Abstract
Natively unfolded or intrinsically disordered proteins (IDPs) are under intense scrutiny due to their involvement in both normal biological functions and abnormal protein misfolding disorders. Polypeptide chain collapse of amyloidogenic IDPs is believed to play a key role in protein misfolding, oligomerization, and aggregation leading to amyloid fibril formation, which is implicated in a number of human diseases. In this work, we used bovine κ-casein, which serves as an archetypal model protein for amyloidogenic IDPs. Using a variety of biophysical tools involving both prediction and spectroscopic techniques, we first established that monomeric κ-casein adopts a collapsed premolten-globule-like conformational ensemble under physiological conditions. Our time-resolved fluorescence and light-scattering data indicate a change in the mean hydrodynamic radius from ∼4.6 nm to ∼1.9 nm upon chain collapse. We then took the advantage of two cysteines separated by 77 amino-acid residues and covalently labeled them using thiol-reactive pyrene maleimide. This dual-labeled protein demonstrated a strong excimer formation upon renaturation from urea- and acid-denatured states under both equilibrium and kinetic conditions, providing compelling evidence of polypeptide chain collapse under physiological conditions. The implication of the IDP chain collapse in protein aggregation and amyloid formation is also discussed.
Introduction
Natively unfolded or intrinsically disordered proteins (IDPs) are contrary to the conventional sequence-structure-function paradigm owing to their structural attributes, which are in contrast to those of the uniquely folded globular proteins (1–4). Bioinformatic analyses indicate that >30% of eukaryotic proteins are disordered (1–5). It has been documented that despite being devoid of a well-defined, ordered 3D structure, IDPs carry out important cellular functions such as molecular recognition, replication, signaling, etc. (3,6–8). Comparison between the amino acid compositions of IDPs and globular proteins has revealed that the sequence and frequent occurrence of disorder-promoting amino acid residues with high flexibility index, low hydrophobicity, and high net charge are accountable for the conformational plasticity observed in IDPs. Also, IDPs can be empirically subclassified into premolten globule, structured molten globule, collapsed disordered ensemble, and extended disordered ensemble, which undergo noncooperative conformational transitions (1–8). Such inherent structural randomness and conformational heterogeneity create a major bottleneck in the structural characterization of IDPs by x-ray crystallography, which has been one of the primary techniques to elucidate high-resolution structure of biomolecules. However, nuclear magnetic resonance (9,10), circular dichroism (CD) (3,4,11), vibrational (12,13), and fluorescence spectroscopic studies (3,14–16) have yielded important structural insights into the dynamic ensembles of the IDPs. Recent single-molecule fluorescence experiments have revealed the dynamic conformational behavior of IDPs, which have intrinsic ability to undergo chain collapse (17–19).
Recent structural investigations of IDPs have gained tremendous importance in amyloid research, since most (misfolded) amyloidogenic proteins adopt a disordered state in their monomeric form, either continuously or transiently, before oligomerization and amyloid formation that is implicated in a variety of neurodegenerative disorders (8,20,21). These proteins include amyloid-β (Alzheimer's), α-synuclein (Parkinson's), polyglutamine (Huntington's), N-terminal disordered segment of prion proteins (mad cow and transmissible spongiform encephalopathy), and functional amyloid proteins such as yeast prion proteins (22,23) and pMel (24) that are unrelated to disease. Some of these amyloidogenic IDPs have been theoretically predicted, and experimentally demonstrated, to undergo a reversible chain collapse from an extended form in good solvents (denaturant) to compact form in poor solvents (water) (17,25–27). The chain-collapse phenomenon of IDPs in water is observed when the favorable interaction between (intramolecular) chain and chain is stronger than that between chain and solvent at an infinitely dilute protein solution in water (25–28). An unstructured collapsed globule ensues when the intrachain interactions are favorable, with a concomitant gain in solvent entropy due to poor chain solvation. At higher protein concentrations, the chain-chain interactions between collapsed, yet disordered, protein molecules can promote intermolecular association leading to unstructured oligomers that conformationally mature into ordered amyloid fibrils (25). Therefore, polypeptide chain collapse is the key early event that plays a pivotal role in protein aggregation leading to amyloid formation. In this work, we used bovine κ-casein as a readily available archetypal IDP that has been previously shown to assemble into amyloid fibrils under carefully controlled laboratory conditions (29–31). Based on the bioinformatic predictions and a variety of spectroscopic tools, we first established that κ-casein undergoes a chain collapse under the native condition and then demonstrated that the excimer formation from κ-casein covalently labeled with two pyrene moieties can be utilized as a simple, convenient, and elegant approach to study the chain collapse of an amyloidogenic IDP under both equilibrium and kinetic conditions.
Materials and Methods
Materials
Bovine κ-casein, guanidinium chloride, urea, dithiothreitol (DTT), iodoacetic acid, ANS (8-anilinonaphthalene-1-sulfonic acid ammonium salt), acrylamide, sodium phosphate (monobasic and dibasic), potassium chloride, sodium citrate, and Tris-HCl were procured from Sigma (St. Louis, MO) and used without any further purification. N-(1-pyrene) maleimide and 5-((2-((iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (IAEDANS) were purchased from Molecular Probes (Eugene, OR) and dimethylsulfoxide (DMSO) was obtained from Merck (Whitehouse Station, NJ) and used as received. Milli-Q water (Millipore, Billerica, MA) was used for the preparation of buffers and other aqueous solutions. The pH of the buffers was checked and adjusted using 1 N HCl or NaOH on a Metrohm (Herisau, Switzerland) 827 pH meter at ∼24°C.
Monomerization of κ-casein
To prepare monomeric κ-casein (reduced and carboxymethylated (RCM)), a previously described procedure (31) was used with slight modifications. Briefly, 1 mM κ-casein solution was prepared in 6 M GdmCl (pH 8, 100 mM Tris-Cl), to which 10 mM DTT was added. To ensure complete reduction (cleavage of intra- and interdisulfide linkages) and denaturation, the reaction mixture was stirred for 1 h at 37°C at a speed of 45 rpm. Next, 50 mM iodoacetic acid (freshly prepared in DMSO) was added into the reaction mixture to react with the thiol groups of the resulting free cysteines, and the mixture was further stirred for another 30 min. After the reaction was complete, traces of free iodoacetic acid were removed and the protein was concentrated using AMICON YM-10 (10 kDa cut-off; Millipore) by washing it extensively with 6 M GdmCl (pH 7, 50 mM phosphate buffer). Accurate concentration of RCM κ-casein was determined by measuring the tryptophan absorbance at 280 nm by UV-Vis spectrophotometer (Perkin Elmer, Waltham, MA) using a molar extinction coefficient of 11,500 M−1 cm−1 (31). RCM κ-casein was prepared freshly before each spectroscopic measurement.
Fluorescence experiments with RCM κ-casein under denatured and native conditions
Freshly prepared RCM κ-casein (6 M GdmCl, pH 7) was incubated separately in i), 8 M urea solution prepared in 5 mM phosphate buffer, pH 7, to form urea-denatured RCM κ-casein; ii), 5 mM KCl-HCl, pH 1.6, to form acid-denatured protein; and iii), 5 mM phosphate buffer, pH 7 (native), to a final protein concentration of 20 μM each for 1 h at room temperature after which the Trp fluorescence intensity and anisotropy of denatured and native κ-casein were measured.
Fluorescence-quenching experiments with RCM κ-casein
To carry out the tryptophan quenching experiments, a stock of 2 M acrylamide was prepared in three sets at pH 7, 5 mM phosphate buffer; pH 1.6, 5 mM KCl-HCl; and 8 M urea prepared in 5 mM phosphate buffer, pH 7. Stock of 1 mM RCM κ-casein (6 M GdmCl, pH 7) was diluted in different concentrations of acrylamide (0–500 mM) to get a final protein concentration of 20 μM at room temperature and the tryptophan fluorescence intensity was recorded as a function of acrylamide concentration under native (pH 7) and denatured (acid- and urea) conditions. The quenching graphs were plotted using OriginPro software, version 8, and fitted using the linear regression equation to estimate the Stern-Volmer constant. The accessibility of tryptophan was determined by using the Stern-Volmer equation, , where F0 and F are the tryptophan fluorescence intensities in the absence and presence, respectively, of quencher, and Ksv is the Stern-Volmer quenching constant.
Fluorescence labeling of κ-casein with IAEDANS
The labeling of the free thiol groups of two cysteines in denatured and reduced κ-casein was carried out in 50 mM phosphate buffer, pH 7. Initially, 1 mM κ-casein solution was prepared in 6 M GdmCl (50 mM phosphate buffer, pH 7), to which an aqueous solution of 10 mM DTT was added. The resulting solution was stirred for 1 h at 37°C at a speed of 45 rpm. Approximately 50 equivalents of IAEDANS, dissolved in dry DMSO, were added into the reduced κ-casein solution and the reaction mixture was stirred again for 1 h at 37°C. After the labeling reaction was complete, the labeled protein was purified and concentrated using AMICON YM-10, whereby the free, unreacted dye was removed. The concentrated protein was diluted in 6 M GdmCl (50 mM phosphate buffer, pH 7), and its concentration was checked by measuring the absorbance at both 280 and 337 nm. The concentration of the labeled protein was determined by subtracting the absorption contribution of AEDANS at 280 nm. The molar extinction coefficients of IAEDANS at 280 nm and 337 nm are 1060 M−1 cm−1 (32) and 6100 M−1 cm−1 (33), respectively.
Fluorescence labeling of κ-casein with N-(1-pyrene) maleimide
Two cysteines in denatured and reduced κ-casein were covalently labeled by pyrene maleimide, a thiol-reactive probe, using a literature protocol (34) with slight modifications. Briefly, to a solution of chemically denatured κ-casein (1 mM prepared in 6 M GdmCl, pH 7, 50 mM phosphate buffer), an aqueous solution of 5 mM DTT was added. The reaction was allowed to stir for 1 h at 37°C at a speed of 45 rpm. After 1 h, 50 mM pyrene maleimide (freshly dissolved in DMSO) was added into the reaction mixture and stirred for another 3 h at 37°C. Once the reaction was complete, the free dye was removed by washing the reaction mixture exhaustively with 6 M GdmCl (50 mM phosphate, pH 7) using AMICON YM-10. The absorbance of filtrates and concentrated labeled protein were measured as a ratio at 280 nm and 340 nm for pyrene-labeled κ-casein. The degree of labeling was determined using a molar extinction coefficient of 40,000 M−1 cm−1 for pyrene at 340 nm (35).
Fluorescence experiments with labeled κ-casein under denatured and native conditions
For experiments under urea-denatured conditions, a stock solution of urea (8 M) was prepared in phosphate buffer (pH 7, 50 mM) and its pH was adjusted to pH 7 at 24–25°C. 1 mM stock solution of labeled (either pyrene or AEDANS) κ-casein (in 6 M GdmCl, pH 7) was diluted 100-fold into varying concentrations of urea (from 8 M to 0 M) to yield a final protein concentration of 10 μM. The solutions were incubated for 1 h at 25°C to ensure denaturation at that particular concentration of urea. Also, 1 mM stock solution of labeled (either pyrene or AEDANS) κ-casein (in 6 M GdmCl, pH 7) was diluted 100-fold into pH 7 and pH 1.6 buffers separately. The urea- and acid denatured as well as the natively unfolded states of labeled κ-casein were monitored by measuring the intensity and anisotropy of covalently attached pyrene and/or AEDANS.
Steady-state fluorescence measurements
All steady-state fluorescence measurements were performed on a Perkin Elmer LS 55 fluorimeter using a 10-mm-pathlength quartz cuvette. The steady-state fluorescence anisotropy (rss) is given as
| (1) |
rss was obtained from the parallel (I‖) and perpendicular (I⊥) intensity components with G-factor correction. An integration time of 30 s was used for anisotropy measurements. The final protein concentration was 10 μM in all experiments except for the tryptophan fluorescence measurements, where it was fixed at 20 μM. The excitation bandpass was fixed at 2.5 nm for all experiments. For Trp fluorescence, λex = 290 nm and λem = 350 nm (bandpass 4 nm). For Trp anisotropy, λex = 300 nm and λem = 350 nm (bandpass 15 nm). For AEDANS fluorescence intensity, λex = 375 nm and λem = 475 nm (bandpass 4 nm). For AEDANS anisotropy, the emission bandpass was 7 nm. For pyrene fluorescence, λex was 340 nm and the spectra were scanned from 350 to 550 nm (bandpass 3.5 nm). All emission spectra were recorded with a scan speed of 10 nm/min and averaged over three scans. The fluorescence resonance energy transfer (FRET) experiments (Trp→AEDANS) were carried out on a Chirascan spectrometer (Applied Photophysics, Leatherhead, United Kingdom).
Stopped-flow fluorescence experiments
The kinetics of conformational changes of RCM and pyrene-labeled κ-casein was monitored by the change in total fluorescence intensity of tryptophan and pyrene excimer, respectively, as a result of pH jump from the acid-denatured state (pH 1.6, 5 mM KCl-HCl buffer) to the native state (pH 7, 50 mM phosphate buffer) with a spectrometer (Chirascan, Applied Photophysics) equipped with a stopped-flow apparatus (Applied Photophysics). Initially, the required volume of chemically denatured protein was equilibrated at pH 1.6 for acid denaturation at room temperature. The acid-denatured protein solution and the refolding buffer of pH 7 were rapidly mixed at a 1:2.5 ratio (dead time ∼1.8 ms) and the renaturation kinetics was monitored. For Trp fluorescence, the final protein concentration was 20 μM. The samples were excited at 290 nm (bandpass 4 nm) and fluorescence was collected using a 320-nm long-pass filter. For experiments with pyrene-labeled κ-casein, the final protein concentration was 10 μM. Pyrene was excited at 340 nm (bandpass 4 nm) and the excimer fluorescence was collected using a 395-nm long-pass filter to separate the contribution from monomer emission. The stopped-flow data were acquired for 0.5 s with 10,000 samples per point and the traces were collected in triplicate five times and averaged to get a satisfactory signal/noise ratio. The baseline was also collected at acidic pH under the same experimental conditions to determine the burst-phase amplitude. The fluorescence traces were fitted to a biexponential function using the Chirascan Pro-Data Software (Applied Photophysics), and the apparent rate constants were estimated. The goodness of the fit was determined by the residual plots.
CD measurements
Far-UV CD spectra of RCM κ-casein (10 μM) were collected under both native and acid-denatured conditions in a quartz cuvette of 2 mm pathlength using a Chirascan CD spectrometer (Applied Photophysics) with a scan range of 200–260 nm. The spectra were recorded with a scan speed of 0.1 nm/s and averaged over five scans. All the CD spectra were buffer-subtracted and smoothened using Pro-Data software provided with the Chirascan CD spectrometer. All the graphs were plotted using the commercially available OriginPro Version 8 software. The mean residue molar ellipticity was calculated as described in the literature (36).
Time-resolved fluorescence measurements
Time-resolved fluorescence decay measurements of the samples were made using a picosecond laser coupled to a time-correlated single-photon-counting setup. A mode-locked frequency-doubled Nd:YAG laser (Spectra Physics, Mountain View, CA) of 1-ps pulse width was used to pump a dye (Rhodamine 6G) laser, and the generated dye-laser pulses were frequency-doubled using a KDP crystal. A wavelength of 295 nm was used for our experiments to excite Trp. The instrument response function (IRF) at 295 nm was collected using a dilute colloidal suspension of dried nondairy whitener. The width of the IRF was ∼40 ps. The Trp fluorescence emission was collected using a microchannel plate photomultiplier (2809u, Hamamatsu, Hamamatsu City, Japan). The emission monochromator was fixed at 350 nm, with a bandpass of 10 nm. A long-pass filter of 345 nm was placed just after the sample to block any scattering from the sample. For anisotropy decay measurements, the emission data were collected at 0° and 90° with respect to the excitation polarization, and the anisotropy decays were analyzed by globally fitting I‖ (t) and I⊥ (t) as follows:
| (2) |
| (3) |
The perpendicular component of the fluorescence decay was corrected for the G-factor of the spectrometer. I(t) is the fluorescence intensity collected at the magic angle (54.7°) at time t. The anisotropy decays were analyzed using a biexponential decay model describing fast and slow rotational correlation times as follows:
| (4) |
where r0 is the intrinsic fluorescence anisotropy; ϕfast and ϕslow are the fast and slow rotational correlation times; and βfast and βslow are the amplitudes associated with fast and slow rotational time.
The global (slow) rotational correlation time (ϕslow) is related to viscosity (η) and molecular volume (V) by the Stokes-Einstein relationship as follows:
| (5) |
| (6) |
where Rh is the hydrodynamic radius of the molecule.
Dynamic light-scattering measurements
The dynamic light-scattering (DLS) experiments were performed on a Delsa Nano C particle analyzer instrument (Beckman Coulter, Brea, CA) at 25°C. Before the DLS experiments, both the phosphate buffer (pH 7) and 6 M GdmCl solution were filtered through 0.22-μm Millipore syringe filters. Freshly prepared RCM κ-casein was added to the filtered pH 7 phosphate buffer and 6 M GdmCl solutions separately to a final protein concentration of 10 μM each. Each data set was a collection of at least 30 scattering intensities. All the data were collected using Delsa Nano software provided with the instrument and were subsequently analyzed using the CONTIN method. The viscosity of 6 M GdmCl was taken from the literature (37).
Results and Discussion
Spectroscopic characterizations of the disordered state
Caseins are relatively small, extremely flexible proteins that belong to the class of IDPs that lack specific secondary structure (3,38). They usually have a tendency to self-associate, leading to micelle formation and amyloid-like deposits in bovine mammary glands (39). Matured signal-peptide cleaved κ-casein is a 169-residue, 18.9-kDa protein (Fig. 1 a) that has a single Trp (residue 76) and two Cys (residues 11 and 88). Our bioinformatic analyses of κ-casein led to the prediction that the protein adopts a disordered and collapsed conformer under physiological conditions (Fig. S1 in the Supporting Material). Naturally occurring κ-casein exists in micellar form due to the presence of intra- and intermolecular disulfide bonds. Therefore, before embarking upon conformational studies under different solution conditions, we first prepared monomeric RCM κ-casein. To do this, we carried out reduction of disulfides followed by carboxymethylation of κ-casein under the denatured condition (see Materials and Methods). ANS-binding experiments suggested that the monomerized form had a much weaker affinity for ANS compared to the micellar form (Fig. S2).
Figure 1.

(a) Amino acid sequence of κ-casein. Trp76 and two Cys are shown with asterisk and underscoring, respectively. (b) Trp fluorescence spectra of 20 μM κ-casein under urea-denatured (dashed line) and native conditions (solid line). (c) Increase in Trp fluorescence anisotropy of κ-casein from the urea-denatured to the native condition.
We next probed the fluorescence properties of the single tryptophan (Trp76), which is located in the central region of the polypeptide chain (Fig. 1 a). The fluorescence emission of Trp is sensitive to the environment (40). Under urea-denatured conditions for RCM κ-casein, the Trp emission spectrum exhibited a band with a maximum at ∼355 nm, which is consistent with exposed Trp under unfolded conditions. Upon transfer to a refolding buffer, the emission maximum underwent a blue shift to ∼347 nm, indicating lower solvent accessibility around Trp76 (Fig. 1 b). In addition to the spectral shift, we observed a small but measurable decrease in the tryptophan fluorescence intensity under native conditions, suggesting a structural change from an expanded to a compact state that results in more efficient quenching of tryptophan fluorescence by proximal amino acid quenchers located on the polypeptide chain. The structural rigidification around Trp is further supported by more restricted rotational motion of Trp, as evidenced by a significant rise in the tryptophan fluorescence anisotropy upon renaturation (Fig. 1 c).
We next measured the Stern-Volmer quenching constant (40) of Trp fluorescence by acrylamide, which again indicated that the native κ-casein is less efficiently quenched, as expected for a protected Trp (Fig. 2 a). Using CD, we showed that the native state is devoid of detectable elements of secondary structure (Fig. 2 b). Taken together, our tryptophan fluorescence and far-UV CD data indicate structural compaction in the native state of κ-casein that remains disordered. The double-wavelength plot ([θ]222 versus [θ]200) analysis (11) of far-UV CD data indicated that the disordered κ-casein appears to assume a premolten-globule-like state (Fig. 2 b). This compact, yet disordered, state can respond to acid denaturation, as indicated by extensive tryptophan fluorescence quenching without much change in secondary structural contents at low pH (Fig. 2). All of the above experiments provide initial experimental support in favor of monomeric κ-casein adopting a compact conformation under the native condition.
Figure 2.
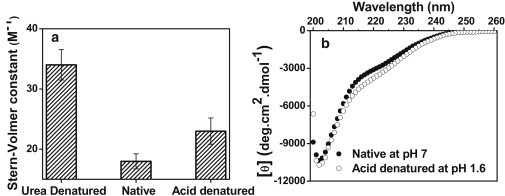
(a) Stern-Volmer quenching constant of Trp of 20 μM κ-casein with acrylamide quencher under different conditions. For Stern-Volmer plots, see Fig. S3. (b) Far-UV CD spectra of 10 μM κ-casein under native (solid circles) and acid-denatured forms (open circles). Under the native condition, [θ]222 and [θ]200 were −2880 and −8880 deg·cm2 dmol−1, respectively.
Chain collapse observed by FRET
The intrinsic tryptophan fluorescence properties are likely to depict a more local structure formation around the tryptophan residue under the native condition. We next investigated whether the chain collapse is a locally restricted phenomenon around the Trp residue or globally felt among other residues along the polypeptide chain. We took advantage of two cysteine residues (Cys11 and Cys88). We labeled them using IAEDANS, a thiol-labeling fluorescent probe that is highly environment-sensitive (40). Fluorescence spectra of the AEDANS-κ-casein conjugate showed a progressive blue shift (490 nm → 480 nm) and a concomitant increase in fluorescence intensity from the denatured to the native state that is consistent with a nonpolar microenvironment of the fluorophore (Fig. 3 a, inset, and Fig. S4). Fluorescence anisotropy of AEDANS also exhibited a distinct increase upon lowering of the denaturant concentration, again suggesting restriction to the environment around the probe (Fig. 3 a). These results prompted us to investigate the FRET from Trp → AEDANS to probe the changes in intrachain distances as a result of compaction under the native condition. Initially, the FRET efficiency from Trp → AEDANS was low under the urea-denatured condition, as expected for a random coil. Upon transferring into the native buffer, an increase in AEDANS (acceptor) fluorescence with a concomitant decrease in Trp (donor) emission indicated an increase in FRET efficiency that revealed a global intrachain compaction under the native condition (Fig. 3 b). Taken together, the aforementioned experiments revealed that the disordered polypeptide chain of κ-casein is likely to be globally collapsed under the native buffer condition.
Figure 3.
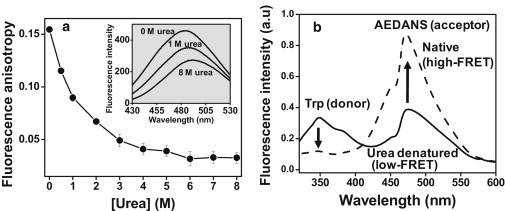
(a) Increase in AEDANS fluorescence anisotropy upon refolding of fluorescently labeled 10-μM κ-casein from the urea-denatured state. (Inset) AEDANS fluorescence spectra as a function of denaturant concentration. (b) FRET between Trp (donor) and AEDANS (acceptor) under denatured and native conditions.
Chain dynamics and dimension observed by time-resolved fluorescence anisotropy and dynamic light scattering
We have carried out picosecond time-resolved fluorescence anisotropy measurements that provide insights into the local and global rotational dynamics of proteins (40,41). Global dynamics is represented by a slow rotational correlation time (ϕslow) that is generally related to the size (hydrodynamic radius) of the protein (Eq. 4). In the denatured form, κ-casein showed two rotational correlation times. The fast rotational correlation time, ϕfast (∼0.2 ns), represents the local dynamics of Trp, whereas ϕslow (∼2.7 ns) could indicate the segmental mobility of the unfolded polypeptide chain (Fig. 4). For an unfolded protein, ϕslow is related to the segmental flexibility and does not depend on the protein size, since the segmental conformation fluctuations depolarize the fluorescence much more rapidly than the global tumbling of the polypeptide chain (41). Upon transferring into the native buffer, ϕslow increased to ∼7 ns, which may correspond to a hydrodynamic radius (Rh) of 1.9 nm for the collapsed globule (Fig. 4 and Eqs. 4–6). This is also corroborated with Rh obtained from our DLS data (1.9 ± 0.3 nm). On the other hand, the DLS data of the denatured form of κ-casein showed an Rh of 4.6 ± 0.7 nm, in accordance with the calculated Rh (∼4.1 nm) for a 169-residue unfolded protein using a previously reported empirical relationship (42). Taken together, time-resolved fluorescence anisotropy and DLS data provide a quantitative description of chain collapse of monomeric κ-casein that undergoes a change in the mean hydrodynamic radius from ∼4.6 nm (denatured) to ∼1.9 nm (native).
Figure 4.

Picosecond time-resolved fluorescence anisotropy decays (r(t)) of Trp in denatured (a) and native (b) κ-casein. The solid line is the biexponential fit (Eq. 4). The recovered rotational correlation times, with associated amplitudes, are ϕfast = 0.2 ± 0.1 ns, βfast = 0.53 ± 0.02, ϕslow = 2.7 ± 0.8, βslow = 0.47 ± 0.02 for denatured κ-casein (a) and ϕfast = 0.2 ± 0.1 ns, βfast = 0.65 ± 0.03, ϕslow = 7.1 ± 0.6 ns, and βslow = 0.35 ± 0.03 for native κ-casein (b).
Pyrene excimer formation upon polypeptide chain collapse
Our next goal was to directly watch the chain collapse using a strategy that allowed us to monitor the changes in the intramolecular distances at short length-scales. One such proximity indicator is pyrene excimer (excited-state dimer), which is observed when two proximal pyrene moieties interact below 5 Å distance within the excited-state lifetime of pyrene (∼100 ns) (40,43). Excimer formation provides a direct handle to monitor conformational changes at a much shorter length-scale compared to the more commonly utilized FRET technique, which offers structural information at much longer length-scales ranging from 20 to 80 Å. Recently, pyrene excimer fluorescence has been successfully utilized to gain important insights into protein conformational changes and aggregation (44–47).
Reduced κ-casein has two free cysteines that are well separated (by 77 residues) in the primary sequence. We labeled the cysteines with two molar equivalents of thiol-reactive pyrene (N-(1-pyrene) maleimide) (see Materials and methods). We conjectured that upon chain collapse, the internal distance between residues 11 and 88 would decrease, as a result of which we would see high excimer fluorescence. To gain information about conformational transitions, we recorded the pyrene fluorescence spectra of urea-denatured protein sequentially transferred to native buffer. Monomer fluorescence emission of pyrene ranges between 370 and 400 nm with vibronic bands, whereas a single excimer band is observed between 450 and 480 nm (Fig. 5 a). As we decreased the urea concentration, the excimer emission band increased at the expense of the monomer emission band. The excimer/monomer ratio increased in a noncooperative fashion from <1 (denatured) to 5 (native), suggesting progressive collapse of the polypeptide chain (Fig. 5 b). We point out also that there was residual excimer intensity present even under the fully denatured condition. This might indicate that the polypeptide chain undergoes rapid nanosecond fluctuations given that the fluorescence lifetime of pyrene is ∼100 ns. Upon renaturation, the pyrene excimer contribution grew significantly, indicating a substantial decrease in inter-pyrene distance arising from chain collapse. To eliminate the possibility of excimer formation by oligomeric aggregates, we performed experiments at different protein concentrations under the native condition and obtained the same excimer/monomer ratio (Fig. S5). These experiments confirmed that excimer formation is solely due to the chain collapse of the monomeric protein. In addition, a gradual blue shift of the excimer band from 475 nm (denatured) to 460 nm (native) is indicative of the change in pyrene microenvironment from polar to nonpolar. Taken together, these results provide direct evidence in favor of chain collapse of disordered κ-casein in water (Fig. 5 c).
Figure 5.
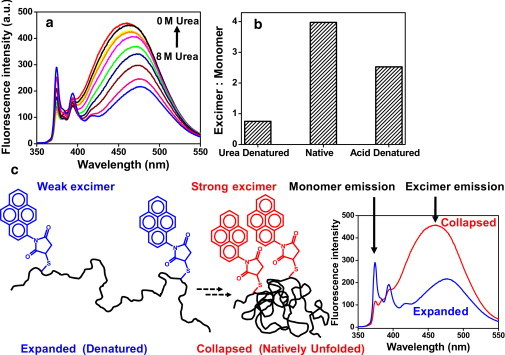
(a) Pyrene excimer formation of dual-labeled 10-μM κ-casein upon renaturation from urea. (b) Excimer/monomer ratio of pyrene under different conditions estimated from the fluorescence spectra shown in a. (c) A schematic of excimer formation upon polypeptide chain collapse.
Kinetics of chain collapse
After establishing the above equilibrium experiments on chain collapse of κ-casein, we directed our efforts to study the kinetics of conformational change from the extended to the collapsed state using stopped-flow pH jump from the acid-denatured (pH 1.6) to the natively unfolded state at neutral pH (pH 7). As discussed previously, natively unfolded κ-casein undergoes pH-induced unfolding (Fig. 2) because of the change in net charge from −1.4 at pH 7 to +17.9 at pH 1.6. Therefore, reversible transitions between compact and expanded can be monitored simply by changing the pH of the solution, without using a high concentration of denaturant. Fig. 6 a shows the steady-state fluorescence spectra of the single tryptophan (Trp76) in carboxymethylated κ-casein at both neutral and low pH, suggesting a significant decrease in Trp emission as a result of acid denaturation (pH 7 → 1.6). Using stopped-flow fluorescence with a pH jump from 1.6 to 7, we observed time-resolved recovery of total Trp fluorescence upon renaturation of acid-denatured κ-casein (Fig. 6 b). The Trp fluorescence kinetics revealed that there is >50% burst phase that could not be resolved within the mixing deadtime of a millisecond or so. This submillisecond kinetic phase is followed by a slow phase with a relaxation time of ∼20 ms. These results indicated that the chain collapse is likely to occur in at least two phases, a fast (submillisecond) phase and a slower phase with a relaxation time of ∼20 ms that increases with the increase in solvent viscosity, as expected for chain collapse. This biphasic relaxation kinetics comprising submillisecond and millisecond components observed for the chain collapse is consistent with previous results on expansion and collapse of a polypeptide (48). We next undertook pH-induced kinetic experiments using dual-pyrene-labeled κ-casein. Fig. 6 c shows the steady-state pyrene emission spectra of pyrene-labeled κ-casein under both neutral and low pH conditions showing an increase in pyrene excimer emission at pH 7 with a concurrent decrease in monomer fluorescence. To selectively monitor the time evolution of pyrene excimer fluorescence upon renaturation kinetics, we used an optical filter (395-nm long-pass) on the emission path (see Materials and Methods). Here, again, as expected, we observed a significant fraction of burst phase along with a slow phase with a relaxation time of ∼20 ms that corroborated with the stopped-flow Trp fluorescence data (Fig. 6 d). The fast component, hidden in the burst phase, could arise due to rapid chain collapse, whereas the 20-ms component could reveal the slow conformational reorganization of the collapsed globules into premolten globules. There was another, much slower component with a small amplitude, which can be attributed to a slow conformational reorganization of premolten-globule conformers comprising a small degree of secondary structural elements. This slow component was not a contribution from aggregation, as confirmed by stopped-flow experiments at different final protein concentrations (Fig. S6).
Figure 6.

(a) Trp fluorescence spectra of 20-μM κ-casein under acid-denatured and native conditions (b) Stopped-flow kinetics of Trp fluorescence upon acid renaturation of κ-casein from pH 1.6 to pH 7.0. An optical filter (320-nm long-pass) was used to collect total Trp fluorescence. The baseline is shown in gray. (c) Pyrene emission spectra of dual-labeled 10-μM κ-casein under acid-denatured and native conditions. (d) Stopped-flow kinetics of pyrene excimer formation upon acid renaturation. The optical filter (395-nm long-pass) used to selectively collect excimer fluorescence in the stopped-flow experiment is shown in c by a dotted line.
Structural and dynamical insights into collapsed IDPs
In this work, using a variety of spectroscopic techniques, we have provided compelling evidence in favor of our hypothesis that a model amyloidogenic IDP, κ-casein, adopts a collapsed, yet disordered, premolten-globule-like conformation under the native condition. This premolten-globule-like disordered state may contain very small elements of secondary structure, as indicated by the CD double-wavelength plot (11), and has a much lower hydrophobicity compared to an archetypal molten globule such as the low-pH form of α-lactalbumin (Fig. S2). We have shown for the first time, to our knowledge, that pyrene excimer formation can be used as a simple, convenient, and elegant tool to study the chain-collapse behavior of IDPs. Since we observed strong excimer fluorescence upon chain collapse, the inter-pyrene distance must approach 5 Å. It is likely that due to backbone dynamics, there is conformational fluctuation faster than the fluorescence lifetime of pyrene (∼100 ns) that may bring the pyrene moieties temporarily to very close proximity. In addition, premolten-globule formation could also contribute to further shortening of the inter-pyrene distance. The contribution of hydrophobic association of the pyrene moieties leading to the chain collapse could be a concern in this type of study. However, this is not a problem in our case, since a variety of fluorescence experiments on the unlabeled protein involving the intrinsic tryptophan fluorescence also indicated the chain-collapse phenomenon. Therefore, we suggest that to use the pyrene excimer methodology, it is important to independently verify the chain-collapse behavior using other methods, such as intrinsic protein fluorescence (spectral position, anisotropy, and quenching).
The observed chain collapse is likely to be largely driven by entropic factors, since water acts as a poor solvent for the polypeptide chain. We believe that the enthalpic contributions may also be important. A closer look at the κ-casein sequence revealed that the well-known disorder-promoting residues proline (P) and glutamine (Q) have more than double (P ∼12% and Q ∼9%) their frequency of occurrence in globular proteins (P ∼5% and Q ∼4%), which is in keeping with the observed conformational flexibility under physiological conditions. The unusually high occurrence of glutamine in κ-casein makes this protein very unique and interesting for aggregation studies, since polyglutamine repeats and glutamine-rich yeast prion have also been shown to undergo chain collapse, which has been implicated in amyloid aggregation (17,26). The chain-collapse phenomenon of IDPs is consistent with the emerging polymer physics perspective of protein aggregation and amyloid formation (25). In this work, we have also shown that chain collapse can be studied by pH-induced renaturation because of the decrease in the net charge of the protein, which allows favorable intrachain interactions through both hydrophobic effect and hydrogen bonding. The collapsed state of the protein is likely to be stabilized by 〉C=O……H−N〈 hydrogen bonding between the side-chain amides of glutamines. Such hydrogen-bonding-mediated chain collapse of disordered peptides has been described previously (27). Recent reports indicate that water serves as a poor solvent for generic polypeptides (49,50). One biologically important amyloidogenic IDP, α-synuclein, has also been demonstrated to adopt a collapsed conformational ensemble in water (51,52).
Conclusions
Overall, our results described in this work clearly demonstrate that water acts as a poor solvent for disordered κ-casein that assumes a collapsed premolten-globule-like conformation. Our results indicate a change in the mean hydrodynamic radius from ∼4.6 nm to ∼1.9 nm upon polypeptide chain collapse. This collapsed conformer may be a reminiscent of an early burst-phase intermediate formed during the folding of many globular proteins. We suggest that at a high protein concentration, polypeptide chains from different protein molecules will interact in an intermolecular fashion and will form oligomers to avoid unfavorable contacts with water molecules. Subsequently, the β-rich amyloidogenic conformers will be sequestered within the oligomers, which will conformationally mature into amyloid fibrils, as described by the nucleated conformational conversion model (23). We believe that our work will find a much broader application in the study of conformational change and polypeptide chain collapse using a combination of biophysical tools. We point out that the pyrene excimer methodology is easy to apply and is applicable to any IDP, either wild-type containing two cysteines or dual-cysteine mutants. We anticipate that the pyrene excimer methodology will serve as a complementary and perhaps better readout of chain collapse than FRET, since the collapse is expected to bring different segments of the polypeptide chain into much closer proximity. In addition, this method involves one-step preparation of the labeled protein, unlike FRET studies, wherein extensive purification is required at each step of both donor and acceptor labeling. In summary, the pyrene excimer methodology offers a robust and convenient spectroscopic tool to study IDPs and can eventually be extended to two-color (monomer/excimer) ratiometric imaging to monitor IDP chain collapse in complex environments such as liposomes and cells.
Acknowledgments
We thank the members of the Mukhopadhyay Lab for critical reading of the manuscript, Prof. G. Krishnamoorthy and Ms. M. Kombrabail (Tata Institute of Fundamental Research, Mumbai) for helping with time-resolved fluorescence measurements, and Dr. Ashish and Ms. S. Singh (Institute of Microbial Technology, Chandigarh) for helping with DLS experiments.
We gratefully acknowledge financial support provided by IISER Mohali and by grants from the Department of Science and Technology (M.B.) and the Council of Scientific and Industrial Research (S.M.).
Supporting Material
References
- 1.Uversky V.N., Dunker A.K. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunker A.K., Lawson J.D., Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 3.Tompa P. CRC Press; Boca Raton, FL: 2010. Structure and Function of Intrinsically Disordered Proteins. [Google Scholar]
- 4.Uversky V.N. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 5.Radivojac P., Iakoucheva L.M., Dunker A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson H.J., Wright P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 7.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 8.Dunker A.K., Silman I., Sussman J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Dyson H.J., Wright P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 10.Dedmon M.M., Lindorff-Larsen K., Dobson C.M. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 11.Uversky V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth E., Syme C.D., Barron L.D. Solution structure of native proteins with irregular folds from Raman optical activity. Biopolymers. 2001;58:138–151. doi: 10.1002/1097-0282(200102)58:2<138::AID-BIP30>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Syme C.D., Blanch E.W., Barron L.D. A Raman optical activity study of rheomorphism in caseins, synucleins and tau. New insight into the structure and behaviour of natively unfolded proteins. Eur. J. Biochem. 2002;269:148–156. doi: 10.1046/j.0014-2956.2001.02633.x. [DOI] [PubMed] [Google Scholar]
- 14.Jeganathan S., von Bergen M., Mandelkow E. Global hairpin folding of tau in solution. Biochemistry. 2006;45:2283–2293. doi: 10.1021/bi0521543. [DOI] [PubMed] [Google Scholar]
- 15.Szilvay G.R., Blenner M.A., Banta S. A FRET-based method for probing the conformational behavior of an intrinsically disordered repeat domain from Bordetella pertussis adenylate cyclase. Biochemistry. 2009;48:11273–11282. doi: 10.1021/bi901447j. [DOI] [PubMed] [Google Scholar]
- 16.Grupi A., Haas E. Segmental conformational disorder and dynamics in the intrinsically disordered protein α-synuclein and its chain length dependence. J. Mol. Biol. 2011;405:1267–1283. doi: 10.1016/j.jmb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay S., Krishnan R., Deniz A.A. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. USA. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreon A.C.M., Gambin Y., Deniz A.A. Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. USA. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreon A.C.M., Moran C.R., Deniz A.A. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods Enzymol. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 20.Uversky V.N. Amyloidogenesis of natively unfolded proteins. Curr. Alzheimer Res. 2008;5:260–287. doi: 10.2174/156720508784533312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turoverov K.K., Kuznetsova I.M., Uversky V.N. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog. Biophys. Mol. Biol. 2010;102:73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheibel T., Lindquist S.L. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat. Struct. Biol. 2001;8:958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- 23.Serio T.R., Cashikar A.G., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 24.Fowler D.M., Koulov A.V., Kelly J.W. Functional amyloid—from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Pappu R.V., Wang X., Crick S.L. A polymer physics perspective on driving forces and mechanisms for protein aggregation. Arch. Biochem. Biophys. 2008;469:132–141. doi: 10.1016/j.abb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crick S.L., Jayaraman M., Pappu R.V. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc. Natl. Acad. Sci. USA. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao A.H., Crick S.L., Pappu R.V. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Möglich A., Joder K., Kiefhaber T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. USA. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorn D.C., Meehan S., Carver J.A. Amyloid fibril formation by bovine milk κ-casein and its inhibition by the molecular chaperones αS- and β-casein. Biochemistry. 2005;44:17027–17036. doi: 10.1021/bi051352r. [DOI] [PubMed] [Google Scholar]
- 30.Ecroyd H., Koudelka T., Carver J.A. Dissociation from the oligomeric state is the rate-limiting step in fibril formation by κ-casein. J. Biol. Chem. 2008;283:9012–9022. doi: 10.1074/jbc.M709928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonil J., Henry G., Putaux J.L. Kinetics of fibril formation of bovine κ-casein indicate a conformational rearrangement as a critical step in the process. J. Mol. Biol. 2008;381:1267–1280. doi: 10.1016/j.jmb.2008.06.064. [DOI] [PubMed] [Google Scholar]
- 32.Atanasiu C., Su T.-J., Dryden D.T. Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–3944. doi: 10.1093/nar/gkf518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson E.N., Weber G. Synthesis and characterization of two fluorescent sulfhydryl reagents. Biochemistry. 1973;12:4154–4161. doi: 10.1021/bi00745a019. [DOI] [PubMed] [Google Scholar]
- 34.Campioni S., Mannini B., Chiti F. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 2010;6:140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 35.Han M.K., Knutson J.R., Brand L. Sugar transport by the bacterial phosphotransferase system. Fluorescence studies of subunit interactions of enzyme I. J. Biol. Chem. 1990;265:1996–2003. [PubMed] [Google Scholar]
- 36.Corrêa D.H.A., Ramos C.H.I. The use of circular dichroism spectroscopy to study protein folding, form and function. J. Biochem. 2009;3:164–173. [Google Scholar]
- 37.Kawahara K., Tanford C. Viscosity and density of aqueous solutions of urea and guanidine hydrochloride. J. Biol. Chem. 1966;241:3228–3232. [PubMed] [Google Scholar]
- 38.Holt C. Structure and stability of bovine casein micelles. Adv. Protein Chem. 1992;43:63–151. doi: 10.1016/s0065-3233(08)60554-9. [DOI] [PubMed] [Google Scholar]
- 39.Reid I.M. Corpora amylacea of the bovine mammary gland. Histochemical and electron microscopic evidence for their amyloid nature. J. Comp. Pathol. 1972;82:409–413. doi: 10.1016/0021-9975(72)90040-0. [DOI] [PubMed] [Google Scholar]
- 40.Lakowicz J.R. 3rd ed. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 41.Saxena A., Udgaonkar J.B., Krishnamoorthy G. Protein dynamics and protein folding dynamics revealed by time-resolved fluorescence. In: Hof M., Hutterer R., Fidler V., editors. Fluorescence Spectroscopy in Biology. Advanced Methods and their Applications to Membranes, Proteins, DNA, and Cells. Springer-Verlag; New York: 2005. pp. 163–179. [Google Scholar]
- 42.Wilkins D.K., Grimshaw S.B., Smith L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 43.Turro N.J. University Science Books; Sausalito, CA: 1991. Modern Molecular Photochemistry. [Google Scholar]
- 44.Krishnan R., Lindquist S.L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drury J.D., Narayanaswami V. Examination of lipid-bound conformation of apolipoprotein E4 by pyrene excimer fluorescence. J. Biol. Chem. 2005;280:14605–14610. doi: 10.1074/jbc.M414019200. [DOI] [PubMed] [Google Scholar]
- 46.Patel A.B., Khumsupan P., Narayanaswami V. Pyrene fluorescence analysis offers new insights into the conformation of the lipoprotein-binding domain of human apolipoprotein E. Biochemistry. 2010;49:1766–1775. doi: 10.1021/bi901902e. [DOI] [PubMed] [Google Scholar]
- 47.Thirunavukkuarasu S., Jares-Erijman E.A., Jovin T.M. Multiparametric fluorescence detection of early stages in the amyloid protein aggregation of pyrene-labeled α-synuclein. J. Mol. Biol. 2008;378:1064–1073. doi: 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 48.Hagen S.J., Eaton W.A. Two-state expansion and collapse of a polypeptide. J. Mol. Biol. 2000;301:1019–1027. doi: 10.1006/jmbi.2000.3969. [DOI] [PubMed] [Google Scholar]
- 49.Tran H.T., Mao A., Pappu R.V. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J. Am. Chem. Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 50.Teufel D.P., Johnson C.M., Neuweiler H. Backbone-driven collapse in unfolded protein chains. J. Mol. Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 51.Morar A.S., Olteanu A., Pielak G.J. Solvent-induced collapse of α-synuclein and acid-denatured cytochrome c. Protein Sci. 2001;10:2195–2199. doi: 10.1110/ps.24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J.C., Langen R., Winkler J.R. α-Synuclein structures from fluorescence energy-transfer kinetics: implications for the role of the protein in Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2004;101:16466–16471. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


