Abstract
Observations of the interior structure of cells and subcellular organelles are important steps in unraveling organelle functions. Microscopy using helium ions can play a major role in both surface and subcellular imaging because it can provide subnanometer resolutions at the cell surface for slow helium ions, and fast helium ions can penetrate cells without a significant loss of resolution. Slow (e.g., 10–50 keV) helium ion beams can now be focused to subnanometer dimensions (∼0.25 nm), and keV helium ion microscopy can be used to image the surfaces of cells at high resolutions. Because of the ease of neutralizing the sample charge using a flood electron beam, surface charging effects are minimal and therefore cell surfaces can be imaged without the need for a conducting metallic coating. Fast (MeV) helium ions maintain a straight path as they pass through a cell. Along the ion trajectory, the helium ion undergoes multiple electron collisions, and for each collision a small amount of energy is lost to the scattered electron. By measuring the total energy loss of each MeV helium ion as it passes through the cell, we can construct an energy-loss image that is representative of the mass distribution of the cell. This work paves the way to use ions for whole-cell investigations at nanometer resolutions through structural, elemental (via nuclear elastic backscattering), and fluorescence (via ion induced fluorescence) imaging.
Introduction
For many years, subcellular components, such as mesoscopic-scale structures buried deep inside a cell, were practically invisible to biologists because of the lack of microscopy techniques with the ability to achieve nanometer resolution while maintaining high spatial resolution through the entire depth of the cell. Recently developed superresolution optical imaging techniques, including photoactivated localization microscopy (1,2), stochastic optical reconstruction microscopy (3,4), stimulated emission depletion (5), and saturated structured-illumination microscopy (6), have overcome the optical diffraction limit and transformed the study of cellular ultrastructure with light microscopy. However, these methods are based on fluorescent techniques, which often have stringent requirements that greatly limit their application. As a result, electron microscopy is still the most widely used technique for high-resolution imaging of subcellular structure, and transmission electron microscopy has already enabled scientists to image thin sections of cells at subnanometer resolution (7,8). However, as a result of the depth-profiling problems of electron microscopy, where multiple scattering is a fundamental limitation to high resolution in thick samples, whole-cell imaging is rarely carried out (8–10). Because it provides deep penetration, x-ray microscopy can overcome this limit, and several groups have obtained whole-cell soft-x-ray structural images with a spatial resolution below 100 nm (11–13). In addition, three-dimensional (3D) soft-x-ray tomography has been suggested as a complementary approach to electron microscopy for observations of whole cells, although it remains to be seen whether high-contrast images can be achieved (14).
Materials and Methods
Sample preparation
Human fetal liver cells (15) were seeded onto silicon nitride windows (100 nm thick) at a density of 10,000 cells/cm2 and allowed to attach for 24 h before fixation in 4% formaldehyde. Samples were then dehydrated through an ethanol gradient followed by critical point drying (16).
Helium ion microscopy: slow helium ions
The helium ion microscope (Orion; Carl Zeiss, Oberkochen, Germany) is based on a single atom field ion emission tip with high helium ion beam brightness (B > 4 × 109 A/cm2.sr) and a very low ion energy spread (<1 eV). Helium ion microscopy (HIM) utilizes electrostatic optics and is capable of achieving subnanometer resolution at the sample surface for helium ion energies of up to 50 keV. The imaging mechanism involves the collection of secondary electrons from a small ionization volume at the sample surface induced by the helium ions. Depending on the material studied, several secondary electrons are generated for each incoming helium ion. The HIM images shown in Fig. 4, a and d, were taken with a 45 keV ion energy and a spatial resolution of ∼5 nm. The images were formed from the detection of induced secondary electrons and constructed as a 2048 × 2048 pixel array.
Figure 4.
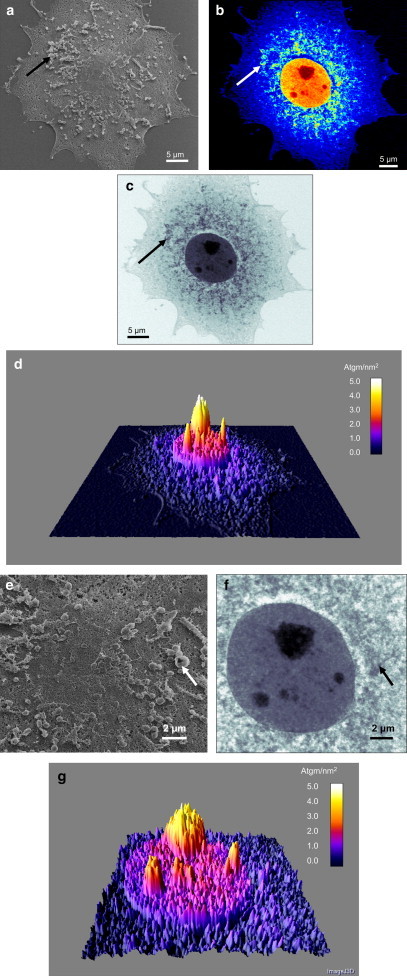
(a) HIM secondary electron image of a human liver cell, showing surface features. Helium ion energy = 45 keV. (b) Helium ion transmission energy-loss images of the same cell, showing structural features common to the surface as well as structural features from within the cell. Helium ion energy = 1.2 MeV. (c) Same image as in b, this time using grayscale to enhance the 3D aspect of the cell. Filamentary structures within the cell, as well as the nucleus, can be observed. (d) Mass image showing a 3D plot of the mass distribution (in units ag/nm2): [1 ag = 10−18 gm]. Helium ion energy = 1.2 MeV. The arrows in a–c correspond to a surface feature that has high contrast in the surface image but exhibits a low-contrast circular structure in the transmission image, implying a hollow structure. (e–g) Higher-magnification images from the region of the cell containing the nucleus. The arrows in e and f correspond to a surface feature that has high contrast in the surface image but has a ring-like structure in the transmission image, once again implying a hollow structure.
HIM: fast helium ions
For imaging of cells with fast helium ions, we used the cell imaging beamline facility (17) at the Centre for Ion Beam Applications (Department of Physics, National University of Singapore). The MeV helium ion beams were produced by a high-brightness High Voltage Engineering Europa Singletron ion accelerator, and focused with the use of a spaced triplet of compact Oxford Microbeams OM52 magnetic quadrupole lenses. We scanned the 1.2 MeV helium ion beam over the sample using electrostatic deflection, and measured the energy-loss signal using a standard silicon particle detector positioned behind the cell. The data were collected and processed via the IONDAQ data acquisition system (17). We recorded the transmission energy-loss images using list mode event-by-event data acquisition, and constructed a 1024 × 1024 pixel array. For each pixel the energies of exactly 15 ions were recorded, and the median energy of these 15 ions was used to construct the final image, thereby reducing statistical noise in the image. In the images depicted in Fig. 4, c and f, dark gray represents the lowest median energy or highest energy loss, whereas light gray represents the highest median energy and thus the lowest energy loss. The median-energy data were also used in the mass plots (see Fig. 4, d and g) in which the mass distribution is color-coded: the highest mass, corresponding to the highest energy loss, is in white, whereas the lowest mass, corresponding to the lowest energy loss, is colored dark blue. We plotted the color-coded mass image as a 3D contour map using the software package ImageJ.
Results and Discussion
To obtain high-resolution images of a cell with any scanning beam microscope, one must be able to produce a sufficiently small probe that can maintain its size as it penetrates the cell, and measure signals emanating from a localized compact region within the sample. In this work, we investigated the use of helium ions as a suitable probe for elucidating cell structure. Fig. 1 gives an indication of the interaction of slow and fast helium ions with respect to a cell.
Figure 1.
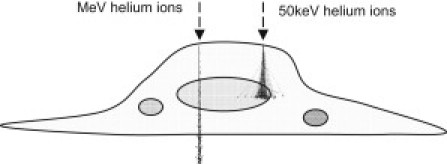
Paths of fast (e.g., ∼MeV) and slow (e.g., ∼50 keV) helium ions through a cell of nominal thickness (1 μm). The ions initially travel in straight lines, losing energy via multiple electron collisions. Toward the end of the range, nuclear collisions and hence large-angle scattering become predominant.
For the case of an MeV ion beam, energy losses are due mainly to multiple electron collisions, and while the energy remains high, there is minimal scattering. Because MeV ions pass through the cell, nuclear collisions are rare and large-angle scattering is minimal. Therefore, images that are formed by detecting the energy loss of transmitted ions do not suffer any significant deterioration in spatial resolution.
In the case of low-energy HIM, the ions do not have the range to pass through the cell, resulting in large-angle scattering within the cell. However, because images are formed by detecting secondary electrons originating near the cell surface, the surface image quality is not affected.
Because of the much smaller de Broglie wavelength, in theory, helium ions can be focused to a much smaller size than either electrons or x-rays. Slow (e.g., 10–50 keV) helium ion beams can now be focused to subnanometer dimensions (∼0.25 nm), and can be used to image the surface of samples by detecting induced secondary electrons originating from a region localized near the surface (<1 nm depth). In this energy range, HIM offers significant advantages compared with electron microscopy for surface imaging, because one can image insulating samples by neutralizing the surface charge using a flood electron gun, thereby eliminating the need for a conducting metallic coating on the sample. Further, the shallow escape depth of the low-energy secondary electrons makes the images extremely surface-sensitive (18–20). The detection of backscattered ions can also be used for image formation, because the backscattered energy is sensitive to the atomic number (Z) of the target atom in the cell. Fast helium ions (e.g., 1–2 MeV) can also be focused to nanodimensions, and because of the increased energy and high mass of the helium ion, this resolution is maintained during penetration of whole cells.
Fig. 2 shows Monte Carlo simulations of fast helium ion trajectories through organic material. If the nominal thickness of a dehydrated cell is taken to be 1 μm, then MeV ions can pass through the cell with most ions maintaining a straight trajectory.
Figure 2.
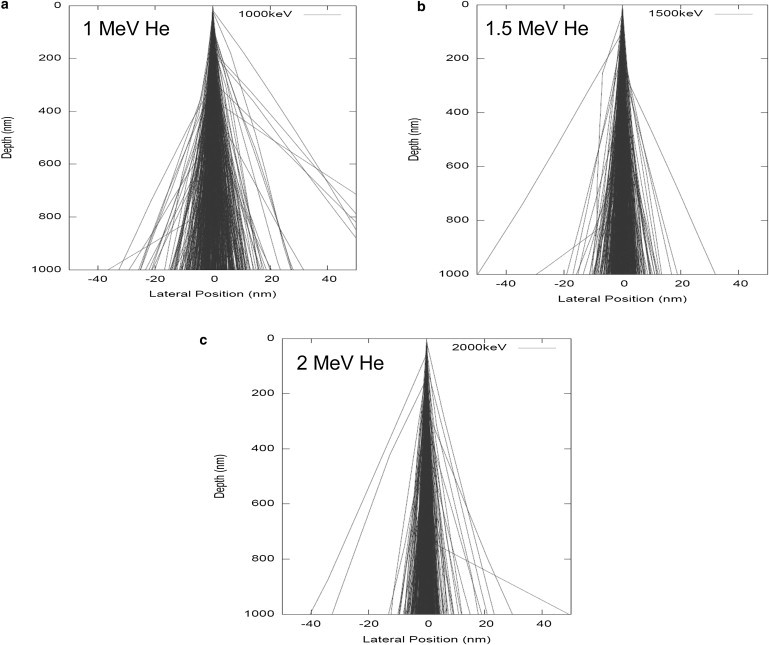
Fast helium ion paths ((a) 1 MeV, (b) 1.5 MeV, and (c) 2 MeV) through 1 μm of biological material calculated using the Monte Carlo simulation code DEEP (21).
At MeV energies, only a small minority of ions suffer large-angle nuclear collisions. Monte Carlo simulations for 500 He ions indicate that 1), for 1 MeV He ions passing through the cell, 45% are contained within a radial distance of 5 nm of the original beam axis, and 80% are contained within 10 nm; 2), for 1.5 MeV He ions, 75% are contained within a radial distance of 5 nm, and 93% are contained within 10 nm; and 3), for 2 MeV He ions, 83% are contained within a radial distance of 5 nm, and 95% are contained within 10 nm. The current state of the art for focusing MeV ions is 30 nm (21), and these simulations indicate that this resolution does not deteriorate significantly as the ions pass through the cell.
Fig. 3 a shows simulations for slow helium ions (50 keV) indicating they do not have the range to pass through a cell of nominal thickness 1 μm, and that their trajectories show significant spreading within the cell (22). Slow helium ions cause slight sputtering of the target material, and induced backscattered secondary electrons are emitted from the surface, which can subsequently be detected to create a high-resolution image of the surface. The nuclear scattering toward the end of the range results in a high degree of lateral and longitudinal straggling, but this does not affect the surface image quality.
Figure 3.
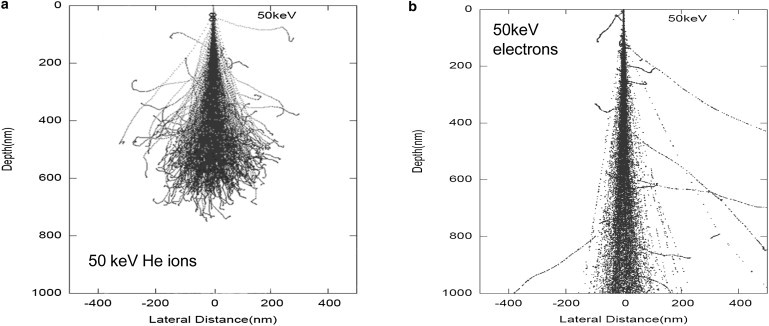
(a) Slow helium ion paths through biological material calculated using the simulation package SRIM (19). In this case, the range for 50 keV helium ions through 1 μm of biological material is ∼600 nm, which is insufficient to pass through a cell of nominal thickness 1 μm. (b) As a comparison, we also used DEEP to simulate the paths of 50 keV electrons through 1 μm of biological material. The energy loss mechanism in this case is mainly electron/electron collisions, resulting in significant large-angle scattering. Note that the lateral dimensions of both plots are 10 times that of Fig. 2, a–c.
In Fig. 3 we have also included, for comparison purposes, the simulated trajectories of an electron beam (50 keV) passing through the cell. In this case, there is a significant broadening of trajectories as the incoming electrons interact with atomic electrons in the sample. The scattering, estimated at >100 nm at the cell exit, precludes high-resolution transmission electron microscopy on thick samples.
As an example of the imaging potential of microscopy using helium ions, we imaged a human liver cell at two different magnifications using both slow and fast helium ions (Fig. 4).
The HIM images in Fig. 4, a and e, were taken at 45 keV ion energy, and many surface features are apparent. Whether these surface features are artifacts or part of the cell is unknown. Images of the same cell obtained with fast helium ions (1.2 MeV) were also acquired at the cell imaging facility (21) of the Centre for Ion Beam Applications (Department of Physics, National University of Singapore). The images depicted in Fig. 4, b, c, and f, are transmission images based on the energy loss of 1.2 MeV helium ions passing through the cell (see Materials and Methods section). These images quite clearly show both surface and internal features, including filamentary structures assumed to be part of the cytoskeleton. Of interest, not all of the surface features shown in the surface images appear with high contrast in the transmission image (see, for example, features marked with arrows in Fig. 4, a–c, e, and f). We assume that in these cases the surface structures are hollow and therefore have lower contrast in the transmission images as compared with the surface images. As expected, however, the nucleus, nucleoli, and various internal structures around the nucleus, which are not seen in the surface image, are observed in the transmission image with high contrast and edge sharpness. The helium ion images in Fig. 4 indicate the complementary nature of imaging using slow and fast helium ions.
Further information can be extracted from the transmission energy-loss data. As the fast helium ions pass through the cell, each ion undergoes multiple collisions with atomic electrons, and at each collision the ion loses a small amount of energy. The link between mass and energy loss can be obtained from the stopping power of the ion in traversing the cell, which can be extracted from known tabulated data. For example, a 1.2 MeV ion passing through a A-150 tissue-equivalent plastic has a stopping power of 2218 MeV cm2/g (23). By measuring the energy loss of each ion at each position in the cell, we can obtain an image that represents the mass distribution within the cell. For example, for the case in which a 1.2 MeV helium ion has lost half of its energy (600 keV) in passing through a cell, the ion will have traversed a calculated tissue-equivalent mass areal density of 2.75 attograms (ag)/nm2. The images in Fig. 4, d and g, represent the mass distribution of the cell, and indicate the high-density difference between the cytoplasm and the nucleus. There is a similar difference in density between the nucleus and the nucleoli. If we assume that we can measure the transmitted α particle beam energy to within 6 keV, which represents the state-of-the-art performance of a silicon particle detector positioned directly behind the cell, then for a 1.2 MeV α particle passing through a cell, the mass/unit area resolution is 0.0275 ag/nm2. This represents an extremely high-contrast mechanism for structural imaging.
Future Prospects
HIM at keV and MeV energies has a high potential for imaging surface and internal structures in whole cells at resolutions that are not attainable with other techniques. HIM at keV energies uses a high brightness field emission ion source that enables resolutions of 0.25 nm to be achieved. By increasing the beam energy to 100 keV in the same column, we may be able to achieve a spatial resolution approaching 0.1 nm. The prototype fast helium microscope operating at MeV energies uses a low-brightness RF ion source, and as such the spatial resolutions are currently limited to ∼30 nm. Work is currently in progress to improve the spatial resolutions of MeV HIM to the nanometer level by increasing the ion source brightness, and this will enable structural imaging of whole cells at nanometer dimensions. By taking several images at varying angles, we will be able to obtain 3D tomographic images at nanometer resolutions because of the increased depth of focus of the ion beams. Further, calculations have indicated that for MeV helium ions the lateral energy density distribution, which is mainly caused by low-energy secondary electron production, is extremely compact and extends only 5–10 nm from the beam axis (24). This compact secondary electron profile creates a localized ionization volume that in turn will excite fluorescence. Because any induced fluorescence created by an ion passing through the cell can be measured by a close-proximity single-photon detector, and the energy loss of each MeV ion passing through the cell can be measured separately by a silicon particle detector positioned behind the cell, simultaneous ultrastructural and fluorescence imaging is feasible. Work is in progress to test simultaneous ultrastructure and fluorescence imaging using transmitted MeV ions (including MeV protons as well as helium ions).
3D fluorescence imaging coupled with simultaneous structural imaging (including quantitative mass profiling) at resolutions <5 nm is a feasible and exciting goal. Ion beam bioimaging represents a clear paradigm shift with respect to electron beam imaging and may offer substantial benefits arising from the heavier mass of the helium ions compared with electrons.
Acknowledgments
This study was supported by the Singapore Bioimaging Consortium, ASTAR grant number SBIC RP C-013/2007.
References
- 1.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 2.Hess S.T., Gould T.J., Zimmerberg J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang B., Wang W., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B., Jones S.A., Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willig K.I., Rizzoli S.O., Hell S.W. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 6.Schermelleh L., Carlton P.M., Sedat J.W. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller S.A., Aebi U., Engel A. What transmission electron microscopes can visualize now and in the future. J. Struct. Biol. 2008;163:235–245. doi: 10.1016/j.jsb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Medalia O., Weber I., Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 9.Lucić V., Förster F., Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 10.Leis A., Rockel B., Baumeister W. Visualizing cells at the nanoscale. Trends Biochem. Sci. 2009;34:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Chao W., Harteneck B.D., Attwood D.T. Soft X-ray microscopy at a spatial resolution better than 15 nm. Nature. 2005;435:1210–1213. doi: 10.1038/nature03719. [DOI] [PubMed] [Google Scholar]
- 12.Huang X., Nelson J., Jacobsen C. Soft X-ray diffraction microscopy of a frozen hydrated yeast cell. Phys. Rev. Lett. 2009;103:198101. doi: 10.1103/PhysRevLett.103.198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang H., Song C., Miao J. Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proc. Natl. Acad. Sci. USA. 2010;107:11234–11239. doi: 10.1073/pnas.1000156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishino Y., Takahashi Y., Maeshima K. Three-dimensional visualization of a human chromosome using coherent X-ray diffraction. Phys. Rev. Lett. 2009;102:018101. doi: 10.1103/PhysRevLett.102.018101. [DOI] [PubMed] [Google Scholar]
- 15.Tan T.M.C., Sit K.H., Wong K.P. Kinetics of sulphate conjugation in extracts of human foetal liver cells in culture. Biochem. Pharmacol. 1988;37:4629–4633. doi: 10.1016/0006-2952(88)90331-0. [DOI] [PubMed] [Google Scholar]
- 16.Minqin R., van Kan J.A., Watt F. Nano-imaging of single cells using STIM. Nucl. Instrum. Methods Phys. Res. B. 2007;260:124–129. [Google Scholar]
- 17.Bettiol A.A., Udalagama C., Watt F. A new data acquisition and imaging system for nuclear microscopy based on a field programmable gate array card. Nucl. Instrum. Methods Phys. Res. B. 2009;267:2069–2072. [Google Scholar]
- 18.Postek M.T., Vladar A.E., McVey S. Helium ion microscopy: a new technique for semiconductor metrology and nanotechnology. Proc. Aip Conf. 2007;931:161–167. [Google Scholar]
- 19.Scipioni L., Stern L.A., Griffin B. Helium ion microscope. Adv. Mater. Process. 2008;166:27–30. [Google Scholar]
- 20.Cohen-Tanugi D., Yao N. Superior imaging resolution in scanning helium-ion microscopy: a look at beam-sample interactions. J. Appl. Phys. 2008;104 063504. [Google Scholar]
- 21.Watt F., Chen X., Bettiol A. The Singapore high resolution single cell imaging facility. Nucl. Instrum. Methods Phys. Res. B. 2011 In press. [Google Scholar]
- 22.Biersack, J. P. The Monte Carle program TRIM. http://www.srim.org.
- 23.NIST. http://physics.nist.gov/PhysRefData/Star/Text/ASTAR.html.
- 24.Udalagama C., Bettiol A.A., Watt F. Stochastic spatial energy deposition profiles for MeV protons and keV electrons. Phys. Rev. B. 2009;80:224107. [Google Scholar]


