at any of the potential residues that could be associated with attenuation (Table 3; prM-156, E-159, NS4B-249, NS5-804, 3′UTR- 10,596, 10,774, 10,799 and 10, 851). Sequencing across these regions failed to identify reversion to the NY99 residues or any genetic polymorphisms at these positions.
Table 3.
Full-genomic sequence differences among West Nile virus strains*
| Position | NY99 | BIRD1461 | BIRD1153 | TM171-03 | TM171-03-pp5 | TM171-03-pp1 |
|---|---|---|---|---|---|---|
| 5′UTR 50 | a | – | – | g | g | g |
| 5′UTR 71 | a | – | – | g | g | g |
| 5′UTR 82 | a | – | – | – | – | g |
| prM-141 | I | – | – | T | T | T |
| prM-156 | V | – | I | – | – | – |
| E-156 | S | – | – | S/P | – | P |
| E-159 | V | A | A | – | – | – |
| NS3-180 | E | D | – | – | – | – |
| NS3-328 | E | K | – | – | – | – |
| NS4A-135 | V | M | – | – | – | – |
| NS4B-245 | I | – | – | V | V | V |
| NS4B-249 | E | – | G | – | – | – |
| NS5-224 | R | – | – | – | G | – |
| NS5-619 | A | S | – | – | – | – |
| NS5-804 | A | – | V | – | – | – |
| NS5-898 | T | – | – | I | I | I |
| 3′UTR 10,408 | c | T | – | – | – | – |
| 3′UTR 10,596 | a | – | g | – | – | – |
| 3′UTR 10,774 | c | – | t | – | – | – |
| 3′UTR 10,799 | a | – | g | – | – | – |
| 3′UTR 10,828 | t | – | – | g | g | g |
| 3′UTR 10,851 | a | g | g | g | g | g |
| 3′UTR 10,989 | g | – | – | a | g | a/g |
Serologic response/protection.
Surviving American crows were bled at 14 dpi and serum samples were tested for neutralizing antibodies as measured by PRNT90 using NY99 wild-type virus. A total of 13 American crows survived challenge with the BIRD1153 (6), TM171-03-pp5 (3), and TM171-03-pp1 (4) variants. With one exception, all serum samples at 14 dpi had at least a 1:40 PRNT90 titer. The one American crow that did not have a PRNT90 titer (< 1:10) had initially been exposed to TM171-03-pp5. However, a detectable viremia (> 1.7 log10 PFU/mL sera) did not develop in this American crow at any time point assayed after initial infection. The PRNT90 titers ranged from 1:40 to 1:1280. All 12 seropositive American crows were protected from challenge with a lethal dose of the NY99 strain and none had any detectable viremia (> 1.7 log10 PFU/mL of serum) post-challenge. The one American crow (AMCR 137) in whom a 14 dpi PRNT90 titer did not develop had typical clinical disease after challenge with NY99 wild-type virus, including a peak viremia of 8.6 log10 PFU/mL of serum on 3 dpi (Figure 3) and died because of the infection on 5 dpi. Interestingly, one of two American crows (AMCR 143) for which no detectable viremias were identified after initial infection with TM171-03-pp5 showed development of a PRNT90 titer of 1:320 and survived challenge with NY99 wild-type virus.
Plaque size analyses.
Small plaque size has been used as a potential marker for murine neuroinvasive attenuation of WNV isolates.20 Mixed plaque morphologies were identified in plaque assays performed for daily viremia samples of American crows infected with the BIRD1153, TM171-03-pp5 and TM171-03-pp1 variants. Therefore, plaque sizes were monitored during the course of infection in American crows to assess any potential modulation in viral phenotype during infection with the BIRD1153, TM171-03-pp5, and TM171-03-pp1 variants that might be associated with clinical outcome of infection. The American crows that were inoculated with the non-neuroinvasive, temperature-sensitive, small plaque variant isolate (BIRD1153 from Texas), showed a 35% mean reduction in the percentage of small plaques in birds that died because of infection (Figure 4A). In contrast, American crows that survived BIRD1153 infection had no significant change in the proportion of viruses with the small plaque phenotype, whereas American crows that survived infection with the E gly(−) variant from Mexico (TM171-03-pp1) had an increase in small plaque percentage from approximately 50% on 2 dpi to greater than 90% on 5 dpi (Figure 4B). No statistically significant differences could be determined because of the considerable variability in plaque phenotypes of virus from American crows that died because of infection with this isolate. Finally, the one viremic American crow survived infection with the TM171-03-pp5 E gly(+) variant from Mexico showed a significantly higher percentage of small plaques on 2, 3, and 6 dpi than the five American crows that died because of infection with this variant (Figure 4C).
Figure 4.

Plaque size comparison of virus derived from the serum of American crows infected with A, the BIRD1153 West Nile virus strain; and B, TM171-03-pp1; and C, pp3 variants. Daily serum samples from all eight American crows were assayed by plaque assay on Vero cell monolayers. Plaque sizes were differentiated at 37°C and graphically displayed on the basis of the clinical outcome of the infection. Bars represent SDs from the mean. Virus from the serum of decedent American crows are designated in black and surviving crows are designated in white.
Viremia response in house sparrows.
Most house sparrows survived infection; only 1 of 6 and 1 of 5 birds succumbed on 5 dpi after inoculation with BIRD1461 and NY99 viruses, respectively. Overall, the amplitude and pattern of the viremia profiles varied significantly among WNV strains/variants (strains: F = 31.9, degrees of freedom = 4, 139, P < 0.0001; strain × time interaction: F = 6.1, degrees of freedom = 20, 139, P < 0.0001; Figure 5). When compared by using an a posteriori Tukey-Kramer test (P > 0.05), the overall mean viremias associated with the various strains could be divided into three statistical group: viremia produced by NY99 was the highest of the groups (4.7 log10 PFU/mL) and peaked on 3 dpi; group 2 comprised BIRD1461, BIRD1153, and TM171-03-pp5 and had broadly overlapping viremias with means of 3.5, 3.0, and 2.5 log10 PFU/mL, respectively, peaking on 2 dpi; and the third and lowest was the viremia produced by the TM171-03-pp1 variant (mean = 1.9 log10 PFU/mL) that was only detectable on 2 dpi. The NY99 strain produced the highest viremia with a peak titer of 7.2 log10 PFU/mL of serum on 3 dpi. The Texas American crow virulent WNV strain BIRD1461 showed a faster onset of viremia than the NY99 strain and generated its peak titer one day earlier (2 dpi). However, peak titer for this virus was approximately 10-fold lower than for the NY99 strain. The peak viremia profiles of both of these viruses were significantly higher than both variants from Mexico but were indistinguishable from that of the BIRD1153 strain.
Figure 5.
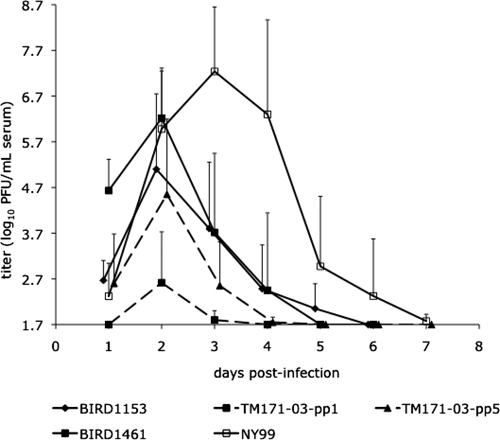
Viremia response (mean ± SD) of six house sparrows inoculated with 1,500 plaque-forming units (PFU) of each respective West Nile virus strain (dashed lines indicate for strains from Mexico). Only positive error bars and offset data points are presented for clarity. Viremias were determined by plaque assay on Vero cells with a detection limit of 1.7 log10 PFU/mL of serum.
Discussion
A major goal of this study was to determine on the basis of a set of representative WNV strains (Table 1) whether virulence in two avian hosts was directly related to virulence in a mouse model of WNV neuroinvasive disease. In addition, we wished to determine whether the isolation of WNV strains in the Americas with reduced virulence in the mouse model indicated possible changes in the transmission dynamics of WNV that would explain apparent differences in the epidemiology of WNV in the Caribbean, Mexico and Central America compared with that in other areas of North America. Identification of the potential attenuating role of mutations in these recent WNV isolates compared with the NY99 North American prototype may provide information on selective criteria associated with the emergence of WNV avian-attenuated phenotypes in tropical or subtropical ecosystems. The results presented indicate that avian and murine virulence and attenuated phenotypes most likely resulted from complex biological host interactions in disparate vertebrate systems.
Loss of glycosylation of the surface E glycoprotein of natural, laboratory-adapted16,22 and site-directed mutants15 has been associated with decreased replication capacity and neuroinvasive properties in a mouse model. Glycosylation status has also been associated with cell type or species specificity for proper viral maturation and release of WNV lineage I and II viruses,23 pH stability, particle assembly, virion maturation, and infectivity in tissue culture systems.23,24 Natural Kunjin viral isolates lacking E glycosylation have been shown to rapidly incorporate a glycosylation motif after serial passage in vertebrate cell culture.24 Transmission electron microscopy has further indicated that the maturation of glycosylation-deficient WNV particles derived from the Sarafend WNV strain on intracellular membranes could explain reduced titers in culture systems.17 These investigators further demonstrated that providing a glycosylated envelope gene in trans was sufficient to abrogate this defective phenotype.17 Despite the aforementioned debilitation in viral replication and loss of murine neuroinvasiveness, nonglycosylated WNV isolates have been found repeatedly from the field.12,25,26
With the exception of a previous study that demonstrated reduced virulence of a plaque-purified WNV lacking a functional glycosylation motif in chicks,18 little is known regarding the role of glycosylation on the modulation of virulence of WNV in natural avian hosts for WNV. Viremia production (American crows/house sparrows) and survival time variation of American crows infected with the TM171-03-pp5 and TM171-03-pp1 variants that differ principally in their glycosylation status and in 5′UTR position 85 (G to A), NS5-224 (Arg to Gly) and 3′UTR position 10989 (A/G mixture to G) mutations indicate the potential for additional attenuating mutations in the genome of the variants from Mexico. Poor peripheral replication presumably reduced the efficiency for dose-dependent neuroinvasion, resulting in murine attenuation of the E-gly(−) variant. Variable peripheral replication in avian hosts appeared to modulate virulence phenotype. However, the results derived from the E gly(−) variant from Mexico indicated the potential for differential tissue tropism or virulence phenotypes of some WNV strains for avian hosts. In contrast to the murine model, in which increased viremia has been directly correlated with neurovirulence, there was a poor correlation between avian death and viremia with the variants from Mexico reported herein, which suggests that these viruses may have increased neurotropism.
Both of the TM171-03 variants and the BIRD1153 strain were significantly attenuated for virulence in the American crow model (as measured by differences in daily viremia at 1–5 dpi, mean peak viremia, viremia onset, mortality rate, and average survival times) compared with the NY99 North American prototype and the Texas BIRD1461 strain. The TM171-03-pp1 and -pp5 viruses both encoded amino acid differences compared with NY99 virus at residues prM-141, NS4B-245, and NS5-89812 (Table 3), and it is likely that one or more of these residues plays a significant role in the reduced avian virulence of these variants. However, reductions in the magnitude and duration of viremia observed for TM171-03-pp1 compared with TM171-03-pp5 (Figure 1) suggested that E glycosylation may have some modulating effect on the virulence phenotype and/or tropism of these variants. Alternatively, other mutations identified in the 5′ and 3′ UTRs of either TM171-03-pp variant or at NS5-225 of TM171-03-pp5 (Table 3) may also have contributed to the differences in viremia profiles observed for these two variants. These potentially confounding additional genetic changes and those in the nonglycosylated plaque purified viruses from Mexico could be responsible for the reduced virulence and viremia observed in this study or in chicks with a plaque-purified glycosylation defective mutant.18 Future reverse genetic studies will be required to specifically assess the role of envelope glycosylation and additional mutations identified herein on avian pathogenesis phenotypes.
Unlike the significant level of attenuation observed in American crows for the BIRD1153 isolate, the E gly(+) strain from Mexico generated mean peak viremias comparable with that of the murine neuroinvasive Texas strain (BIRD1461) in house sparrows (Figure 5). This finding could indicate the relative selective importance of different avian hosts for emergence of alternative WNV genotypes. Significantly higher viremias were identified at multiple time points in house sparrows inoculated with NY99 virus compared with the BIRD1461 strain, and differences were only observed at 1 dpi in American crows, which highlights the interspecies incongruence in virulence potential for WNV strains in alternative avian hosts.
The BIRD1461 and BIRD1153 strains differed at eight nucleotide positions (Table 3). Of these differences, only the NS4B-249 and NS5-619 amino acids and 10,596, 10,774, and 10,799 3′UTR nucleotide substitutions were unique to the BIRD1153 strain (i.e., not present in the NY99 viral genome). Incorporation of the NS4B BIRD1153 mutation in combination with either the NS5 or the three 3′UTR BIRD1153 substitutions into the NY99 virus genetic backbone were required to elicit a temperature-sensitive, small plaque, murine non-neuroinvasive strain.27 Interestingly, none of the potential genetic determinants have been identified in the variants from Mexico, which indicates that there are multiple molecular mechanisms for the attenuation of these variants through independent, polygenic pathways.
Previous studies have demonstrated that small numbers of genetic substitutions can have a dramatic effect on avian virulence phenotypes. For instance, infection with a WNV strain from Kenya (KN-3829)28 that differed from the NY99 WNV strain by only 11 amino acids29 produced a mortality rate of only 20–30% in American crows and has demonstrated significantly reduced serum viremias compared with the NY99 strain.30 None of the amino acid substitutions identified between the WNV strain from Kenya and the NY99 strain29 have been identified in either of the variants from Mexico or the BIRD1153 murine non-neuroinvasive virus investigated in these studies. This finding was not particularly surprising given the fact that the KN-3829 strain has a virulent, neuroinvasive phenotype in mice.14 In this study, strains that were non-neuroinvasive in mice still caused significant mortality in avian species, indicating that variable pathogenic mechanisms of virulence and attenuation are present in these disparate vertebrate models and that the murine model may not accurately predict virulence in avian hosts.
Viremia and mortality appear to be highly correlated in the American crow virulence model of WNV infection. This finding was evident for infection with the BIRD1461, BIRD1153, and the TM171-03-pp5 strains/variants, in which the mean peak viremias of all decedent American crows exceeded 8.4 log10 PFU/mL of serum and the mean peak viremias for survivors never exceeded 6 log10 PFU/mL of serum. This model is consistent with a hypothesis that once peripheral replication in the avian host reaches a particular threshold (> 6 log10 PFU/mL of serum), invasion of the central nervous system or other vital organ systems occurs through a non-specific passive process. However, the E gly(−) variant from Mexico (TM171-03-pp1) did not demonstrate this viremia response/mortality relationship. Interestingly, peak viremias of crows that died because of infection with this variant were in some cases lower than those identified for survivors (Figure 2). Only one American crow in this infection group showed development of a viremia > 6 log10 PFU/mL of serum. Nonetheless, four of eight crows died because of infection with this plaque-purified virus, and all four decedents had nonglycosylated WNV isolated from brain tissue at the time of death as determined by sequence analyses.
To investigate this viremia magnitude/mortality relationship, peak viremias were plotted as a function of percent mortality and a regression analysis was performed to describe this relationship deriving a best-fit line (y = 0.059X + 3.143, R2 = 0.702). On the basis of this relationship, and assuming that each of the viruses exhibited similar virulence (i.e., survivorship was exclusively a determinant of viremia magnitude), an estimate of a peak viremia of 6.1 log10 PFU/mL of serum would result in a 50% mortality rate within the infection group. Further investigations of differences in increased viremia duration and differential tissue tropisms are needed to further evaluate this theory. However, the finding of a 50% mortality rate for which three of the four decedent American crows infected with the TM171-03-pp1 E gly(−) variant from Mexico showed development of a peak viremia well below the > 6 log10 PFU/mL of serum hypothetical threshold (3.6, 3.4, 4.4 log10 PFU/mL of serum, respectively) suggest that there could be differences in tissue tropism for some of these strains/variants.
Removal of the TM171-03-pp1 data point from the regression analyses generated a best-fit line (y = 0.048X + 4.71) with a much greater R2 = 0.895, which further supports differential tropisms of this variant from Mexico. This second regression line subsequently increased the hypothetical circulating titer required to result in 50% mortality from 6.1 to 7.1 log10 PFU/mL of serum and was consistent with estimates reported for a variety of avian species.31 When viremia parameters of survivors and decedents were compared, viremia magnitude, onset, and daily magnitudes (1–6 dpi) were all significantly different (P < 0.0001). On the basis of these preliminary findings, further studies investigating the alternative pathogenicity of the variants from Mexico that yield low viremias are warranted.
Numerous reports have provided serologic evidence of WNV transmission in the Western hemisphere south of the United States.10,32–38 Despite this fact, with the exception of sites adjacent to the United States border,39 WNV was isolated only once in Mexico from a diseased raven in a Tabasco zoo.11 This paucity of viral isolations could be the result of a combination of reduced avian mortality, reduced viral loads within avian hosts, or limited mosquito testing. In addition to observation of reduced avian mortality in Mexico, the Caribbean, and Central and South America, serosurveys of equids have demonstrated evidence of spillover transmission32,34,35,40–42 without documented equine disease, until reaching the temperate latitudes of Argentina.43
A number of theories could explain reduced virulence. These theories include 1) circulation of more attenuated strains in Latin America that have been the result of differential selection in local avian or mosquito fauna; 2) founder's effects from rare introduction events from viremic migratory birds; 3) herd immunity afforded by serologic cross-protection by heterologous flaviviruses44 circulating in these geographic locations; or 4) potential dilution effect of the greater avian species diversity in the tropics that could confound the identification of mortality in highly susceptible species, as has been prevalent in temperate regions of North America.45,46
Data in this study indicate that the small plaque variant isolated from southeastern Mexico, in addition to being non-neuroinvasive in mice, was also considerably attenuated in its ability to replicate in two avian species. In particular, seven of eight American crows infected with the E gly(−) variant from Mexico did not have viremia titers greater than 4.4 log10 PFU/mL. On the basis of dose-response data for common WNV mosquito vectors within the United States,47 mosquito oral infection rates would be expected to be quite low at these titers, which would limit viral transmission potential (and overall fitness) of these avian attenuated strains/variants. The glycosylation-competent, large plaque–forming variant from Mexico produced serum viremias greater than 5 log10 PFU/mL in six of the eight American crows, which indicated that this virus would be much more likely to be infective and subsequently to be transmitted by competent mosquito vectors. Specific vector competence data for mosquito species from Latin America will be needed to further assess this hypothesis.
The fact that the TM171-03 isolate constituted a mixture of two plaque variants12 is intriguing and could indicate that differential selective factors were associated with the presence of two distinctive viral populations in the infected raven. Highly susceptible and temperate region prevalent avian hosts (American crows and house sparrows) were used for these virulence studies and could afford a radically different selective environment from that found in sympatric avian hosts in Latin America. Preliminary data have indicated that the TM171-03 isolate (mixed plaque isolate) was capable of eliciting significant viremias in resident house sparrows, clay-colored thrushes, and great-tailed grackles from Mexico (Komar N and Estrada-Franco JG, unpublished data), indicating that viremia potential could be significantly modulated by avian species and/or intraspecies geographic variants or the potential presence of the mixed viral populations. Host competence studies using sympatric avian hosts with additional isolates from areas lacking human, animal, and avian mortality will be needed to fully assess the role of alternative WNV genotypes with altered disease presentation in Latin America.
ACKNOWLEDGMENTS
We thank Vincent Martinez and Brian Carroll (Center for Vector-borne Diseases) for providing excellent technical support for this project. Trapping of American crows was performed under U.S. Fish and Wildlife Scientific Collecting Permit MB-032526, and experimental inoculations of crows were performed under UC Davis IACUC protocol 1127. Collection and infection of house sparrows was performed under UC Davis IACUC Protocol 11184, U.S. Geological Survey Federal Bird Banding Permit 22763, California Resident Scientific Collection Permit 801049-02 from the State of California Department of Fish and Game, and Federal Fish and Wildlife Permit MB082812-0.
Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of The Centers for Disease Control and Prevention.
Footnotes
Financial support: This study was supported by National Institutes of Health grants AI061822, AI055607, and AI67847; Pacific Southwest Regional Center for Excellence (PSWRCE) U54 AI065359; and Centers for Disease Control and Prevention grant CI000235.
Authors' addresses: Aaron C. Brault, Division of Vector-borne Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, E-mail: abrault@cdc.gov. Stanley A. Langevin, Wanichaya N. Ramey, Ying Fang, Christopher M. Barker, and William K. Reisen, Center for Vector-borne Diseases and Department of Pathology, Microbiology and Immunology, School of Veterinary Medicine, University of California, Davis, CA, E-mails: salangevin1@gmail.com, wramey@berkeley.edu, ylfang@ucdavis.edu, cmbarker@ucdavis.edu, and arbo123@pacbell.net. David W. C. Beasley and Alan D. T. Barrett, Departments of Pathology and Microbiology and Immunology, Center for Emerging Infectious Diseases and Biodefense, Institute for Human Infections and Immunity, University of Texas Medical Branch, Galveston, TX, E-mails: dwbeasle@utmb.edu and abarrett@utmb.edu. Todd A. Sanders, U.S. Fish and Wildlife Service–Division of Migratory Bird Management, Portland, OR, E-mail: todd_sanders@fws.gov. Richard A. Bowen, Department of Biomedical Sciences, Colorado State University, Fort Collins, CO, E-mail: rbowen@colostate.edu.
References
- 1.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, Hall RA, MacKenzie JS, Cropp CB, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 2.van Regenmortel MHV. In: Virus Taxonomy. Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses. van Regenmortel MH, Fauquet CM, Bishop DH, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeogh DJ, Pringle CR, Wickner RB, editors. San Diego: Academic Press; 2000. pp. 3–16. (Introduction to the species concept in virus taxonomy). [Google Scholar]
- 3.Malkinson M, Banet C. The role of birds in the ecology of West Nile virus in Europe and Africa. Curr Top Microbiol Immunol. 2002;267:309–322. doi: 10.1007/978-3-642-59403-8_15. [DOI] [PubMed] [Google Scholar]
- 4.Steele KE, Linn MJ, Schoepp RJ, Komar N, Geisbert TW, Manduca RM, Calle PP, Raphael BL, Clippinger TL, Larsen T, Smith J, Lanciotti RS, Panella NA, McNamara TS. Pathology of fatal West Nile virus infections in native and exotic birds during the 1999 outbreak in New York City, New York. Vet Pathol. 2000;37:208–224. doi: 10.1354/vp.37-3-208. [DOI] [PubMed] [Google Scholar]
- 5.Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerg Infect Dis. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brault AC, Langevin SA, Bowen RA, Panella NA, Biggerstaff BJ, Miller BR, Komar N. Differential virulence of West Nile strains for American crows. Emerg Infect Dis. 2004;10:2161–2168. doi: 10.3201/eid1012.040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinney RM, Huang CY, Whiteman MC, Bowen RA, Langevin SA, Miller BR, Brault AC. Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol. 2006;87:3611–3622. doi: 10.1099/vir.0.82299-0. [DOI] [PubMed] [Google Scholar]
- 8.Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, Ramey WN, Panella NA, Holmes EC, Powers AM, Miller BR. A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet. 2007;39:1162–1166. doi: 10.1038/ng2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langevin SA, Brault AC, Panella NA, Bowen RA, Komar N. Variation in virulence of West Nile virus strains for house sparrows (Passer domesticus) Am J Trop Med Hyg. 2005;72:99–102. [PubMed] [Google Scholar]
- 10.Fernandez-Salas I, Contreras-Cordero JF, Blitvich BJ, Gonzalez-Rojas JI, Cavazos-Alvarez A, Marlenee NL, Elizondo-Quiroga A, Lorono-Pino MA, Gubler DJ, Cropp BC, Calisher CH, Beaty BJ. Serologic evidence of West Nile virus infection in birds, Tamaulipas State, Mexico. Vector Borne Zoonotic Dis. 2003;3:209–213. doi: 10.1089/153036603322662192. [DOI] [PubMed] [Google Scholar]
- 11.Estrada-Franco JG, Navarro-Lopez R, Beasley DW, Coffey L, Carrara AS, Travassos da Rosa A, Clements T, Wang E, Ludwig GV, Cortes AC, Ramirez PP, Tesh RB, Barrett AD, Weaver SC. West Nile virus in Mexico: evidence of widespread circulation since July 2002. Emerg Infect Dis. 2003;9:1604–1607. doi: 10.3201/eid0912.030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beasley DW, Davis CT, Estrada-Franco J, Navarro-Lopez R, Campomanes-Cortes A, Tesh RB, Weaver SC, Barrett AD. Genome sequence and attenuating mutations in West Nile virus isolate from Mexico. Emerg Infect Dis. 2004;10:2221–2224. doi: 10.3201/eid1012.040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett AD. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Beasley DW, Davis CT, Whiteman M, Granwehr B, Kinney RM, Barrett AD. Molecular determinants of virulence of West Nile virus in North America. Arch Virol Suppl. 2004:35–41. doi: 10.1007/978-3-7091-0572-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Beasley DW, Whiteman MC, Zhang S, Huang CY, Schneider BS, Smith DR, Gromowski GD, Higgs S, Kinney RM, Barrett AD. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol. 2005;79:8339–8347. doi: 10.1128/JVI.79.13.8339-8347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirato K, Miyoshi H, Goto A, Ako Y, Ueki T, Kariwa H, Takashima I. Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J Gen Virol. 2004;85:3637–3645. doi: 10.1099/vir.0.80247-0. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Bhuvanakantham R, Howe J, Ng ML. The glycosylation site in the envelope protein of West Nile virus (Sarafend) plays an important role in replication and maturation processes. J Gen Virol. 2006;87:613–622. doi: 10.1099/vir.0.81320-0. [DOI] [PubMed] [Google Scholar]
- 18.Murata R, Eshita Y, Maeda A, Maeda J, Akita S, Tanaka T, Yoshii K, Kariwa H, Umemura T, Takashima I. Glycosylation of the West Nile virus envelope protein increases in vivo and in vitro viral multiplication in birds. Am J Trop Med Hyg. 2010;82:696–704. doi: 10.4269/ajtmh.2010.09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beasley DW, Li L, Suderman MT, Barrett AD. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology. 2002;296:17–23. doi: 10.1006/viro.2002.1372. [DOI] [PubMed] [Google Scholar]
- 20.Davis CT, Beasley DW, Guzman H, Siirin M, Parsons RE, Tesh RB, Barrett AD. Emergence of attenuated West Nile virus variants in Texas, 2003. Virology. 2004;330:342–350. doi: 10.1016/j.virol.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, Garcia S, Lothrop B. Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. J Med Entomol. 2008;45:494–508. doi: 10.1603/0022-2585(2008)45[494:pwnvta]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers TJ, Halevy M, Nestorowicz A, Rice CM, Lustig S. West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J Gen Virol. 1998;79:2375–2380. doi: 10.1099/0022-1317-79-10-2375. [DOI] [PubMed] [Google Scholar]
- 23.Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherret JH, Mackenzie JS, Khromykh AA, Hall RA. Biological significance of glycosylation of the envelope protein of Kunjin virus. Ann NY Acad Sci. 2001;951:361–363. doi: 10.1111/j.1749-6632.2001.tb02719.x. [DOI] [PubMed] [Google Scholar]
- 25.L'Vov DN, Shchelkanov M, Dzharkenov AF, Galkina IV, Kolobukhina LV, Aristova VA, Al'khovskii SV, Prilipov AG, Samokhvalov EI, Deriabin PG, Voronina AG, Vasil'ev AV, Bezzhonova OV, L'Vov DK. Population interactions of West Nile virus (Flaviviridae, Flavivirus) with arthropod vectors, vertebrates, humans in the middle and low belts of Volga delta in 2001–2006 [in Russian] Vopr Virusol. 2009;54:36–43. [PubMed] [Google Scholar]
- 26.Adams SC, Broom AK, Sammels LM, Hartnett AC, Howard MJ, Coelen RJ, Mackenzie JS, Hall RA. Glycosylation and antigenic variation among Kunjin virus isolates. Virology. 1995;206:49–56. doi: 10.1016/s0042-6822(95)80018-2. [DOI] [PubMed] [Google Scholar]
- 27.Davis CT, Galbraith SE, Zhang S, Whiteman MC, Li L, Kinney RM, Barrett AD. A combination of naturally occurring mutations in North American West Nile virus nonstructural protein genes and the 3′ untranslated region alter virus phenotype. J Virol. 2007;81:6111–6116. doi: 10.1128/JVI.02387-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller BR, Nasci RS, Godsey MS, Savage HM, Lutwama JJ, Lanciotti RS, Peters CJ. First field evidence for natural vertical transmission of West Nile virus in Culex univittatus complex mosquitoes from Rift Valley Province, Kenya. Am J Trop Med Hyg. 2000;62:240–246. doi: 10.4269/ajtmh.2000.62.240. [DOI] [PubMed] [Google Scholar]
- 29.Charrel RN, Brault AC, Gallian P, Lemasson JJ, Murgue B, Murri S, Pastorino B, Zeller H, de Chesse R, de Micco P, de Lamballerie X. Evolutionary relationship between Old World West Nile virus strains. Evidence for viral gene flow between Africa, the Middle East, and Europe. Virology. 2003;315:381–388. doi: 10.1016/s0042-6822(03)00536-1. [DOI] [PubMed] [Google Scholar]
- 30.Brault AC, Langevin SA, Bowen RA, Panella NA, Biggerstaff BJ, Miller BR, Nicholas K. Differential virulence of West Nile strains for American crows. Emerg Infect Dis. 2004;10:2161–2168. doi: 10.3201/eid1012.040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler SS, Barker CM, Fang Y, Armijos MV, Carroll BD, Husted S, Johnson WO, Reisen WK. Differential impact of West Nile virus on California birds. Condor. 2009;111:1–20. doi: 10.1525/cond.2009.080013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blitvich BJ, Fernandez-Salas I, Contreras-Cordero JF, Marlenee NL, Gonzalez-Rojas JI, Komar N, Gubler DJ, Calisher CH, Beaty BJ. Serologic evidence of West Nile virus infection in horses, Coahuila State, Mexico. Emerg Infect Dis. 2003;9:853–856. doi: 10.3201/eid0907.030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupuis AP II, Marra PP, Kramer LD. Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg Infect Dis. 2003;9:860–863. doi: 10.3201/eid0907.030249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorono-Pino MA, Blitvich BJ, Farfan-Ale JA, Puerto FI, Blanco JM, Marlenee NL, Rosado-Paredes EP, Garcia-Rejon JE, Gubler DJ, Calisher CH, Beaty BJ. Serologic evidence of West Nile virus infection in horses, Yucatan State, Mexico. Emerg Infect Dis. 2003;9:857–859. doi: 10.3201/eid0907.030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattar S, Edwards E, Laguado J, Gonzalez M, Alvarez J, Komar N. West Nile virus antibodies in Colombian horses. Emerg Infect Dis. 2005;11:1497–1498. doi: 10.3201/eid1109.050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz L, Cardenas VM, Abarca M, Rodriguez T, Reyna RF, Serpas MV, Fontaine RE, Beasley DW, Da Rosa AP, Weaver SC, Tesh RB, Powers AM, Suarez-Rangel G. Short report: serological evidence of West Nile virus activity in El Salvador. Am J Trop Med Hyg. 2005;72:612–615. [PubMed] [Google Scholar]
- 37.Komar O, Robbins MB, Klenk K, Blitvich BJ, Marlenee NL, Burkhalter KL, Gubler DJ, Gonzalvez G, Pena CJ, Peterson AT, Komar N. West Nile virus transmission in resident birds, Dominican Republic. Emerg Infect Dis. 2003;9:1299–1302. doi: 10.3201/eid0910.030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komar O, Robbins MB, Contreras GG, Benz BW, Klenk K, Blitvich BJ, Marlenee NL, Burkhalter KL, Beckett S, Gonzalvez G, Pena CJ, Peterson AT, Komar N. West Nile virus survey of birds and mosquitoes in the Dominican Republic. Vector Borne Zoonotic Dis. 2005;5:120–126. doi: 10.1089/vbz.2005.5.120. [DOI] [PubMed] [Google Scholar]
- 39.Deardorff E, Estrada-Franco J, Brault AC, Navarro-Lopez R, Campomanes-Cortes A, Paz-Ramirez P, Solis-Hernandez M, Ramey WN, Davis CT, Beasley DW, Tesh RB, Barrett AD, Weaver SC. Introductions of West Nile virus strains to Mexico. Emerg Infect Dis. 2006;12:314–318. doi: 10.3201/eid1202.050871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phoutrides E, Jusino-Mendez T, Perez-Medina T, Seda-Lozada R, Garcia-Negron M, Davila-Toro F, Hunsperger E. The utility of animal surveillance in the detection of West Nile virus activity in Puerto Rico, 2007. Vector Borne Zoonotic Dis. 2011;11:447–450. doi: 10.1089/vbz.2010.0011. [DOI] [PubMed] [Google Scholar]
- 41.Morales-Betoulle ME, Morales H, Blitvich BJ, Powers AM, Davis EA, Klein R, Cordon-Rosales C. West Nile virus in horses, Guatemala. Emerg Infect Dis. 2006;12:1038–1039. doi: 10.3201/eid1206.051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pupo M, Guzman MG, Fernandez R, Llop A, Dickinson FO, Perez D, Cruz R, Gonzalez T, Estevez G, Gonzalez H, Santos P, Kouri G, Andonova M, Lindsay R, Artsob H, Drebot M. West Nile virus infection in humans and horses, Cuba. Emerg Infect Dis. 2006;12:1022–1024. doi: 10.3201/eid1206.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morales MA, Barrandeguy M, Fabbri C, Garcia JB, Vissani A, Trono K, Gutierrez G, Pigretti S, Menchaca H, Garrido N, Taylor N, Fernandez F, Levis S, Enria D. West Nile virus isolation from equines in Argentina, 2006. Emerg Infect Dis. 2006;12:1559–1561. doi: 10.3201/eid1210.060852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Hovav E, Powers AM, Lin M, Dorman KS, Platt KB, Bartholomay LC, Soto V, Beaty BJ, Lanciotti RS, Blitvich BJ. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- 45.Ostfeld RS. Biodiversity loss and the rise of zoonotic pathogens. Clin Microbiol Infect. 2009;15((Suppl 1)):40–43. doi: 10.1111/j.1469-0691.2008.02691.x. [DOI] [PubMed] [Google Scholar]
- 46.Swaddle JP, Calos SE. Increased avian diversity is associated with lower incidence of human West Nile infection: observation of the dilution effect. PLoS ONE. 2008;3:e2488. doi: 10.1371/journal.pone.0002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reisen WK, Fang Y, Martinez VM. Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. J Med Entomol. 2005;42:367–375. doi: 10.1093/jmedent/42.3.367. [DOI] [PubMed] [Google Scholar]


