Abstract
Transcriptional memory of transient signals can be imprinted on living systems and influence their reactivity to repeated stimulations. Although they are classically ascribed to structural chromatin rearrangements in eukaryotes, such behaviors can also rely on dynamic memory circuits with sustained self-amplification loops. However, these phenomena are either of finite duration, or conversely associated to sustained phenotypic changes. A mechanism is proposed, in which only the responsiveness of the target gene is durably reset at a higher level after primary stimulation, using the celebrated but still puzzling vitellogenesis memory effect. The basic ingredients of this system are: 1), a positive autoregulation of the estrogen receptor α gene; 2), a strongly cooperative action of the estradiol receptor on vitellogenin expression; and 3), a variant isoform of the estradiol receptor with two autonomous transcription-activating modules, one of which is signal-independent and the other, signal-dependent. Realistic quantification supports the possibility of a multistationary situation in which ligand-independent activity is unable by itself to prime the amplification loop, but can click the system over a memory threshold after a primary stimulation. This ratchet transcriptional mechanism can have developmental and ecotoxicological importance and explain lifelong imprinting of past exposures without apparent phenotypic changes before restimulation and without need for persistent chromatin modifications.
Introduction
Transcriptional memory of eukaryotic cells is generally attributed to structural chromatin changes such as DNA methylation, histone methylation (1), or subnuclear DNA localization (2), but an alternative possible mechanism relies on dynamic circuits with positive feedbacks (3,4) that are capable of generating sustained responses after transient stimulations. However, these responses are either of limited duration (restricted to the lifetime of the signal-dependent target, such as permease for the lactose operon (3)) or indefinitely clicked at maximal level for an autonomously active amplification target. In this case, the phenotype remains modified after signal withdrawal. A system is presented in this study in which the responsiveness of the target gene to future stimulations is readjusted in the absence of visible phenotypic change. This phenomenon can be mediated by a ligand-dependent transcription factor (TF) and maintained after ligand removal, in a latent manner and without chromatin alteration. It has been derived from the famous vitellogenesis memory effect, which has long been established but remains puzzling.
Estrogen-dependent vitellogenesis is a process allowing the massive accumulation of nutrient egg proteins for the future embryos in egg-laying vertebrates such as fishes, amphibians, or birds. Before the identification of the estrogen receptor (ER) that mediates this response, it already appeared clear that vitellogenin (Vg) accumulates much more vigorously during secondary estrogen stimulation in chicken (5), Xenopus laevis (6), or the fish rainbow trout (7). This effect is striking because the level of Vg returns to zero between the successive stimulations. It has first been investigated at the level of the Vg gene structure; but if DNA demethylation and chromatin opening (as monitored by nuclease hypersensitivity) are indeed correlated with Vg expression, this long-term memory effect cannot be imputed to the maintenance of these modifications (8). An alternative explanation is proposed here, in which a single TF, a variant of ER, is sufficient for both recording and keeping alive transcriptional memory in the context of egg-laying vertebrate hepatocytes.
Materials and Methods
Quantification of molecule numbers
ER and Vg mRNAs
Total RNA samples were quantified, denatured, spotted, and UV-cross-linked to transfer membrane. RNA dots were hybridized to radioactive ER or Vg cDNA. The mRNA cell contents were then quantified as pg per μg of total RNA and converted into molecule copy number per cell, based on the molecular mass of the rtERs and Vg mRNAs and on the average trout hepatocyte content in total RNA.
ER protein cell content
The ER protein number was estimated indirectly by counting the high affinity binding sites for estradiol in trout hepatocytes. Liver nuclear extracts were incubated with [3H]-17-β-estradiol (Amersham, Little Chalfont, Buckinghamshire, UK), with or without 100-fold excess of cold hormone, to determine specific binding by subtraction. Free and bound hormone was separated by two successive treatments with charcoal/Dextran and radioactivity was measured by liquid scintillation.
Rates estimations
mRNA degradation rates
RNAs were extracted from cell aliquots at different time periods after transcription block using actinomycin D, and ER and Vg mRNAs were quantified as described above.
Nuclear run-on experiment
Nuclei were isolated at 0°C from cultured hepatocytes and resuspended in transcription buffer containing ribonucleotides and [α32P]-UTP. Transcription completion was then allowed to proceed for 30 min at 26°C. Radioactive RNAs were phenol-extracted, ethanol-precipitated, redissolved, and used as probes for hybridization to linear ER, Vg, and actin cDNAs, previously fixed on nylon membranes.
Clearance of estradiol-17β in rainbow trout
After injection of 17β-estradiol in a male trout, circulating estradiol was measured by radioimmunoassay.
Transient expression assays
β-Galactosidase assays in budding yeast
Yeast cells were stably transformed with the β-galactosidase reporter plasmid driven by the estrogen-responsive element of the rtER gene, with or without hCOUP-TF1 and with either rtERs or hER expression plasmids. β-Galactosidase activity in response to the indicated treatments was determined by spectrophotometry.
Luciferase assays
After plasmid transfection and transient expression, cells were harvested and the firefly luciferase activity was determined and normalized with β-galactosidase whose expression is driven by a β-actin-promoter. In the experiment of comparison of the transcriptional strengths of the fish ER gene promoter and the cytomegalovirus (CMV) promoter, these promoters directed the expression of firefly luciferase and were measured by comparison with a thymidine kinase promoter-Renilla luciferase control expression plasmid, using the Dual-Glo Luciferase Assay System (Promega, Madison, WI).
Additional detailed Materials and Methods and the parameter values used for numerical application, are available in the Supporting Material.
Results
The model proposed to explain the vitellogenesis memory effect will be quantified using new and previously published data obtained with the fish rainbow trout, but it can apply to other oviparous vertebrates. Studies on vitellogenesis are facilitated by the capacity of male animals to undertake vitellogenesis in response to estrogen administration, making it possible to avoid interferences with endogenous hormones.
The vitellogenesis memory effect is a cellular phenomenon
To establish whether the vitellogenesis memory effect is a cellular level phenomenon, it was first necessary to determine whether it can be evidenced in trout hepatocytes ex vivo. As primary cultures of trout hepatocytes do not divide and cannot be maintained for long periods, hepatocytes were prepared from male animals previously treated or not with estradiol one month ago. Once spread in monolayer in culture dishes, they were incubated with estradiol to compare the kinetics of Vg expression. As shown in Fig. 1, strong differences of the initial rates of accumulation were observed, depending on whether the cells were prepared from animals pretreated or not with estradiol. The effect is obvious for Vg as well as for Vg mRNA (Fig. 1). These observations indicate that the vitellogenesis memory effect is retained in cells separated from animals and does not require cross talks with other organs such as the nervous system. In the monolayer culture conditions used here to treat the cells with hormone soon after extraction, hepatocytes rapidly dedifferentiate and the expression of Vg progressively declines, as visible in Fig. 1. In the rest of the study, the primary hepatocytes were allowed to slowly organize tridimensionally into aggregates, to recover a more stable gene expression (see the Supporting Material).
Figure 1.
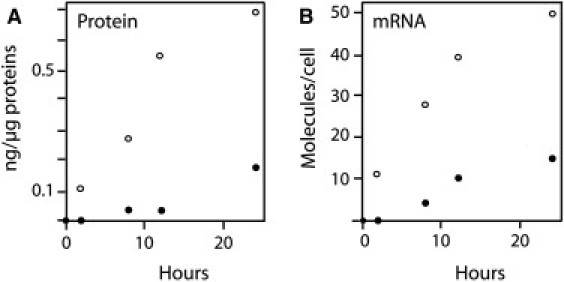
Vitellogenesis memory effect is preserved in cultured hepatocytes. Hepatocytes were prepared from male rainbow trout previously treated with estradiol one month ago (open circles) or hormone-naïve (solid circles). The time course of accumulation of Vg (A) and Vg mRNA (B) was measured after addition of 1 μM estradiol in the culture medium.
Basis of the model
The model is built on four lines of observation:
-
1.
A general feature of vitellogenesis evidenced in chicken (9), X. laevis (10), and rainbow trout (11) is the autoregulation of ER, mediated by a strong DNA-binding site for ER, present in the promoter of ER genes. This observation led to the admitted scenario in which estrogen treatment results in the upregulation of ER and that this receptor is rate-limiting in Vg gene transcription.
-
2.
An interesting observation is that the expression of ER remains high after a single transient hormone administration, long after the arrest of Vg synthesis (12,13).
-
3.
There is a cooperative response of Vg to ER, preventing Vg expression at low levels of ER (our results).
-
4.
Finally, an element essential for this mechanism is the existence in the liver of all egg-laying vertebrates tested so far, of a particular ER variant whose precise role is not yet understood, but which turns out to be particularly well suited to explain the vitellogenesis memory effect.
The central actor: a variant ER with two independent transactivation domains
Because Vg is expressed in an estrogen-dependent manner in the liver which contains a particular isoform of ERα named rtERs (14), we reasoned that this isoform could be precisely involved in the intracellular recording of the memory effect. Interestingly, in birds whose eggs also include white, the yolk protein vitellogenin is still synthesized in the liver but the white egg protein gene ovalbumin is expressed in the oviducts. Ovalbumin synthesis is also estrogen-dependent but not subject to memory effects and accordingly in chicken, liver contains the short ER isoform whereas the oviducts contain the long isoform (15). The short ERα found in the liver of egg-laying vertebrates singularly lacks the N-terminus present in long ERα isoforms. This domain, called A, coordinates the two transcription activating activities of ER named AF1 and AF2. AF2 is ligand-dependent and can be selectively poisoned by the partial antagonist 4-hydroxytamoxifen (4-OHT) (16). The activity of AF1, observed only in certain cellular contexts and in certain gene promoter configurations, has been shown to be strongly upregulated upon phosphorylation after interaction with the orphan nuclear receptor COUP-TFI (17). The hormone-dependence of AF1 is due to an inhibitory intramolecular interaction between the distal domains of the long ER isoform, which is broken upon ligand binding to AF2 (18,19). To compare the capacity of the short and long ER isoforms to stimulate transcription in the absence of hormone, we transfected them, together with COUP-TFI and a LacZ reporter gene driven by the estrogen-responsive element (ERE), into yeast cells. These cells were selected as a convenient tool because they are naturally devoid of nuclear receptors, but contain appropriate coactivators. As shown in Fig. 2 A, an estrogen-independent activity is clearly obtained for the short isoform whereas for the long one, the AF1 activity is subordinated to the presence of hormone and to AF2.
Figure 2.
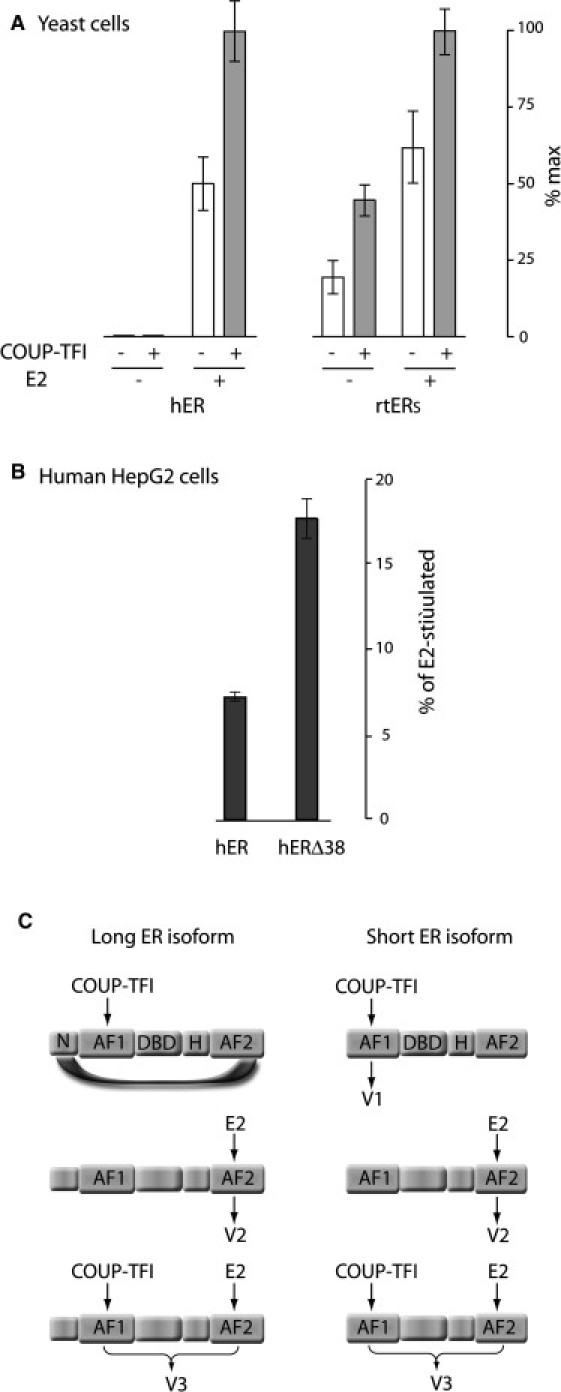
Role of the N-terminal domain of ER absent from the liver rtER isoform. (A) Relative transcriptional activity in yeast cells, of a single integrated β-galactosidase reporter gene construct whose expression is driven by the region of the rtER gene promoter carrying the palindromic ER-binding site. Cells were transformed or not with human COUP-TFI and treated or not with 10 nM E2 before measuring the activity of the β-galactosidase reporter enzyme. (B) Relative estrogen-independent activities in human cells, of human ER either unmodified (hER) or deleted form its N-terminal A domain (hERΔ38). (C) Transcriptional interplay between ER AF1 and AF2 activities in the long or short ERα isoforms, assumed in this model. In the long forms, AF1 is subordinated to the presence of hormone through ligand-induced breaking of an inhibitory distal interaction between the N-terminal domain and the AF2 domains of ER. In the liver-specific short form, the amino-terminal domain involved in the distal interaction is absent, so that AF1 can work autonomously.
To confirm that the high estrogen-independent activity of trout liver-specific ER isoform is due to its liver-specific N-terminal truncation, we verified that the estrogen-independent activity of human ER (hER) can be also increased after deletion of its N-terminal domain. As shown in Fig. 2 B, in human HepG2 cells, this deletion (Δ38) increases the basal transcriptional activity of hER from 7 to 18% of its maximal hormone-stimulated activity. Based on the results obtained with yeast cells that do not contain COUP-TF1 contrary to human cell lines, three possible maximum transcription activities of ER, termed here V1, V2, and V3, can be rationally defined depending on the cellular contexts and on the ER isoform (Fig. 2 C). V1 is the maximum transcription initiation rate caused by AF1 alone (i.e., measured in AF1-permissive contexts in the absence of hormone). V2 is the maximum transcription initiation rate caused by AF2 alone. V3 is the maximum transcription initiation rate expected from the synergistic action of AF1 and AF2 (i.e., measured in AF1-permissive contexts in presence of saturating hormone). AF1 and AF2 activities are roughly independent and additive in yeast (Fig. 2 A), but have been shown to recruit cooperatively certain coactivators in mammalian cells (20,21). Different modes of ER activity are observed with respect to the combination V1, V2, and V3, including the two following ones:
-
1.
V1 = 0 and V3 >> V2. For the long isoform, the modification induced by COUP-TFI has only a potentializing role on hormone action, but no apparent effect in the absence of hormone (17). This synergistic action of COUP-TFI and estradiol clearly results from transactivation cooperativity because it is observed with reporter constructs containing a single ER-binding site (17), ruling out the involvement of classical DNA-binding cooperativity.
-
2.
V3 ≥ V1 + V2. rtERs can transactivate a promoter fragment of the trout ERα gene carrying an ERE (11), suggesting that it can participate in the autoregulatory loop of ER in the liver. Given that rtERs can stimulate Vg expression exclusively in presence of E2, but can stimulate its own expression in both E2-dependent and -independent manners, one can propose the system schematized in Fig. 3. In the phenomenon of dynamic memory based on this organization, the primary estrogen stimulation triggers the ER loop, which could be maintained to a certain level after estrogen withdrawal by the AF1 activity alone. In turn, in absence of primary stimulation, AF1 is unable to prime the autoregulatory loop.
Figure 3.
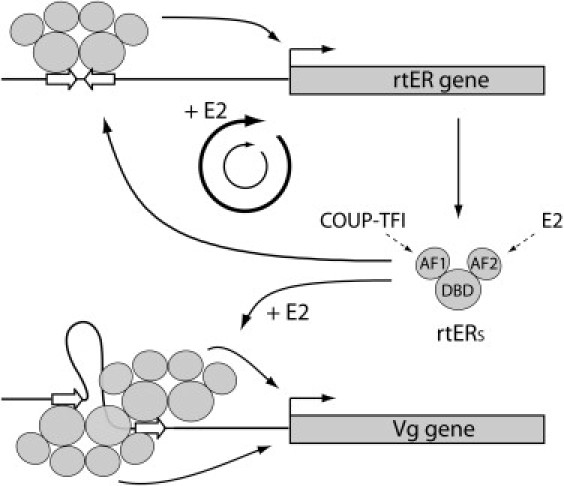
Molecular actors involved in estrogen-dependent vitellogenin synthesis. The rtER gene is subject to an autoamplification loop in which two ER molecules dimerize and bind to a palindromic ERE present in its own gene promoter. Each ER monomer contains an AF1 and an AF2 domain mediating the transcriptional effects of phosphorylation and E2, respectively. The expression of the vitellogenin gene (Vg) is strictly dependent on the presence of E2 and necessitates high concentrations of hormone-liganded rtERs for binding to imperfect ER half sites (open arrows). In turn, rtER expression is stimulated by E2 treatment and also, to a lower extent by unliganded ER, which allows to maintain the production of rtER at a certain level (thin cycle) after primary stimulation (bold cycle).
Formulation
Rate equations are generally considered as no longer appropriate and are replaced by discrete stochastic treatments when thermal fluctuations affect certain molecules present in low copy number (such as the ER mRNA). However, in this study, deterministic differential equations were retained as an approximation because Vg is secreted by the liver and taken up by the ovaries, so that its global production is averaged at the tissue level. For the same reason, biochemical techniques were preferred to single-cell approaches for the accurate determination of average kinetic parameters. In the absence of precise data on certain points, several hypotheses were assumed:
-
1.
Unmodified ER dimers are not supposed to reach DNA. Unliganded ER can bind to DNA in vitro; but in vivo, chromatin immunoprecipitation assays using ER-expressing cells reproducibly showed a strong enrichment of the ER DNA-binding activity upon hormone stimulation (22), possibly related to the desequestration of ER from anchor proteins (heat shock proteins and others). In addition, the low amount of chromatin-bound unliganded ER can correspond to active phosphorylated ER (23).
-
2.
The two partners of a given dimer are considered as symmetrically modified by either phosphorylation, hormone ligation, or both, considering that two tightly interacting protomers should be submitted to the same conformational modifications to share a common dimerization surface.
The expression of Vg genes is strictly dependent on the presence of hormone. The trout Vg gene promoter contains imperfect ER-binding elements and multiple half-sites. This organization resembles that of X. laevis Vg B1 described in Martinez and Wahli (24), whose promoter includes two nonconsensual sites with low affinity for ER. Assuming that ER equilibrates rapidly with DNA compared to the slower dynamics of gene product fluctuations, a continuous equation in which the production is driven by the cooperative fixation of two liganded ER dimers is
| (1) |
where RH is the hormone-liganded ER; VV is the maximal transcription initiation rate of the Vg gene, mV is the Vg mRNA; rmV is its first-order rate of removal; and the terms βv (concentration−2) are the products of DNA-binding and dimerization constants. Kc (unitless) is the cooperative enhancement factor of the affinity of ER for DNA once the first site is occupied. This type of equation can be extended to the cooperative fixation of more ER molecules and can be, depending on the parameters, approximated in the form of Hill functions.
The transcriptional activation of a reporter construct containing the ERE present in the ER gene promoter, by unliganded rtERs (Fig. 2 A), could be related to the high affinity of ER for this DNA target site, to which both hormone-bound and phosphorylated-only ER can bind, so that different sources of transcription activation are expected. The AF1 transcriptional activity of ER is activated by phosphorylation (P), whereas AF2 is activated by the hormone (H). The different forms of ER dimers should share time for interacting with this DNA target site (25). Hence,
| (2) |
where RP0 is phosphorylated only, RH0 is hormone-liganded only, and RPH is both phosphorylated and hormone-bound. The term rmR is the rate of removal of the ER mRNA mR; the terms βR are the products of dimerization and DNA-binding constants of ER, which depend on its modification state; and V1, V2, and V3 are the rates defined earlier.
An additional level of complexity of this model is that the mRNA degradation rates are not constant, but depend on estrogens (26). This ER mRNA stabilization during hormone stimulation enhances the autoamplification loop of ER gene expression. The lower stabilization of the ER mRNA relatively to that of Vg (Fig. 4 A) could be physiologically related to the respective roles of the derived proteins, considering that Vg is a secreted end-product inactive in the cell, whereas ER is a signal sensor that should remain reactive to conditions changes. This atypical property complicates the treatment of the network and, given the poor knowledge of mRNA stabilization machineries, it obliges us to use primary trout hepatocytes despite their technical inconvenience and forbids the possibility to convey the vitellogenesis memory effect into heterologous cellular contexts in a synthetic biology attempt.
Figure 4.
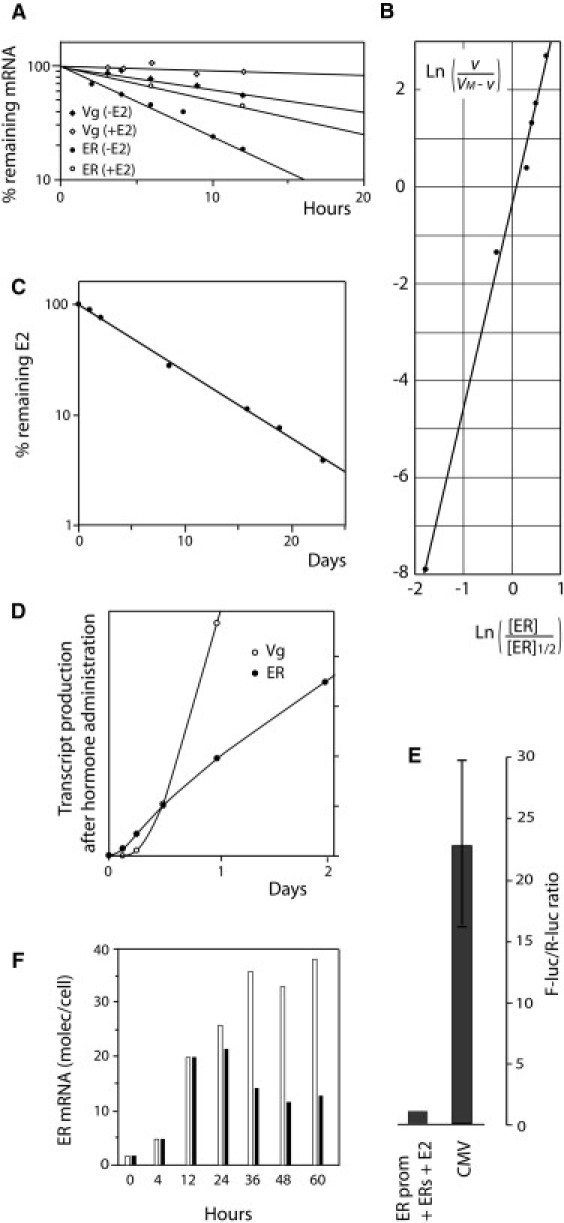
Quantification experiments. (A) Degradation rates of ER and Vg mRNA in cultured trout hepatocytes (circles, ER; diamonds, Vg; open symbols, without hormone; solid symbols, with 1 μM hormone). (B) Cooperative ER concentration-dependence of the Vg gene transcription rate (v). (C) Rate of clearance of estradiol in vivo in a male rainbow trout, determined by radio-immunoassay. (D) Transcript accumulation for Vg (open circles) and ER (solid circles), determined by simultaneous run-on analyses of the ER and Vg gene transcription rates in nuclei isolated from hormone-stimulated liver cells. (E) Comparative maximal expression in HeLa cells, of the CMV promoter and of the zebrafish ER gene promoter in presence of zERs and of 1 μM E2. HeLa cells were transfected with the short isoform of zebrafish ER and a F-Luc reporter gene driven by the zebrafish ER gene promoter. (F) Selective poisoning of the AF2/LBD domain using 4-OHT in cultured trout hepatocytes. Estradiol was added at time 0 and 4-OHT was added (solid histograms) or not (open histograms) 12 h later.
The mechanism proposed in the literature to underlie Vg mRNA stabilization by E2 is the hormone-dependent synthesis of a shield RNA-binding protein named vigillin, preventing the action of a polysomal endo-RNase (27). If the shield RNA-binding protein S is abundant relatively to the Vg mRNA, one has
| (3) |
where βV3 is the affinity of S for the Vg mRNA (mV), and rmV0 and rmVH are the degradation rates of mV in the absence and presence of hormone, respectively.
In Eqs. 1 and 2, the concentrations of the different forms of ER are not directly measurable, but can be recovered from observable quantities in a probabilistic manner, under the quasi-equilibrium assumption. Because the diffusing hormone is not limitative and equilibrates rapidly with ER, the proportion of hormone-bound ER in the cell, PH, corresponds to
| (4) |
where KdH is the dissociation equilibrium constant between hormone and ER. Similarly, let PP be the fraction of phosphorylated ER band. If the modifications of ER by hormone binding and COUP-TFI-mediated phosphorylation are independent events, the concentrations of the different ER forms used in Eqs. 1 and 2 can all be expressed as functions of total ER (RT), which will itself be expressed as a function of its mRNA:
| (5a) |
| (5b) |
| (5c) |
| (5d) |
| (5e) |
In all these formulations, the modification of ER, through phosphorylation or hormone binding, is assumed to precede dimerization. The available data are unclear about the hierarchy of ER ligation and dimerization, but in fact, these events could be inverted with the same quantitative results under the quasi-equilibrium assumption. The principle of microreversibility simplifies the hierarchy of coupled phenomena, which cannot be separated from a near-equilibrium perspective. ER ligation and dimerization belong to a single pentagonal cycle defined by the following set of equilibrium constants,
| (6a) |
| (6b) |
| (6c) |
| (6d) |
| (6e) |
where R is the ER monomer and RF is the free monomer. In the absence of free energy inputs, microscopic reversibility implies K2K12 = K3K4K5. Hence, considering that dimerization occurs before or after hormone ligation, is equivalent.
The total concentration of ER results from the translation/degradation balance,
| (7) |
where tR and rR are the mRNA translation and protein degradation rates, respectively. The ratio tR/rR was found to be relatively constant in this system and will be thereafter written α. It is quantified for better precision as the steady-state concentration ratio between the ER protein and ER mRNA in the presence of saturating hormone. We considered that although the hormone arrival is rapid, its decrease is slower, particularly in vivo, and could contribute, to some extent, to the sustained level of ER observed after primary stimulation (13). Indeed, when hormone production or administration stops, PH does not abruptly fall to zero in vivo but becomes a logistic function of time,
| (8) |
where [H]0 is the starting hormone concentration (considered to be lower than the capacity of steroid-binding plasma proteins), KdH is the constant of dissociation between the hormone and the receptor, and c is the first-order hormone clearance rate. To determine if the model is compatible with realistic parameters, we introduced in the equations the molecule numbers and interaction constants estimates described in the Supporting Material. They can be summarized as follows.
Quantification
Counting molecules
ER and Vg mRNA molecule numbers were deduced by comparison of linear hybridization signals to avoid possible biases due to nonlinear amplification approaches and to reverse-transcription efficiency. Although many techniques exist for quantifying mRNA, counting molecules is much more difficult for proteins. The classical immunoblotting experiments are at most useful in comparative approaches but a gap of knowledge exists with respect to the absolute protein numbers. Fortunately, a technical opportunity is offered for the case of a steroid-binding protein, by the possibility granted to count the hormone-binding sites within the cell.
Rates determination
The determination of degradation and production rates of mRNAs are technically facilitated in eukaryotic cells by the spatial separation between these processes, taking place in the cytoplasm and nucleus, respectively. To determine the cytoplasmic mRNA degradation rate, the afflux of fresh mRNA molecules was prevented by the transcription inhibitor actinomycin-D. Estradiol induced a strong stabilization of presynthesized Vg mRNA (half-life of 3.5 days; rate of 0.2 day−1) (Fig. 4 A). The relative transcription initiation rates are measured in run-on experiments; cells were broken and nuclei were isolated and allowed to complete the transcriptions initiated in intact cells.
As no RNA degradation and no RNA synthesis initiations are possible in isolated nuclei (because of the nonrenewal of components from the cytoplasm), only extensions of nascent transcripts frozen before the separation between nuclei and cytoplasm are visualized. The results represent the density of active RNA polymerases on the considered genes, which directly reflects the transcription initiation rate (28) (Fig. 4 D). Based on the effect of 4-OHT that selectively inactivates AF2 (Fig. 4 F) and on the activities of ER in yeast (Fig. 2 A), we approximated that for rtERs, V3 = V1 + V2 and V1 = V2.
Predicted evolutions
ER expression
Positive autoregulatory loops can provide multiple stationary states that are reached when the production and destruction of given products compensate each other. These values are the roots of a quadratic equation resulting here from the conjugation of Eqs. 2 and 7 (see Appendix A). The stationary values of [mR] in absence of hormone are shown in Fig. 5 A. In the absence of hormonal stimulation in naïve animals, parameters are such that the lower solution cannot be spontaneously reached when starting from low or noisy ER gene expression lower than a certain level, set at eight molecules/cell based on our data. But after an initial rise of ER mRNA triggered by hormone pretreatment, this mRNA no longer disappears because of a persistent loop maintained by unliganded ER. It decreases to the upper solution that can be envisioned as a memory threshold, and afterwards remains at this value through a ratchetlike mechanism.
Figure 5.
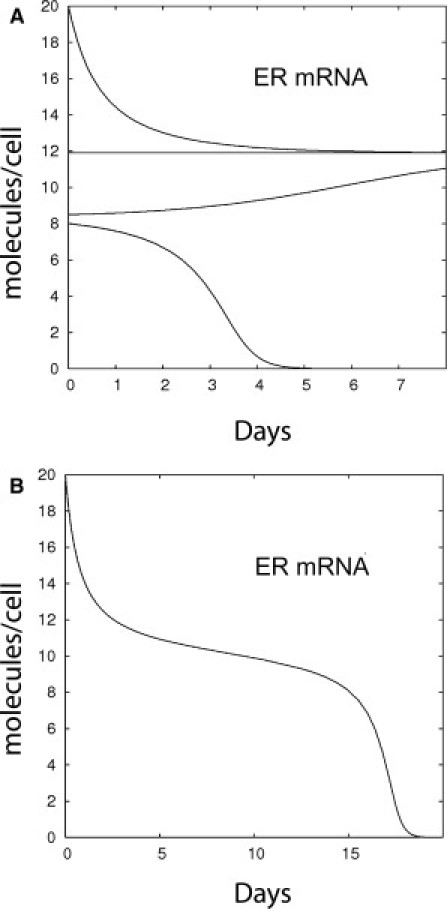
Evolution of the cellular content in ER mRNA. (A) Simulation without hormone and starting from various initial conditions. The memory threshold at 12 molecules/cell is reached only if the initial number of molecules exceeds eight molecules per cell. (B) Particular case of short-term memory obtained near the ghost bifurcation.
Accordingly, as shown in Fig. 4 F, if 1 μM 4-OHT is added to E2-stimulated hepatocytes to inactivate AF2 after crossing this threshold, the following decrease of ER mRNA molecule number stabilizes at a higher level, in line with the 3.2-fold reduction of the number of ER proteins observed after 4-OHT treatment in X. laevis (29). Different behaviors are expected depending on the delicate balance between all the parameters. When the two solutions are close together, the risk of priming the autoamplification loop in virgin animals is further diminished but the risk of memory loss is high. In turn, when the two solutions are far away enough, the memory is strongly stabilized and resistant to stochastic fluctuations of mRNA concentrations. In addition, undesirable priming of the positive feedback is prevented by the difference between the numbers of mRNA molecules, before stimulation (2.5 molecules/cell) and the threshold level (eight molecules/cell). A critical situation is obtained near the so-called “ghost bifurcation” (30). In this configuration, as shown in Fig. 5 B, the level ER mRNA does not immediately drop and the vitellogenesis memory effect is programmed for a certain delay T (see Appendix B), corresponding, in this case, to
| (9) |
Such a situation could be obtained, for example, in the case of insufficient phosphorylation of ER, just lower than
| (10) |
For example, with the numerical estimates used here, if the fraction of phosphorylated ER decreases from 10% to 9.8%, memory is lost after a delay of T ≈ 9 days. However, the slow hormone decrease observed in vivo (Fig. 4 C), could prolong further the sustained level of ER after primary stimulation.
Vg expression
Plotting the response of Vg to ER in Hill coordinates reveals a strong degree of cooperativity, with a Hill coefficient of 4.2 (Fig. 4 B), suggesting that more than two ER dimers regulate Vg expression. This strong cooperativity, coupled to the relative destabilization of the Vg mRNA in the absence of hormone, allows that this mRNA disappears between two stimulations. As shown in Fig. 6, the lower affinity and higher cooperativity of action of ER on the Vg gene, relatively to the ER gene, make the responses of these two genes not parallel, with a delay in the decrease (Fig. 6 A) and the accumulation (Fig. 6 B) of Vg transcripts. This delay is strongly shortened in prestimulated cells when starting from the memory level, of 12 molecules of ER mRNA per cell on average (Fig. 6 C). Hence, a vitellogenesis memory effect can indeed be predicted using our model.
Figure 6.
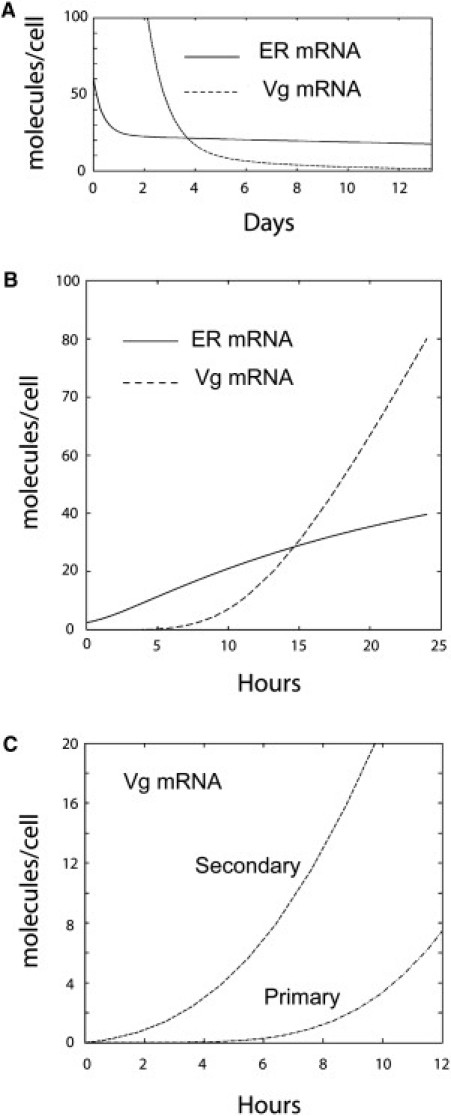
Comparative predicted evolutions of ER and Vg mRNA cell contents. (A) ER and Vg mRNA evolution after hormone withdrawal, with a clearance rate of 0.084/day and starting from the stationary plateau values of ER and Vg. (B) Evolution of ER and Vg mRNA after hormone addition and starting from 2.5 molecules/cell of ER mRNA. (C) Comparative kinetics of accumulation of the Vg mRNA in hormone-naïve cells (primary stimulation) and previously stimulated cells (secondary stimulation).
Discussion
We propose a model of dynamic memory that can contribute to the vitellogenesis memory effect, without recourse to structural chromatin rearrangements and that is possible with realistic kinetic parameters and molecule numbers. Chromatin modifications are associated to vitellogenin expression but could have helper rather than decisional roles, as suggested in Burch and Evans (8). A pivotal aspect of our model is the involvement of a positive feedback, whose importance has long been pointed out in bacterial systems such as the lactose operon, in which the permease derived from this operon accelerates the entry of the inducer, thereby increasing its own synthesis (3).
The induced state can tolerate dilution of permease and survive cell division or a transient decrease of the inducer concentration, but this kind of memory is restricted to the period of stability of the presynthesized permease. This founder model has then been extended to many cases of direct positive feedback situations in which a TF activates its own gene (4,31,32). But in eukaryotic cells, heritable structural chromatin modifications are always put forward to explain transcriptional memory. The idea that transcriptional memory should be structurally engraved in chromosomes is predominant, but, to our knowledge, it is not really supported by any recent observations showing that structural epigenetic modifications are, for example, unstable and continuously reconfigured, and thus poorly appropriate for long-term memory at the scale of months or years.
Examples of positive feedbacks, associated to bistability, hysteresis, and memory, strongly accumulate in gene network modeling studies, suggesting that they have fundamental roles in the structuring of biochemical systems. However, the case presented in this study has some distinctive original features: 1), it ensures the memory of gene responsiveness rather that the sustained expression of Vg; 2), the same component, rtERs, is involved both in sensing the signal and preserving its memory; and 3), the system is largely controlled at the level of mRNA stability, whereas most reported feedback systems are tuned at the level of production (transcription and translation rates).
In this respect, the trout liver appears to contain very specific machineries because estrogen treatments generally lead to ER mRNA destabilization in other cellular contexts. In hepatocytes, the inverse regulation by estradiol of the stability of vitellogenesis-related and unrelated mRNAs, could be interpreted as a mechanism redirecting liver expression machineries during the egg-production period. This essential regulation of ER and Vg mRNA stability allows that every mRNA molecule contributes lastingly to the massive Vg synthesis and shortens the recovery from vitellogenesis after estrogen withdrawal. In addition to this accessory loop linking the stability of the ER mRNA to estrogen exposure, the minimal system described here can be embedded in a more densely reticulated circuitry, ensuring its robustness by coordinating the different values. For example, the degree of ER phosphorylation could be continuously readjusted through some unidentified fast negative loop to prevent the large effect shown above, caused by a slight modification of ER phosphorylation while keeping unchanged the other parameters.
Some cooperativity is necessary for positive feedback bistability (33). The mechanism of cooperativity retained here is ER dimerization, which means that either the dimerization strength or the concentration of activated ER should be limiting enough. Additional mechanisms are involved in the strongly cooperative Vg gene expression. Although trout Vg expression is strictly and strongly estrogen-dependent, its gene is devoid of high-affinity ERE. We propose that this seemingly paradoxical situation is a condition to generate a nonhyperbolic response with a threshold effect. Given the weakness of the individual ER target sites present in the Vg gene promoter and the reported capacity of two ER dimers to interact and organize into a tetramer (34,35), the mechanism of modulated self-assembly recently proposed to regulate transcription both precisely and flexibly (36), is a particularly good candidate for contributing to the control of Vg gene expression. Such a strong cooperativity contributes both to the delay of the Vg gene response to hormone and to the memory effect.
The delayed response of Vg to estrogens, with respect to that of ER, allows that Vg expression starts only when the amplification of ER is unambiguously engaged. This property can be envisioned as a noise-reduction mechanism preventing inappropriate Vg expression upon unwanted bursts of hormone. With respect to stochastic behaviors, modeling dynamic memory using differential equations is questionable for systems containing low-copy-number molecules, because averaging a collection of cells is not equivalent to a macroscopic system in the case of threshold effects. The deterministic approach has, however, been used here as an approximation, in absence of strongly nonlinear phenomena and because the memory concerns the accumulation of a secreted gene product whose blood concentration derives from a lot of cells. This situation could explain why Vg is not directly synthesized by oocytes, but taken up from a common pool, to avoid heterogenous Vg contents between individual eggs.
The transcription rates calculated here can appear relatively low. One should, however, keep in mind that only productive transcription is retained in these results. Considering the high proportion of defective transcription elongation and transcript maturation, for example by defective splicing, the total number of initiations could be much higher. Notably, the number of ER transcripts produced from the fish ER gene promoter is reached within 1 h by the CMV promoter, as determined by single-cell imaging (37). This difference is consistent with the relative strengths of the CMV viral promoter and the zebrafish ER gene promoter activated by zERs in presence of hormone, compared in transient expression assays (Fig. 4 E). These infrequent transcription initiation events can forbid stochastic transcriptional bursts to occur, thereby preventing unintentional crossing of the memory threshold.
Many additional actors involved in the vitellogenesis memory effect are likely to be missing in this study. For example, the basal ER mRNA content that we observed in absence of hormone is not predicted by the autoregulatory schemes used here. This low expression could be ensured by the proximal rtER gene promoter, which includes a TATA box capable of generating spontaneous transcriptional activity (38). Such a nonregulated activity could be useful for priming the amplification loop upon hormone addition. The minimal model can work using a combination of realistic parameters, which opens the possibility that in addition to DNA demethylation, dynamic mechanisms can also be involved.
This model can have important biological roles, in addition to providing an alternative explanation of the vitellogenesis memory effect. Considering the generalized presence of two transactivation domains in nuclear receptors and the various splicing variants reported in the literature for this family of TFs, one cannot exclude that such mechanisms could participate to the delayed effects of transient exposures to xenohormones. Our study provides an additional mechanism possibly underlying such phenomena, complementary to the classical explanation of structural chromosome marking.
Acknowledgments
This work was supported by University of Rennes 1, Action Défits Scientifiques Émergents. F.N.-B. holds a Fellowship from the French Ministère de la Recherche.
Appendix A: Bistability Analysis in the Absence of Hormone
The stationary states and their stability are particularly important in absence of estrogens, and can be analyzed by adimensioning the system. If setting
and
the possible stationary states of the system should solve
| (11a) |
and u can be 0 or can be a root of
| (11b) |
Hence, there are one or three solutions depending on the value of μ. If 0 ≤ μ < 1/2, the three solutions are
if μ > 1/2, u = 0 is the unique solution.
To analyze the stability of the solutions through the linear approach, we consider a small perturbation δu of a stationary solution u∗ (i.e., u = u∗ + δu).
For the solution u∗ = 0,
Hence, δu decreases exponentially and consequently the solution u∗ = 0 is always stable for every positive u.
For the solution
| (12) |
the right term is then always negative for 0 ≤ μ < 1/2, and the solution is stable in this range.
For the solution
the right term is positive and this solution is unstable for every μ between 0 ≤ μ < 1/2.
These results can be gathered in a bifurcation diagram showing the stationary states as a function of μ (see Fig. S1 in the Supporting Material) drawn with a plain line when stable and a dashed line when unstable. The bifurcation at μ = 0.5 is compatible with the estimated parameters listed in Table S1 in the Supporting Material, which give 11.9 ER mRNA molecules per cell.
Appendix B: Ghost Bifurcation
Dynamic memory to estrogen exposure, through the model proposed here, can be of limited duration for given sets of parameters, which can be examined using the adimensioned parameters defined above. When μ ≥ 0.5, only the solution u∗ = 0 exists and is stable; nevertheless, around the value u = 1, the variation rate du = dτ is close to 0 and the system spends some time in the vicinity of that value, before falling to the stationary solution u = 0. The time spent near u = 1 can be estimated (30), using a development of the second order on u and μ = 1 + v and μ = 1/2 + r2:
| (13) |
Hence,
| (14) |
The delay to reach u = 0 when the initial condition is u >> 1, can be estimated by integrating Eq. 14 between +∞ and 0,
| (15) |
Consequently, the time to reach the stationary state is approximately
| (16) |
Supporting Material
References
- 1.Thomassin H., Flavin M., Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20:1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brickner J.H. Transcriptional memory: staying in the loop. Curr. Biol. 2010;20:R20–R21. doi: 10.1016/j.cub.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Novick A., Weiner M. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smolen P., Baxter D.A., Byrne J.H. Frequency selectivity, multistability, and oscillations emerge from models of genetic regulatory systems. Am. J. Physiol. 1998;274:C531–C542. doi: 10.1152/ajpcell.1998.274.2.C531. [DOI] [PubMed] [Google Scholar]
- 5.Beuving G., Gruber M. Induction of phosvitin synthesis in roosters by estradiol injection. Biochim. Biophys. Acta. 1971;232:529–536. doi: 10.1016/0005-2787(71)90607-1. [DOI] [PubMed] [Google Scholar]
- 6.Baker H.J., Shapiro D.J. Rapid accumulation of vitellogenin messenger RNA during secondary estrogen stimulation of Xenopus laevis. J. Biol. Chem. 1978;253:4521–4524. [PubMed] [Google Scholar]
- 7.Le Guellec K., Lawless K., Tenniswood M. Vitellogenin gene expression in male rainbow trout (Salmo gairdneri) Gen. Comp. Endocrinol. 1988;71:359–371. doi: 10.1016/0016-6480(88)90264-x. [DOI] [PubMed] [Google Scholar]
- 8.Burch J.B., Evans M.I. Chromatin structural transitions and the phenomenon of vitellogenin gene memory in chickens. Mol. Cell. Biol. 1986;6:1886–1893. doi: 10.1128/mcb.6.6.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mester J., Baulieu E.E. Nuclear estrogen receptor of chick liver. Biochim. Biophys. Acta. 1972;261:236–244. doi: 10.1016/0304-4165(72)90334-0. [DOI] [PubMed] [Google Scholar]
- 10.Perlman A.J., Wolffe A.P., Tata J.R. Regulation by estrogen receptor of vitellogenin gene transcription in Xenopus hepatocyte cultures. Mol. Cell. Endocrinol. 1984;38:151–161. doi: 10.1016/0303-7207(84)90113-8. [DOI] [PubMed] [Google Scholar]
- 11.Le Dréan Y., Lazennec G., Valotaire Y. Characterization of an estrogen-responsive element implicated in regulation of the rainbow trout estrogen receptor gene. J. Mol. Endocrinol. 1995;15:37–47. doi: 10.1677/jme.0.0150037. [DOI] [PubMed] [Google Scholar]
- 12.Westley B., Knowland J. Estrogen causes a rapid, large and prolonged rise in the level of nuclear estrogen receptor in Xenopus laevis liver. Biochem. Biophys. Res. Commun. 1979;88:1167–1172. doi: 10.1016/0006-291x(79)91531-6. [DOI] [PubMed] [Google Scholar]
- 13.Barton M.C., Shapiro D.J. Transient administration of estradiol-17 β establishes an autoregulatory loop permanently inducing estrogen receptor mRNA. Proc. Natl. Acad. Sci. USA. 1988;85:7119–7123. doi: 10.1073/pnas.85.19.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menuet A., Anglade I., Kah O. Tissue-specific expression of two structurally different estrogen receptor α isoforms along the female reproductive axis of an oviparous species, the rainbow trout. Biol. Reprod. 2001;65:1548–1557. doi: 10.1095/biolreprod65.5.1548. [DOI] [PubMed] [Google Scholar]
- 15.Griffin C., Flouriot G., Gannon F. Two functionally different protein isoforms are produced from the chicken estrogen receptor-α gene. Mol. Endocrinol. 1999;13:1571–1587. doi: 10.1210/mend.13.9.0336. [DOI] [PubMed] [Google Scholar]
- 16.Metzger D., Losson R., Chambon P. Promoter specificity of the two transcriptional activation functions of the human estrogen receptor in yeast. Nucleic Acids Res. 1992;20:2813–2817. doi: 10.1093/nar/20.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Métivier R., Gay F.A., Pakdel F. Formation of an hER α-COUP-TFI complex enhances hER α AF-1 through Ser118 phosphorylation by MAPK. EMBO J. 2002;21:3443–3453. doi: 10.1093/emboj/cdf344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Métivier R., Petit F.G., Pakdel F. Function of N-terminal transactivation domain of the estrogen receptor requires a potential α-helical structure and is negatively regulated by the A domain. Mol. Endocrinol. 2000;14:1849–1871. doi: 10.1210/mend.14.11.0546. [DOI] [PubMed] [Google Scholar]
- 19.Métivier R., Stark A., Gannon F. A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol. Cell. 2002;10:1019–1032. doi: 10.1016/s1097-2765(02)00746-3. [DOI] [PubMed] [Google Scholar]
- 20.Benecke A., Chambon P., Gronemeyer H. Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 2000;1:151–157. doi: 10.1093/embo-reports/kvd028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Métivier R., Penot G., Pakdel F. Synergism between ERα transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol. Endocrinol. 2001;15:1953–1970. doi: 10.1210/mend.15.11.0727. [DOI] [PubMed] [Google Scholar]
- 22.Lin C.Y., Ström A., Liu E.T. Discovery of estrogen receptor α-target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González L., Zambrano A., Aranda A. Activation of the unliganded estrogen receptor by prolactin in breast cancer cells. Oncogene. 2009;28:1298–1308. doi: 10.1038/onc.2008.473. [DOI] [PubMed] [Google Scholar]
- 24.Martinez E., Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J. 1989;8:3781–3791. doi: 10.1002/j.1460-2075.1989.tb08555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel D. How transcription factors can adjust the gene expression floodgates. Prog. Biophys. Mol. Biol. 2010;102:16–37. doi: 10.1016/j.pbiomolbio.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Brock M.L., Shapiro D.J. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell. 1983;34:207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 27.Dodson R.E., Shapiro D.J. Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 28.Smale S.T. Nuclear run-on assay. Cold Spring Harb. Protoc. 2009 doi: 10.1101/pdb.prot5329. [DOI] [PubMed] [Google Scholar]
- 29.Riegel A.T., Aitken S.C., Schoenberg D.R. Differential induction of hepatic estrogen receptor and vitellogenin gene transcription in Xenopus laevis. Endocrinology. 1987;120:1283–1290. doi: 10.1210/endo-120-4-1283. [DOI] [PubMed] [Google Scholar]
- 30.Strogatz S.H. Perseus Books; Cambridge, MA: 1994. Nonlinear Dynamics and Chaos: with Applications to Physics, Biology, Chemistry, and Engineering. [Google Scholar]
- 31.Becskei A., Séraphin B., Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maamar H., Raj A., Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry J.L., Adler F.R. How to make a biological switch. J. Theor. Biol. 2000;203:117–133. doi: 10.1006/jtbi.2000.1068. [DOI] [PubMed] [Google Scholar]
- 34.Tanenbaum D.M., Wang Y., Sigler P.B. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. USA. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massaad C., Coumoul X., Barouki R. Properties of overlapping EREs: synergistic activation of transcription and cooperative binding of ER. Biochemistry. 1998;37:6023–6032. doi: 10.1021/bi972445e. [DOI] [PubMed] [Google Scholar]
- 36.Vilar J.M., Saiz L. Control of gene expression by modulated self-assembly. Nucl. Acids Res. 2011 doi: 10.1093/nar/gkr272. 0:gkr272v1-gkr272 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darzacq X., Shav-Tal Y., Singer R.H. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy K.F., Adams R.M., Collins J.J. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


