Abstract
Eukaryotic gene activation requires selective unfolding of the chromatin fiber to access the DNA for processes such as DNA transcription, replication, and repair. Mutation/modification experiments of linker histone (LH) H1 suggest the importance of dynamic mechanisms for LH binding/dissociation, but the effects on chromatin's unfolding pathway remain unclear. Here we investigate the stretching response of chromatin fibers by mesoscale modeling to complement single-molecule experiments, and present various unfolding mechanisms for fibers with different nucleosome repeat lengths (NRLs) with/without LH that are fixed to their cores or bind/unbind dynamically with different affinities. Fiber softening occurs for long compared to short NRL (due to facile stacking rearrangements), dynamic compared to static LH/core binding as well as slow rather than fast dynamic LH rebinding (due to DNA stem destabilization), and low compared to high LH concentration (due to DNA stem inhibition). Heterogeneous superbead constructs—nucleosome clusters interspersed with extended fiber regions—emerge during unfolding of medium-NRL fibers and may be related to those observed experimentally. Our work suggests that fast and slow LH binding pools, present simultaneously in vivo, might act cooperatively to yield controlled fiber unfolding at low forces. Medium-NRL fibers with multiple dynamic LH pools offer both flexibility and selective DNA exposure, and may be evolutionarily suitable to regulate chromatin architecture and gene expression.
Introduction
The two-meter-long DNA in eukaryotic cells is severely condensed for storage inside the micron-sized nuclei. This tight packing is achieved by folding of a compact array of nucleoproteins, or nucleosomes, joined by open DNA segments (DNA linkers) into the 30-nm-wide chromatin fiber (1). The nucleosome consists of a histone octamer (two copies each of H2A, H2B, H3, and H4) with 147 basepairs (bps) of DNA wrapped ∼1.75 turns around it (2–4). The nucleosome irregular charge distribution and contoured surface favor electrostatic interactions with other nucleosomes and DNA linkers that help keep the fiber folded (4,5). Furthermore, 10 highly positively charged and flexible tails (two N-terminal domains from each histone dimer plus two additional C-terminal domains from dimer H2A) extend from the nucleosome surface and mediate interactions with different chromatin regions (3,4). A fifth type of linker histone (LH) protein (H1 or H5) binds the linker DNA at its entry/exit point to the nucleosome, screening the DNA/DNA electrostatic repulsion and allowing closer contact within the fiber.
The detailed structure of the 30-nm fiber has been a puzzle for over three decades (6,7), and several models have been proposed (one-start, two-start, etc.). In the solenoid helix (one-start), consecutive nucleosomes are in closest contact and joined by bent DNA linkers (8). In the zigzag model (two-start), alternate nucleosomes are in closest contact and DNA linkers are relatively straight (9,10). Recent experiments and modeling have put forth the notion of a variable heteromorphic chromatin architecture (11–13) that reconciles both models. That is, one fiber can contain both straight and bent linkers, especially at divalent ion conditions (11).
Understanding the kinetics and thermodynamics of chromatin compaction and unfolding is essential for interpreting gene function. Eukaryotic gene activation requires decondensation of specific sections of the chromatin fiber to access the DNA for processes such as DNA transcription, replication, and repair. Deciphering the forces required to unfold the chromatin fiber is challenging because they depend on many factors, which include the ionic environment, the binding of linker histones and of remodeling factors, the chemical composition of the chromatin components, and the nucleosome repeat length (NRL; 147 bp of nucleosomal DNA plus length of linker DNA). Moreover, it has been demonstrated that these factors should be analyzed collectively. For example, LH occupancy decreases with the NRL (14) and LH's structural effect is negligible for short-NRL (<182 bp) arrays but strong for medium-NRL (191–209 bp) fibers (13,15).
During the past 10 years, single-molecule experiments have been developed and applied to examine forces needed to unfold polymers, including the chromatin fiber (see reviews in Chien and van Noort (16) and Lavelle et al. (17)). Such forces mimic cellular forces exerted by molecular machines, like RNA and DNA polymerases of up to 35 pN (18). Chromatin unfolding has been revealed by atomic force microscopy (19), optical tweezers (20–24), and magnetic tweezers (25) experiments. Although much progress has been made, modeling represents a necessary complement to the experimental analysis to provide structural and mechanistic views of the unfolding process. Indeed, many groups have applied various modeling approaches to study chromatin aspects under tension (e.g., (26–31)).
It has been shown that LH forms complex networks of interactions that likely regulate chromatin fiber folding (for an excellent review, see Raghuram et al. (32)). The manner by which LH triggers chromatin folding is only transparent in the zigzag architecture, where LH forms rigid DNA stems that reduce the separation angle of entering and exiting DNAs and bring the nucleosome cores closer together (33). In the rigid DNA stem model, the linker histones are fixed to the nucleosome near its dyad position and establish contacts with the entering and exiting DNA linkers. However, fluorescent-recovery-after-photobleaching techniques showed that rather than remaining permanently fixed to their nucleosome cores, LH molecules tend to dissociate from their binding sites, diffuse away, and then rebind to another viable site (34–36). The binding affinity between LH molecules and nucleosome cores determines the fraction of time that LH molecules remain free or bound to the chromatin fiber. Low LH/core binding affinity produces high LH mobility and rapid diffusion rates, whereas high LH/core binding affinity is associated with low LH mobility and slow diffusion rates.
The LHs of higher organisms are made of three domains: a short N-terminal region, a central globular domain, and a highly charged C-terminal domain. The binding affinity of LH is cooperatively determined by H1's C-terminal domain and two binding sites located on opposite sites of H1's globular domain (36). Deletions of any of these three key binding elements reduce the binding affinity of LH, with deletion of the C-tail producing the strongest effect (36,37). In addition, several posttranslational modifications of the histones H1 and their nucleosome binding sites modulate LH's binding affinity (38). For example, both acetylation of core histones and phosphorylation of H1 decrease the binding affinity of LH to the nucleosome core and increase the concentration of free H1 (39,40). The binding affinity of LH decreases significantly when the NRL is shortened; H1 binding affinity is twice the value in 188-bp arrays compared to 161-bp species and negligible in 154-bp arrays (37). Furthermore, a correlation between the absence of DNA methylation and decreased LH mobility has been observed (41).
The complexity of LH's dynamic binding behavior and its effects on chromatin's unfolding extend further. Fluorescent-recovery-after-photobleaching experiments revealed that, within a single chromatin fiber, two different dynamic populations of LH coexist: a fast binding state (high LH binding affinity/fast rebinding rate) and a slow binding state (low LH binding affinity/slow rebinding rate with higher probability of diffusion away from the fiber) (42). These two LH populations could result from different phosphorylation conditions or different arrangements of the C-tail (32). How these two binding populations of LH affect chromatin fiber structure remains unknown.
Here we use our coarse-grained model of chromatin (see Fig. S1 in the Supporting Material) (12,13,43–46) to investigate systematically how different LH binding affinities alter the mechanism of chromatin unfolding. Specifically, we use modeling to mimic single-molecule stretching experiments and analyze the effects of NRL, LH concentration, and LH binding affinity in the unfolding mechanism of chromatin fibers in monovalent salt; separate studies for 209-bp fibers divalent conditions (R. Collepardo-Guevara and T. Schlick, in preparation) suggested the importance of LH mobility and heteromorphic unfolding intermediates. Prior model applications have clarified that the formation of extended fibers at low salt is driven by repulsion among DNA linkers, and that the secondary folding of chromatin at higher salt is determined by histone-tail mediated internucleosome interactions (44). We also suggested that H4 histone tails are the most important for internucleosomal interactions, while H3 tails are crucial for screening the electrostatic repulsion between entering/exiting DNA linkers (47).
Combined with cross-linking experiments and electromagnetic visualization in the Grigoryev lab, modeling has revealed that, in the presence of magnesium ions and LH, medium-NRL fibers adopt a heteromorphic structure with mostly zigzag features accented with a small percentage of bent linkers (11). Further studies showed that chromatin structure is sensitively affected by the NRL: short-NRL fibers form narrow two-start structures regardless of LH presence; medium-NRL fibers form compact zigzag structures in the presence of LH and looser zigzag and solenoid arrangements without LH; and long-NRL (>218 bp) fibers are more heteromorphic and thus less compact (13).
Here our analyses suggest that chromatin's unfolding mechanism and stiffness depend sensitively on the NRL and the binding affinity of LH. The stiffness and the detailed unfolding mechanism of medium-NRL fibers is determined by the kinetic characteristics of the LH/core bond. Indeed, whereas the presence of permanently fixed LH (maximum binding affinity) greatly stiffens the stretching response of medium-NRL arrays, dynamic LH (moderate binding affinity) yields softer fibers. Our results also show that the unfolding effects of fast and slow LH binding populations include different desirable characteristics for the control of DNA expression. The combined studies thus suggest that a medium NRL and the presence of multiple dynamic LH pools might operate together to facilitate selective DNA exposure at natural cellular pulling forces.
Materials and Methods
To elucidate the effect of the NRL and LH binding in the stretching resistance and unfolding mechanism of chromatin, we use Monte Carlo simulations of our mesoscale model to study 24-core oligonucleosomes with two different DNA linker lengths (173 bp and 209 bp) at high monovalent salt (0.15 M) and room temperature (293 K) under constant applied forces between 0 and 40 pN in the z-direction, as detailed below. The Supporting Material contains further information on the model, sampling approach, and analysis tools.
For each condition studied, we start trajectories from representative simulation structures for fibers with LH at zero pulling force. These initial structures have zigzag features and were obtained from converged simulations started from both idealized zigzag and solenoid forms (13). By construction, we do not consider the effects of nucleosome repositioning or nucleosome unwrapping during DNA extension; experiments have suggested that nucleosome unwrapping occurs at high forces (between 20 and 40 pN) (21).
Chromatin mesoscale model
Our mesoscale model (see Fig. S1) (11–13,43–45,47–50) treats the nucleosome core, histone tails, linker DNA, linker histone proteins, and physiological environment (water and ions in solution) with different coarse-grained strategies. Full details are provided in the Supporting Material and cited works.
In brief, each nucleosome, including the histone octamer without protruding tails and the 147 bp of DNA wound around it, is modeled as an irregularly-shaped electrostatic charged object through the discrete-charge-optimization algorithm (43,50), which defines 300 Debye-Hückel charges on the nucleosome crystal structure (Protein Data Bank entry 1KX5). Additionally, we attach 10 flexible histone tails to their idealized positions in the nucleosome crystal structure. Each histone tail is modeled as a flexible chain of charged beads with parameters that reproduce properties of the atomistic histone tails via Brownian dynamics simulations (45). Attached to each nucleosome, other than the first, are two DNA linkers (exiting and entering), which are modeled as elastic wormlike chains of spherical beads (51,52) (see Fig. S2 a). Each DNA linker bead has an internal force field comprising of stretching, bending, and twisting terms and carries a salt-dependent negative charge that mimics the electrostatic potential of linear DNA (53).
An LH molecule can be rigidly or dynamically attached to the nucleosome core in its dyad position. Our LH model has been developed based on the rat H1d LH structure of Bharath et al. (54,55) predicted by fold recognition and molecular modeling. From the three H1d domains, we neglect the short (33 residues) relatively uncharged N-terminal region. We model the central globular domain (76 residues) with one bead, and the highly charged C-terminal region (110 residues) with two beads (47). The charges of the three LH beads are approximated by fitting the full atomistic potential obtained by solving the nonlinear Poisson-Boltzmann equation through the discrete-charge-optimization algorithm (50). We have recently refined our LH model (R. Collepardo-Guevara and T. Schlick, in preparation) by introducing moderate bending and stretching flexibility to the LH beads through semistiff harmonic potentials, and by allowing the LH beads to interact electrostatically with all chromatin components. This approach permits the LH beads to adopt optimized positions near the dyad axis. We analyze the effects of LH concentration by varying the number of LH proteins (0 or 1) that are rigidly attached to each core a priori. Additionally, we model fast and slow LH dynamic binding/unbinding behavior by including a Monte Carlo move (LH on and off move), as developed in R. Collepardo-Guevara and T. Schlick (in preparation) and described below.
Our model treats the water around the oligonucleosome implicitly as a continuum with a dielectric constant of 80. The screening of electrostatic interactions due to the presence of monovalent ions in solution (0.15 M NaCl) is treated using a Debye-Hückel potential with an inverse Debye length at 0.15 M NaCl of 1.27 nm−1 (47). The charges assigned to the core, linker DNA, linker histone, and histone tails thus depend on the salt concentration. The different chromatin components interact electrostatically with one another. To prevent overlap between chromatin components, each component is assigned an effective excluded volume through a Lennard-Jones potential. As already discussed in Arya and Schlick (47), our mesoscale model neglects ion-ion correlations, specific protein interactions (hydrogen bonding), and solvation effects. Although charge-correlation effects are likely to be important in chromatin in a solution with a high concentration of monovalent salt and especially with divalent ions, an accurate description of such effects is currently computationally unfeasible for large oligonucleosomes because it would require explicit treatment of ions, and the determination of their positions within chromatin for each Monte Carlo configuration. Further model details, including specific expressions for the oligonucleosome energy and all parameters, are provided in the Supporting Material.
Monte Carlo simulations
Monte Carlo (MC) simulations with five different MC moves (global pivot; local translation; local rotation; tail regrowth; and LH reorganization) (47) (R. Collepardo-Guevara and T. Schlick, in preparation) are used to sample the ensemble of oligonucleosomes at constant temperature. In addition, dynamic LH binding/unbinding is simulated as developed in R. Collepardo-Guevara and T. Schlick (in preparation) by an optional MC move called the “LH on-and-off” move; this move proposes a trial conformation in which a randomly chosen LH is either detached (if bound) from its parent core with a probability pd, or reattached (otherwise) with a probability pa. The trial configuration is then accepted or rejected using the Metropolis criteria. The values of the dissociation-and-diffusion (pd) and association (pa) probabilities determine the relative LH binding affinities in our chromatin fibers. Here, we compare two cases:
-
1.
pa >> pd (i.e., pa =1 and pd = 0.25), which mimics a high binding affinity (rebinding occurs much faster than diffusion to infinity); and
-
2.
pa = pd (here pa = pd =1), which models a low binding affinity (diffusion away from the fiber or rebinding to another core are equally likely to occur).
Full details and the description of the other moves are presented in the Supporting Material.
Single-molecule stretching
To mimic the extension experiments, we fix the geometric center position of the first nucleosome core to its initial position and apply a constant force on the z-direction to the last nucleosome in the oligonucleosome chain (see Fig. S2 b). This is accomplished by adding an external stretching energy term, Epull, to the total oligonucleosome energy. The stretching energy term is calculated at each MC step as
| (1) |
where Fpull is the external stretching force, zk is the z-coordinate of core k, and Δz is the so-called end-to-end distance that measures the distance in the z-direction between the geometric centers of the first (k = 1) and last (k = Nc) cores.
Data collection
We simulate fibers with short (173 bp) and medium (209 bp) NRL. For NRL = 173 bp, we analyze fibers: 1), without LH; and 2), with one fixed LH/core. For NRL = 209 bp, we study fibers: 3), without LH; 4), with one fixed LH/core; 5), with 0.5 fixed LH/core; 6), with 0.25 fixed LH/core; 7), with dynamic LH in a fast binding mode; and 8), with dynamic LH in a slow binding mode. For each of the eight conditions, we run a set of simulations at pulling forces ranging between 0 pN and 20 pN, or between 0 pN and 40 pN, as required to achieve complete unfolding. Every experimental set (NRL, LH characteristics, and pulling force) includes 12 simulation trajectories that cover the mean DNA twist angle and two DNA twist deviations (±12°) from the mean twist to mimic natural variations, by four independent MC trajectories each, as done previously (13). Each simulation trajectory was run for up to 50 × 106 MC steps and the last 5 × 106 steps were used for statistical analysis. Convergence has been demonstrated previously by the behavior of the total energy and local descriptors of the fiber geometry (13). Additionally, here we demonstrate convergence well before 45 × 106 MC steps by the behavior of the end-to-end distance in Fig. S3.
Results
We analyze the effects of LH binding through force-extension curves combined with configurational snapshots at key points along the curve, internucleosome interaction plots, and electrostatic interaction energies. The so-called force-extension or force-distance (F-D) curve measures chromatin fiber extension under an applied force, and its slope characterizes the resistance of the fiber to unfolding.
LH has modest effect on unfolding for short-NRL fibers
Before we examine dynamic LH binding, we investigate the effect of NRL on the unfolding mechanism of chromatin fibers with and without LH. Short-NRL fibers exhibit simple ladderlike structures, in which the nucleosomes, by construction, only establish strong interactions with cores separated by one or two DNA linkers (i ± 1 and i ± 2). This simplicity makes interpretation of unfolding straightforward from the F-D curves and simulation snapshots. Fig. 1 compares the F-D behavior of short-NRL (173 bp) arrays without LH and with one LH molecule permanently attached to each core.
Figure 1.
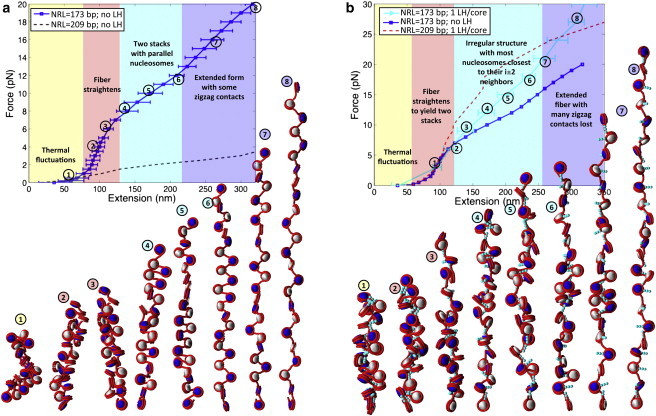
F-D curve of 24-unit 173-bp oligonucleosome chains at 0.15 M monovalent salt, and space-filling models based on MC stretching simulation snapshots of the same systems: (a) arrays without LH and (b) arrays with LH. (White and navy) Alternating nucleosomes. (Red) Wrapped DNA. (Turquoise) LH (see Fig. S1 in the Supporting Material). In the curve, different background colors and text describe the different stretching responses that the chromatin fiber exhibits as the pulling force increases, as revealed by the simulation snapshots. (Circled numbers) Relation of each snapshot with a point along the F-D curve.
The stretching elasticity modulus is defined as the force needed to produce a twofold extension from the equilibrium resting length (equilibrium length at zero pulling force) (20); we estimate the resting length of short-NRL fibers with/without LH to be ∼69 nm. The F-D curves reveal that the elasticity modulus of short-NRL fibers without LH is σ ∼ 8 pN, and only slightly larger, σ ∼ 9 pN, with fixed LH. This small change can be explained by inhibited DNA stem formation in short-NRL fibers, because the 173-bp linkers are shorter than LH, which roughly corresponds to the length of 30 bp (55). This behavior agrees with previous modeling that shows a tendency of short-NRL arrays to form equally compact zigzag conformations in the presence or absence of LH, as well as inability of short DNA linkers to form full DNA stems (13). Our observations are also consistent with recent experiments that show that short-NRL species have a reduced LH binding affinity because their DNA linkers are too short to establish critical interactions with the H1 C-tail; such interactions may induce folding of the H1 C-tail that is otherwise disordered (37).
A more interesting effect of LH's presence arises when comparing the unfolding simulation snapshots in Fig. 1. Two force regimes in the F-D curves are evident.
In the first regime (2–6 pN), both fibers behave similarly: the F-D curves have maximal slopes and the fibers straighten to maintain their ladderlike zigzag organization with the cores oriented with their largest surface area perpendicular to the fiber axis (13). A perpendicular orientation of nucleosomes facilitates intense internucleosome interactions and explains the strong resistance to stretching.
In the second force regime (above 6 pN), behavior is LH-dependent. Without LH, a decreased F-D slope (lower stretching resistance) coincides with formation of an extended zigzag array with the cores oriented parallel to the fiber axis; here the fiber opens in an accordion-like manner as the force increases. In this unfolding mechanism, all DNA regions become equally accessible, which may be favorable for binding of remodeling factors or template-directed mechanisms. In contrast, when LH is present, an uneven fiber opening occurs that makes some regions of the DNA more accessible than others. Given that short-NRL fibers are associated with smaller concentrations of LH (14), selective positioning of the few LH proteins along the fiber might thus help inhibit certain regions of the genome.
How does the chromatin unfolding process relate to changes in the stabilizing internucleosome interaction energy? Fig. 2 presents the average nucleosome/nucleosome electrostatic interaction energy per core, considering all nucleosomal and tail-bead charges, as a function of the pulling force. For all fibers, the strength of the stabilizing internucleosome interaction energy is maximal at zero pulling force, where the structure is most compact, and approaches zero as the force increases and the fiber unfolds. However, the actual magnitude of the zero-force internucleosome energy and the rate at which it weakens with the pulling force are different for each fiber structure.
Figure 2.
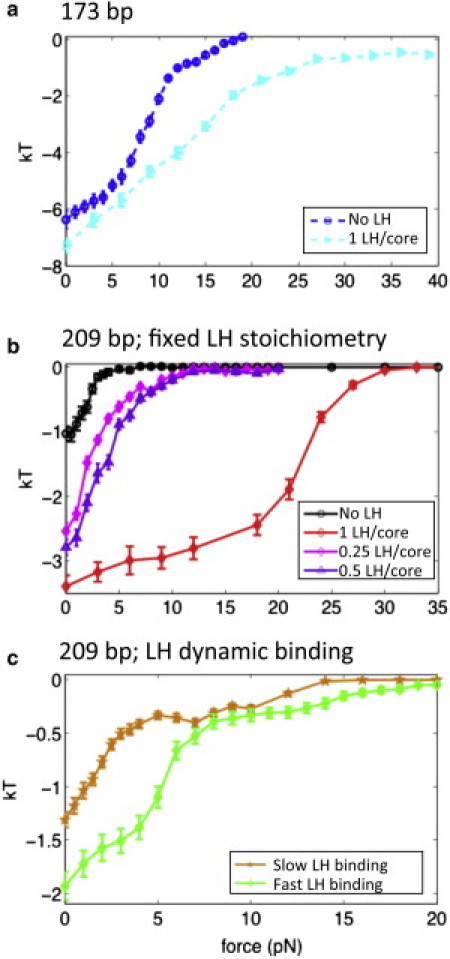
Internucleosome interaction energy per core for 24-unit oligonucleosome fibers with two different NRLs—173 bp (a) and 209 bp (b and c), at 0.15 M monovalent salt. The figures are for no LH, 1 LH/core, 0.5 LH/core, 0.25 LH/core, and dynamic LH (fast and slow binding).
As expected from their stretching responses, the internucleosome interaction energy of short-NRL fibers (Fig. 2 a) is only mildly affected by presence of LH. At zero pulling force, short-NRL fibers with and without LH are both stabilized by a strong internucleosome interaction energy (∼6 kBT and ∼7 kBT for fibers without and with LH, respectively), which agrees with their compact ladderlike structures with stacks of perpendicular cores. After the perpendicular-to-parallel nucleosome orientation transition at 6 pN, the internucleosome energy of fibers without LH decreases more rapidly than that of fibers with LH, in agreement with LH's small stiffening effect revealed above. Hence, short-NRL fibers are stiff because their folded state is stabilized by intense internucleosome interactions among parallel nucleosomes, and their short linkers limit nucleosome reorientation to stable, extended arrays.
Fixed LH binding frustrates medium-NRL chromatin fiber unfolding
In Fig. 3 we examine the stretching behavior of medium-NRL (209 bp) chromatin without LH and with one LH permanently fixed to each nucleosome core at its dyad position; Fig. S4 considers additional LH/core ratios (0.5 LH/core and 0.25 LH/core). Corresponding internucleosome interaction energies and patterns (see the Supporting Material) are shown in Fig. 2 b and Fig. S5, a–c.
Figure 3.
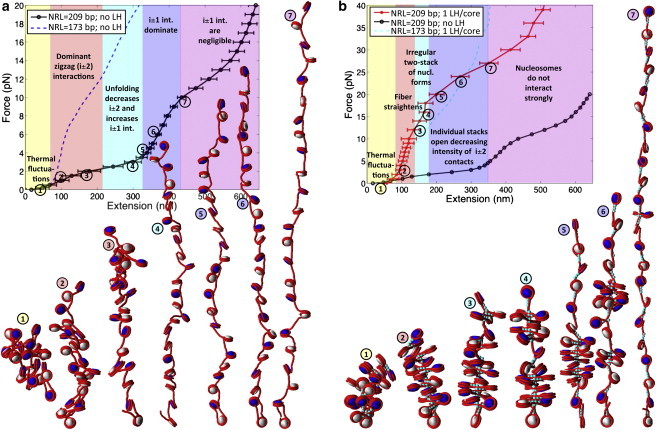
F-D curve of 24-unit 209-bp oligonucleosome chains at 0.15 M monovalent salt, and space-filling models based on MC stretching simulation snapshots of the same systems: (a) arrays without LH and (b) arrays with one permanently fixed LH/core. (White and navy) Alternating nucleosomes. (Red) Wrapped DNA. (Turquoise) LH (see Fig. S1). In the curve, different background colors and text describe the different stretching responses that the chromatin fiber exhibits as the pulling force increases, as revealed by the simulation snapshots. (Circled numbers) Relation of each snapshot with a point along the F-D curve.
Overall, the analyses reveal that the unfolding response of medium-NRL fibers is strongly influenced by the presence of LH. Fibers without LH have a resting length of ∼74 nm and an elasticity modulus of 2 pN. Fibers with one fixed LH/core—with resting length of ∼68 nm—have an elasticity modulus of σ ∼ 10 pN, five times higher!
The unfolding mechanism is also highly influenced by the presence/absence of rigidly fixed LH. Medium-NRL arrays without LH exhibit an irregular unfolding behavior. Below 4 pN, the F-D curve is linear and has its minimal slope; a significant extension thus results from a small force increase. Here, the fiber forms irregular conformations in which the nucleosomes are perpendicular to the fiber axis and mainly interact with their i ± 2 neighbors. At ∼4 pN, an increased F-D slope signals a structural transition. A similar change in slope was previously reported by Cui and Bustamante (20) in the presence of LH and a moderate ionic concentration. Our snapshots (Fig. 3) and force-dependent internucleosome interaction patterns (see Fig. S5 a) clarify that this change corresponds to the transition into an array of perpendicular nucleosomes that resembles a 10-nm string with dominant i ± 1 interactions (single-stack conformation). As for the short-NRL case (Fig. 1), the perpendicular orientation of nucleosomes is responsible for the enhanced fiber rigidity. The force we predict for this transition is consistent with previous single-molecule stretching experiments of medium-NRL chromatin fibers, which reported that forces <5 pN trigger a transition from the 30-nm fiber into a 10-nm string (20,25). Above 10 pN, the fiber adopts a beads-on-a-string form, and the intensity of i ± 1 interactions decreases; here the curve is no longer linear due to overstretching of the DNA. Our model cannot be used in this regime where nucleosome unwrapping is expected to occur before DNA overstretching.
Unfolding of fibers with one fixed LH/core is determined by the presence of rigid DNA stems at every nucleosome position. Below 20 pN, these stems remain unperturbed, which constrains the entering and exiting DNA linkers and stabilizes a compact zigzag organization. At forces above 20 pN, a change in slope in the F-D curve coincides with the partial disruption of individual DNA stems and formation of interesting heteromorphic conformations, that we term superbeads-on-a-string. These heteromorphic structures combine extended fiber sections, in which the DNA is fully exposed, with superbeads or compact clumps, in which the DNA is inaccessible to the cellular machinery. These superbeads have strong i ± 2 contacts (see Fig. S5 b) and are stabilized by the presence of LH proteins that screen the repulsion among DNA linkers. The fiber eventually forms a fully extended string at ∼32 pN. As shown in Fig. S4 and Fig. S5 c, lower LH stoichiometries decrease significantly the stiffness of medium-NRL fibers and stabilize two-stack unfolding conformations with a limited presence of superbeads.
The internucleosome interaction energy for medium-NRL fibers with 1 LH/core, 0.5 LH/core, 0.25 LH/core, and without LH is ∼3.4, ∼2.7, ∼2.5, ∼1 kBT, respectively (Fig. 2 b); such values are consistent with the value (∼3 kBT) measured experimentally for medium-NRL fibers with LH in the absence of magnesium ions (20). The decrease of this energy with a reduced LH/core ratio underscores the significant compaction effect of LH for medium-NRL fibers. A higher LH concentration also reduces the slope at which this energy weakens with the pulling force, which proposes a correlation between fiber stiffness and the number of rigid DNA stems in fiber.
Hence, the unfolding mechanism of medium-NRL fibers is determined by interactions between the DNA and LH. This is consistent with other modeling (12,13) and experimental (15) studies that demonstrate that a notable compaction effect of LH occurs for medium-NRL fibers.
Fast and slow LH dynamic pools cooperate to facilitate fiber opening via heteromorphic intermediates
Capturing the dynamic nature of LH is crucial for a thorough understanding of LH's effects in the unfolding mechanism of chromatin. H1 molecules participate in both fast and slow binding interactions, with ∼28% of H1 molecules within a chromatin fiber binding dynamically in a slow state, and ∼71% in a fast state (36).
We now analyze medium-NRL arrays with LH that exhibit two different rebinding scenarios: 1) high LH/core affinity, where unbinding is followed by fast hopping and rebinding; and 2) low LH/core binding affinity, where LH molecules have the same probability of rebinding as diffusing to infinity. Our choice of parameters produces equilibrium ensembles of oligonucleosome conformations where fast binding corresponds to ∼0.8 LH/core and slow binding to ∼0.5 LH/core on average (see Monte Carlo Sampling in the Supporting Material).
As expected, dynamic LH binding dramatically reduces the magnitude of the forces required to stretch medium-NRL fibers and also affects their detailed unfolding mechanism (Fig. 4). The strong softening effect of slow dynamic LH binding is caused by the destabilization of the rigid DNA stems, and the subsequent local and temporary increase of the DNA/DNA repulsion. This DNA stem destabilization makes fibers with dynamic LH only slightly stiffer than fibers without LH, and much softer than fibers with one fixed LH/core.
Figure 4.
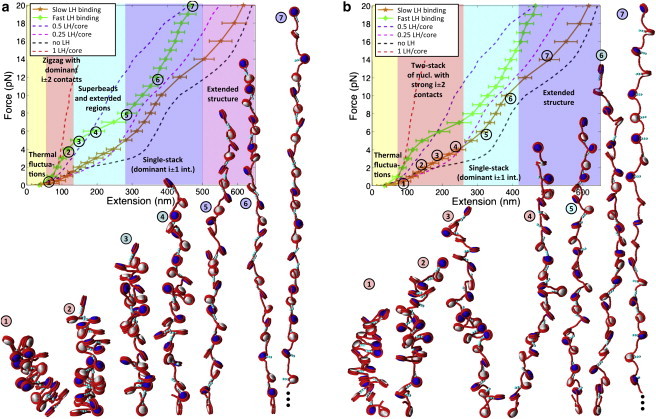
F-D curve of 209-bp 24-unit oligonucleosome chains at 0.15 M monovalent salt with: (a) fast and (b) slow LH binding. Space-filling models based on the same MC stretching simulations. (White and navy) Alternating nucleosomes. (Red) Wrapped DNA. (Turquoise) LH (see Fig. S1). In the curve, different background colors and text describe the different stretching responses that the chromatin fiber exhibits as the pulling force increases, as revealed by the simulation snapshots. (Circled numbers) Relation of each snapshot with a point along the F-D curve.
The softening effect due to dynamic LH binding/unbinding, compared to fixed LH, decreases as the LH/core binding affinity increases: the elasticity modulus of fibers with fast and slow LH binding populations are reduced to σ ∼ 5 pN and σ ∼ 2 pN, respectively, from ∼10 pN for 1 fixed LH/core fibers. As mentioned above, fibers with a fast LH binding population exhibit a 30% higher average concentration of LH molecules than their slow LH binding counterparts. This suggests that fast LH binding increases the lifetime and concentration of rigid DNA stems and thus the overall stiffness of the chromatin fiber. Despite the increasing amount of single-molecule experimental and simulation data for the chromatin fiber, side-by-side comparisons are not straightforward given the different experimental conditions examined in each study. However, in Fig. S6 we compare our results to those reported by Cui and Bustamante (20). Both studies were performed for medium-NRL fibers (24-core 209-bp in our work versus 280-core 210-bp in the experiment), with LH, and at monovalent salt only (0.15 M NaCl in our work versus 0.04 M NaCl in the experiment). Because Cui and Bustamante report curves corresponding to a conserved ratio of ∼1 LH/core, the comparison is made with our fixed LH and high LH/core affinity cases. Despite the differences in the conditions, the figure confirms that our simulated curves lie within the range of the experimental release and stretch curves.
The fast and slow dynamic LH binding scenarios produce different unfolding conformations. Fibers with fast LH binding preserve a compact zigzag architecture up to 4 pN (see Fig. S5 d), and within 4–8 pN, they unfold via the superbeads-on-a-string arrays that expose the linker DNA material selectively. The superbeads are not as pronounced as those in fibers with fixed LH, or fibers with dynamic LH and divalent ions (analyzed in R. Collepardo-Guevara and T. Schlick, in preparation); this is due to the decreased screening of the DNA by LH in monovalent salt compared to divalent ion conditions. Above 8 pN, the superbeads extend, yielding a single-stack conformation with dominant i ± 1 contacts.
Fibers with a slow LH rebinding population form unfolding intermediates with only a marginal presence of superbeads below 3 pN; the low LH binding affinity further decreases the screening of DNA repulsion and destabilizes the compact superbead structures. Above 3 pN, the unfolding stabilizes a single-stack conformation with strong i ± 1 interactions (see Fig. S5 e). Above 15 pN, an extended beads-on-a-string conformation forms.
Note that on its own, the F-D curve of fibers with a slow LH rebinding population does not reveal the two-stack to single-stack structural transition; the conserved slope above 3 pN indicates that the F-D curve is not sufficient to dissect all structural changes during fiber unfolding. Details of these configurations, however, are suggested by our snapshots.
The maximal strength of the internucleosome interaction energy is lower for the slow than the fast LH binding cases: ∼1.4 versus ∼1.9 kBT (Fig. 2 c), and both values are small compared to fibers with fixed LH, ∼3.4 kBT. Dynamic LH binding/unbinding thus promotes unfolding, with slow rebinding after dissociation enhancing this effect.
Therefore, slower LH binding might be crucial to reduce the forces needed to unfold the chromatin fiber and make them consistent with the stalling forces of molecular motors that operate on chromatin, whereas fast LH binding stabilizes the superbead constructs. The latter might have an important biological role: the enhancement of screening of DNA repulsion promotes compact clumps in the superbeads-on-a-string structures.
Discussion and Conclusions
Our simulations of 24-unit oligonucleosomes subject to pulling forces with different variations of LH content and binding affinity (fast and slow binding) have revealed the following:
-
1.
Fiber stiffening factors include short NRL (due to hampered fiber reorganization), permanently fixed LH (due to rigid stem formation), increased number of LH (due to higher stem presence), and higher LH binding affinity (due to stem stabilization).
-
2.
Correspondingly, fiber softening factors include longer NRL (due to more facile stacking rearrangements and enhanced flexibility), reduced LH concentration (due to inhibited DNA stem formation), and slower dynamic LH binding rates (due to DNA stem destabilization).
-
3.
Without LH, short-NRL fibers unfold in a regular accordion-like manner and medium-NRL arrays follow a disordered unfolding pattern.
-
4.
Medium-NRL fibers with rigidly fixed LH or a fast dynamic LH population form superbeads-on-a-string unfolding intermediates that combine fully extended regions with compact clumps.
-
5.
Not all the structural transitions that occur during chromatin fiber unfolding alter the shape of the F-D curve; notably, the transition into a one-stack conformation in fibers with slow dynamic LH conserves the slope of the F-D curve and is only revealed by the complementary modeling analysis.
Important biological implications emerge from these observations. Although short-NRL fibers make chromatin unfolding more challenging for molecular motors, once their unfolding force is reached, they expose most of their DNA simultaneously to the transcription and replication machinery. This feature might be advantageous for simple organisms with short life spans, as it might facilitate access to all protein sequences and therefore allow higher reproduction rates (13). The low sensitivity to LH addition/depletion in the stiffness of short-NRL fibers agrees with various modeling (12,13) and in vitro experimental studies (15) that demonstrate that the structure and compaction of short-NRL fibers is not significantly altered by LH addition. For example, experiments also show that LH reduction causes minor phenotypical changes in simple organisms (56,57), and that short DNA linkers cannot establish critical interactions with the H1 C-tail and that this effect reduces LH/core binding affinity (37). Despite this low sensitivity, our work suggests that LH addition to short-NRL fibers has a subtle mechanistic effect: introduction of irregularity in the DNA exposure. Thus, although LH does not play an important structural role in short-NRL fibers, it might restrict access to certain DNA regions when the fiber opens.
Our data also suggest that LH dynamic behavior is important for regulation of chromatin unfolding and selective DNA expression. Fixed LH-core binding produces rigid DNA stems that render medium-NRL fibers too stiff to be accessible to the cellular machinery. That is, the 10 pN required to produce a twofold extension for one fixed LH/core fibers is higher than typical forces of some eukaryotic molecular machines, such as RNA polymerase II, which ceases to transcribe at 7.5 ± 2 pN (compared to 35 pN for its prokaryotic counterpart) (58). In contrast, dynamic LH binding/unbinding destabilizes the rigid DNA stems and strongly softens the chromatin fiber, reducing the force needed to initiate fiber unfolding by a factor of up to 5.
Desirable effects for gene regulation were revealed for both fast and slow LH dynamic binding populations. Whereas slow binding populations reduce fiber stiffness to allow DNA access by normal molecular motor forces (a twofold extension is now reached at only ∼2 pN), fast LH binding promotes superbead constructs that might facilitate selective DNA exposure. In addition, these heteromorphic unfolding conformations might represent transition states between different fiber forms. Similar superbead forms have been visualized experimentally in digested medium-NRL chromatin fibers with LH (59,60), which adopt partially unfolded conformations. This suggests that the fast and slow LH binding pools, observed simultaneously in chromatin in vivo (42), might act cooperatively to yield controlled chromatin fiber unfolding at low forces. Thus, medium-NRL fibers with multiple dynamic LH pools might be evolutionarily suitable for gene regulation because they offer both flexibility and selective DNA exposure.
During unfolding, the mechanical constraints imposed by the pulling machinery may contribute to the global destabilization of the LH-core bound state (23), increasing the pool of LH molecules that follow slow rebinding. This effect suggests an additional way in which the simultaneous presence of fast and slow LH binding states collaborate to allow optimal DNA storage and accessibility: at zero pulling force, the high LH-core binding affinity state might be favored, yielding a compact chromatin array ideal for DNA storage; under normal stretching forces, LH molecules might exhibit a reduced binding affinity and favor a more open chromatin structure required for facile DNA access.
Through changes in the structure of LH, posttranslational modifications of the LH-core binding sites, or the presence of competing binding factors, the cellular machinery could fine-tune the rates of binding of LH and induce unfolding of certain chromatin regions over others. In particular, it has been suggested that phosphorylation of the C-terminal tail of LH (40), hyperacetylation of the N-terminal tails of core histones, and the presence of HMG binding factor reduce the binding rate of H1 to the core (39,61,62). This great variety of mechanisms to control the relative presence of pools of fast and slow LH binding molecules can determine chromatin's fiber propensity to opening under tension as well as details of resulting unfolding mechanisms. Further single-molecule experimental and modeling studies that explore the effects of covalent modifications to LH and its binding sites in the stretching response of chromatin will undoubtedly help provide additional information regarding such processes. Model improvements, for example to represent H1 in greater resolution, can also be envisioned.
Finally, our results help associate trends observed in experimental F-D curves with structural transitions and rearrangements. The modeling snapshots and internucleosome interaction patterns at different unfolding stages reveal that zigzag fibers can form both single-stack and two-stack structures upon unfolding; therefore, a single-stack unfolding intermediate is not evidence for an initial solenoid structure as suggested previously (25). In addition, whereas a compact solenoid architecture would require a high internucleosome interaction energy (∼14 kBT (25)) to counteract the strong DNA-DNA repulsion caused by high linker DNA bending, our most compact chromatin fibers are maintained by weaker internucleosome energies (∼7 kBT for short-NRL fibers with one fixed LH/core, and ∼3.4 kBT for the medium-NRL counterparts). Such values are adequate for two-start structures with straight DNA linker and moderate DNA-DNA repulsion, and consistent with the 3 kBT value of Cui and Bustamante (20).
Overall, the results presented here highlight the complex role of LH and the intricate network of interactions it triggers and impacts during chromatin fiber unfolding. Control of the dynamic nature of LH emerges as a straightforward mechanism for modulation of chromatin fiber opening. We suggest an expansion from the traditional view of LH as a repressor for gene expression to a regulator of different dynamic LH binding modes for facilitating selective chromatin unfolding. The studies also underscore the importance of considering the highly dynamic nature of chromatin to understand gene regulation. Together, local and global changes in the binding states and concentrations of LH molecules and the values of the NRL produce strong variations in chromatin's fiber stiffness, its unfolding mechanism, and DNA accessibility.
Acknowledgments
We are grateful to Dr. Grigoryev for his insightful comments. Computing support from the New York University HPC USQ and Cardiac Clusters is acknowledged.
This work was supported by National Science Foundation grant MCB-0316771 and National Institutes of Health grant R01 GM55164 to T.S. Acknowledgment is also made to the donors of the American Chemical Society (award PRF39225-AC4) Petroleum Research Fund and Philip Morris USA, and to Philip Morris International. R.C.-G. gratefully acknowledges funding from the Schlumberger Faculty for the Future Program.
Supporting Material
References
- 1.van Holde K. Springer-Verlag; New York: 1988. Chromatin. [Google Scholar]
- 2.Finch J.T., Lutter L.C., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 3.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 5.Chodaparambil J.V., Barbera A.J., Luger K. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodcock C.L. Chromatin architecture. Curr. Opin. Struct. Biol. 2006;16:213–220. doi: 10.1016/j.sbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.van Holde K., Zlatanova J. Chromatin fiber structure: where is the problem now? Semin. Cell Dev. Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Finch J.T., Klug A. Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc. Natl. Acad. Sci. USA. 1981;78:1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodcock C.L., Frado L.L., Rattner J.B. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J. Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlick T., Perisić O. Mesoscale simulations of two nucleosome-repeat length oligonucleosomes. Phys. Chem. Chem. Phys. 2009;11:10729–10737. doi: 10.1039/b918629h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perišić O., Collepardo-Guevara R., Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 15.Routh A., Sandin S., Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien F.-T., van Noort J. 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr. Pharm. Biotechnol. 2009;10:474–485. doi: 10.2174/138920109788922128. [DOI] [PubMed] [Google Scholar]
- 17.Lavelle C., Victor J.-M., Zlatanova J. Chromatin fiber dynamics under tension and torsion. Int. J. Mol. Sci. 2010;11:1557–1579. doi: 10.3390/ijms11041557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bustamante C., Cheng W., Mejia Y.X. Revisiting the central dogma one molecule at a time. Cell. 2011;144:480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leuba S.H., Yang G., Bustamante C. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. USA. 1994;91:11621–11625. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y., Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 2000;97:127–132. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennink M.L., Leuba S.H., Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 22.Brower-Toland B.D., Smith C.L., Wang M.D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pope L.H., Bennink M.L., Marko J.F. Single chromatin fiber stretching reveals physically distinct populations of disassembly events. Biophys. J. 2005;88:3572–3583. doi: 10.1529/biophysj.104.053074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roopa T., Shivashankar G.V. Direct measurement of local chromatin fluidity using optical trap modulation force spectroscopy. Biophys. J. 2006;91:4632–4637. doi: 10.1529/biophysj.106.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruithof M., Chien F.-T., van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 26.Katritch V., Bustamante C., Olson W.K. Pulling chromatin fibers: computer simulations of direct physical micromanipulations. J. Mol. Biol. 2000;295:29–40. doi: 10.1006/jmbi.1999.3021. [DOI] [PubMed] [Google Scholar]
- 27.Schiessel H., Gelbart W.M., Bruinsma R. DNA folding: structural and mechanical properties of the two-angle model for chromatin. Biophys. J. 2001;80:1940–1956. doi: 10.1016/S0006-3495(01)76164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Haïm E., Lesne A., Victor J.-M. Chromatin: a tunable spring at work inside chromosomes. Phys. Rev. E. 2001;64:051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 29.Mergell B., Everaers R., Schiessel H. Nucleosome interactions in chromatin: fiber stiffening and hairpin formation. Phys. Rev. E. 2004;70:011915. doi: 10.1103/PhysRevE.70.011915. [DOI] [PubMed] [Google Scholar]
- 30.Aumann F., Lankas F., Langowski J. Monte Carlo simulation of chromatin stretching. Phys. Rev. E. 2006;73:041927. doi: 10.1103/PhysRevE.73.041927. [DOI] [PubMed] [Google Scholar]
- 31.Stehr R., Kepper N., Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys. J. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghuram N., Carrero G., Hendzel M.J. Molecular dynamics of histone H1. Biochem. Cell Biol. 2009;87:189–206. doi: 10.1139/O08-127. [DOI] [PubMed] [Google Scholar]
- 33.Bednar J., Horowitz R.A., Woodcock C.L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misteli T., Gunjan A., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 35.Lever M.A., Th'ng J.P., Hendzel M.J. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 36.Stasevich T.J., Mueller F., McNally J.G. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 2010;29:1225–1234. doi: 10.1038/emboj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caterino T.L., Fang H., Hayes J.J. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl terminal domain. Mol. Cell Biol. 2011 doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 39.Perry C.A., Annunziato A.T. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp. Cell Res. 1991;196:337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]
- 40.Dou Y., Bowen J., Gorovsky M.A. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 2002;158:1161–1170. doi: 10.1083/jcb.200202131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert N., Thomson I., Bickmore W.A. DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell Biol. 2007;177:401–411. doi: 10.1083/jcb.200607133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrero G., Crawford E., de Vries G. Characterizing fluorescence recovery curves for nuclear proteins undergoing binding events. Bull. Math. Biol. 2004;66:1515–1545. doi: 10.1016/j.bulm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q., Beard D.A., Schlick T. Constructing irregular surfaces to enclose macromolecular complexes for mesoscale modeling using the discrete surface charge optimization (DiSCO) algorithm. J. Comput. Chem. 2003;24:2063–2074. doi: 10.1002/jcc.10337. [DOI] [PubMed] [Google Scholar]
- 44.Arya G., Zhang Q., Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arya G., Schlick T. Efficient global biopolymer sampling with end-transfer configurational bias Monte Carlo. J. Chem. Phys. 2007;126:044107. doi: 10.1063/1.2428305. [DOI] [PubMed] [Google Scholar]
- 47.Arya G., Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reference deleted in proof.
- 49.Beard D.A., Schlick T. Computational modeling predicts the structure and dynamics of chromatin fiber. Structure. 2001;9:105–114. doi: 10.1016/s0969-2126(01)00572-x. [DOI] [PubMed] [Google Scholar]
- 50.Beard D.A., Schlick T. Modeling salt-mediated electrostatics of macromolecules: the discrete surface charge optimization algorithm and its application to the nucleosome. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 51.Allison S., Austin R., Hogan M. Bending and twisting dynamics of short linear DNAs. Analysis of the triplet anisotropy decay of a 209 base pair fragment by Brownian simulation. J. Chem. Phys. 1989;90:3843–3854. [Google Scholar]
- 52.Heath P.J., Gebe J.A., Schurr J.M. Comparison of analytical theory with Brownian dynamics simulations for small linear and circular DNAs. Macromolecules. 1996;29:3583–3596. [Google Scholar]
- 53.Stigter D. Interactions of highly charged colloidal cylinders with applications to double-stranded. Biopolymers. 1977;16:1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- 54.Bharath M.M., Chandra N.R., Rao M.R. Prediction of an HMG-box fold in the C-terminal domain of histone H1: insights into its role in DNA condensation. Proteins. 2002;49:71–81. doi: 10.1002/prot.10204. [DOI] [PubMed] [Google Scholar]
- 55.Bharath M.M., Chandra N.R., Rao M.R.S. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 2003;31:4264–4274. doi: 10.1093/nar/gkg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen X., Yu L., Gorovsky M.A. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 57.Shen X., Gorovsky M.A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 58.Galburt E.A., Grill S.W., Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 59.Zentgraf H., Franke W.W. Differences of supranucleosomal organization in different kinds of chromatin: cell type-specific globular subunits containing different numbers of nucleosomes. J. Cell Biol. 1984;99:272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caño S., Caravaca J.M., Daban J.R. Highly compact folding of chromatin induced by cellular cation concentrations. Evidence from atomic force microscopy studies in aqueous solution. Eur. Biophys. J. 2006;35:495–501. doi: 10.1007/s00249-006-0057-7. [DOI] [PubMed] [Google Scholar]
- 61.Rao J., Bhattacharya D., Shivashankar G.V. Trichostatin-A induces differential changes in histone protein dynamics and expression in HeLa cells. Biochem. Biophys. Res. Commun. 2007;363:263–268. doi: 10.1016/j.bbrc.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 62.Catez F., Brown D.T., Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


