Abstract
Recent studies demonstrated that the efficiency, rate and yield of prion amplification in vitro could be substantially improved by supplementing Protein Misfolding Cyclic Amplification (PMCA) with Teflon beads [Gonzalez-Montalban, et al. (2011) PLoS Pathog. 7, e1001277]. Here we employed the new PMCA format with beads (PMCAb) to gain insight into the mechanism of prion amplification. Using a panel of six hamster prion strains, the effect of beads on amplification was found to be strain-specific, with the largest improvements in efficiency observed for strains with the highest conformational stability. This result suggests a link between PrPSc conformational stability and its fragmentation rate and that beads improved amplification by assisting fragmentation. Furthermore, while exploring the PrPSc-independent bead effect mechanism, a synergy between the effects of RNA and beads on amplification was observed. Consistent with previous studies, amplification of all six hamster strains tested here was found to be RNA-dependent. Under sonication conditions used for PMCA, large RNA molecules were found to degrade into smaller fragments of a size that was previously shown to be the most effective in facilitating prion conversion. We speculate that sonication-induced changes in RNA size distribution could be one of the rate-limiting steps in prion amplification.
In the past ten years, protein misfolding cyclic amplification (PMCA) has become an important tool in prion research (1). A number of studies have provided strong evidence that fully infectious authentic prion structures can be generated de novo or amplified in PMCA (2–4). When seeded with experimental prion strains, PMCA reactions preserve strain-specific features (5;6). PMCA has been employed for elucidating cross-species transmission barriers (6;7), studying strain adaptation and interference (7;8), exploring co-factors involved in conversion (9–12) , investigating the role of prion protein glycosylation (13), and developing prion detection assays (14–17).
While the range of application of the PMCA technique has been expanding, the mechanism responsible for prion amplification remains hypothetical. PMCA consists of two alternating steps: sonication and incubation. The sonication step is presumably responsible for breaking large PrPSc particles into smaller fragments, whereas the incubation step is presumably required for the growth of small PrPSc particles through recruitment of PrPC molecules (1). The rate-limiting steps in prion amplification in PMCA and the reasons behind substantially different amplification rates for different prion strains remain unknown.
In previous studies, we found that prion amplification efficiency could be significantly improved by supplementing PMCA reactions with Teflon beads (18). Because beads improve the rate and yield of prion conversion, identification of the reaction steps affected by beads can provide new information about the prion amplification mechanism. To explain the positive effect of beads, several hypothetical mechanisms have been proposed: (i) beads optimize the efficiency of PrPSc fragmentation; (ii) beads increase the accessibility and/or slow down degradation of PrPC during PMCA reactions; or (iii) beads increase the accessibility of cofactors, specifically, RNA. The current work utilizes PMCA with beads (PMCAb) for exploring the mechanism of prion amplification.
Experimental Procedures
Reagents
A panel of six hamster prion strains was used: Hyper, ME7H, Drowsy and 139H scrapie brain homogenates were kindly provided by Richard Bessen (Montana State University, Bozeman, MT); 263K and 10% NBH were kindly provided by Robert Rohwer (Veterans Affair Maryland Health Care System, Baltimore, MD); SSLOW scrapie brain homogenate was prepared using animals from the second passage of SSLOW with an incubation time to clinical disease 481±4 days (19). Teflon beads (2.38 mm diameter, McMaster-Carr, Los Angeles, CA) were used in all PMCAb reactions or sonication procedures.
Protein misfolding cyclic amplification
Healthy hamsters were euthanized and immediately perfused with PBS, pH 7.4, supplemented with 5 mM EDTA. Brains were dissected, and 10% brain homogenate (w/v) was prepared using ice-cold conversion buffer and glass/Teflon tissue grinders cooled on ice and attached to a constant torque homogenizer (Heidolph RZR2020). The brains were ground at low speed until homogeneous, then 5 additional strokes completed the homogenization. The composition of conversion buffer was as previously described (2): Ca2+-free and Mg2+-free PBS, pH 7.5, supplemented with 0.15 M NaCl, 1.0% Triton and 1 tablet of Complete protease inhibitors cocktail (Roche, Cat. # 1836145) per 50 ml of conversion buffer. The resulting 10% normal brain homogenate (NBH) in conversion buffer was used as the substrate in PMCA or PMCAb reactions. To prepare seeds, 10% scrapie brain homogenates in PBS were serially diluted 102- to 105-fold, as indicated, in the conversion buffer and 10 μl of the dilution were used to seed amplification in 90 μl of NBH. Samples in 0.2 ml thin-wall PCR tubes (Fisher, Cat. # 14230205) were placed in a floating rack inside a Misonix S-4000 microplate horn filled with 350 ml water. Two coils of rubber tubing attached to a circulating water bath were installed to maintain 37°C inside the sonicator chamber. The standard sonication program consisted of 30 sec sonication pulses delivered at 50% to 60% power efficiency applied every 30 min during a 24 hour period. For PMCAb three Teflon beads were placed into the 0.2 ml tubes first, then NBH and seeds were added. To analyze PMCA or PMCAb products, 15 μl of each sample were supplemented with 2.5 μl SDS and 2.5 μl proteinase K (PK), to a final concentration of SDS and PK of 0.25% and 50 μg/ml, respectively, followed by incubation at 37°C for 1 hour. The digestion was terminated by addition of SDS-sample buffer and incubated for 10 min at 100°C. Samples were loaded onto NuPAGE 12% BisTris gels, transferred to PVDF membrane, stained with 3F4 antibody, and the Western blot was developed with a Supersignal West Pico Chemiluminescent Substrate kit (Thermo Scientific, Rockford, IL).
Analysis of conformational stability
10% brain homogenates were diluted 10 times into conversion buffer, then supplemented with an equal volume of GdnHCl solution in PBS to a final concentration of GdnHCl ranging from 0.4 to 4 M, and incubated at room temperature for 1 h. Next, nine volumes of 2% sarkosyl in PBS were added to all samples followed by 1 h incubation at room temperature, then the samples were treated with 20 μg/ml PK for 1 h at 37°C with shaking. The digestion was stopped by 2 mM PMSF, and the proteins were precipitated in four volumes of ice-cold acetone, overnight incubation at −20°C, and subsequent 30 min centrifugation at 16,000×g. Pellets were dried for 30 min, re-suspended in 1×SDS-sample buffer, loaded into NuPAGE 12% bisTris gels, then transferred to PVDF membrane, and stained with 3F4 antibody.
Preparation of RNA-depleted normal brain homogenate
50ul of 10mg/ml RNase A (Sigma, Cat. # R4875) was added to 5ml of 10% Syrian hamster NBH to a final RNase concentration of 100ug/ml. To prepare mock-digested NBH, 50ul of RNA-free water was added to 5 ml of 10% NBH. Both mixtures were incubated at 37ºC for one hour under gentle rotation, then total RNA was purified and analyzed using gel electrophoresis as described below.
Purification of total RNA and RNA analysis
The fresh livers of C57 mice were stored at 4° C overnight in 10 V of RNAlater Solution (Ambion , Cat. # AM7020) to allow the solution to penetrate the liver tissue and protect RNA from degradation. Then supernatant was removed and livers were store at −80° C. For purification livers were taking out from −80° C, quickly wiped to remove the excess of RNAlatter, homogenized in 10 V of TRI Reagent Solution (Ambion, Cat. # AM9738), and total RNA was purified according to the manufacturer’s procedure. Purified RNA was diluted with DEPC-treated RNase free water to a final concentration of 500 ug/ml and subjected to a sonication procedure identical to that used in PMCA (30 sec sonication pulses applied every 30 min during a 24 hour period). For the analysis of RNA fragmentation, gel electrophoresis was performed using 1.2% Agarose Gel in TBE buffer; each lane was loaded with 6 ug of RNA. As an alternative method, the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano kit were used according to the manufacturer’s procedure (Agilent Technologies, CA). To avoid RNA degradation due to possible contamination, plastic tubes, beads, and tweezers were treated with 1% DEPC for 2 hours at 37ºC and then autoclaved.
RESULTS
Effect of beads is strain-specific
To test whether the effect of beads in improving the amplification efficiency is generic, we employed a panel of hamster strains with a broad range of conformational stabilities including 263K, Hyper (HY), SSLOW, ME7H, Drowsy (DY) and 139H (Fig. 1). Each strain was subjected to two PMCA rounds in the absence or presence of beads using three seed dilutions (Fig. 2). Three serial seed dilutions were used because of individual strain-specific differences in amplification sensitivity and because beads were previously found to be most effective at high seed dilutions (18). This experiment revealed the following rank order with respect to the impact of beads on improving the efficiency of amplification (starting from a strain for which beads had the highest impact): 263K>SSLOW>HY>ME7H ≈ DY>139H (Fig. 2a,b,c,d,e,f). Beads were found to have the largest effect on amplification of 263K, HY and SSLOW, a mild effect on amplification of ME7H and DY, and no detectible improvement on 139H (Fig. 1f).
FIGURE 1. Analysis of conformational stability of six hamster strains.
Scrapie brain homogenate from animals inoculated with SSLOW (panel a), 263K (panel b), HY (panel c), ME7H (panel d), DY (panel e) or 139H (panel f) was incubated with increasing concentrations of GdnHCl from 0.4 to 4 M for 1 h, then diluted out of GdnHCl and digested with PK. Undigested brain material exposed to 0.4 M GdnHCl is provided as a reference.
FIGURE 2. Effect of beads on amplification of six hamster strains.
263K (panel a), SSLOW (panel b), HY (panel c), ME7H (panel d), DY (panel e) or 139H (panel f) scrapie brain materials were diluted 102-107-fold into 10% NBH as indicated and subjected to two serial PMCA rounds in the absence or presence of beads. The material amplified in the first round was diluted 10-fold into 10% NBH for the second round. Prior to electrophoresis, samples were digested with PK. Undigested 10% NBH is provided as a reference.
Correlation between PrPSc conformational stability and the impact of beads on amplification efficiency
To test whether the strain-specific differences in efficiency of beads is attributed to differences in PrPSc physical properties, conformational stability and strain-specific amplification rates were analyzed for the six hamster strains listed above. Consistent with previous studies, the conformational stability assay showed that SSLOW, 263K and HY were the most stable strains, ME7H was moderately stable, while DY and 139H were the least stable strains (Fig. 1). The highest improvements in amplification efficiency were observed for the strains with the highest conformational stability, whereas the strains with low stability showed very minor or no improvement in amplification rate. Taken together, these results support the hypothesis that beads facilitate fragmentation of PrPSc.
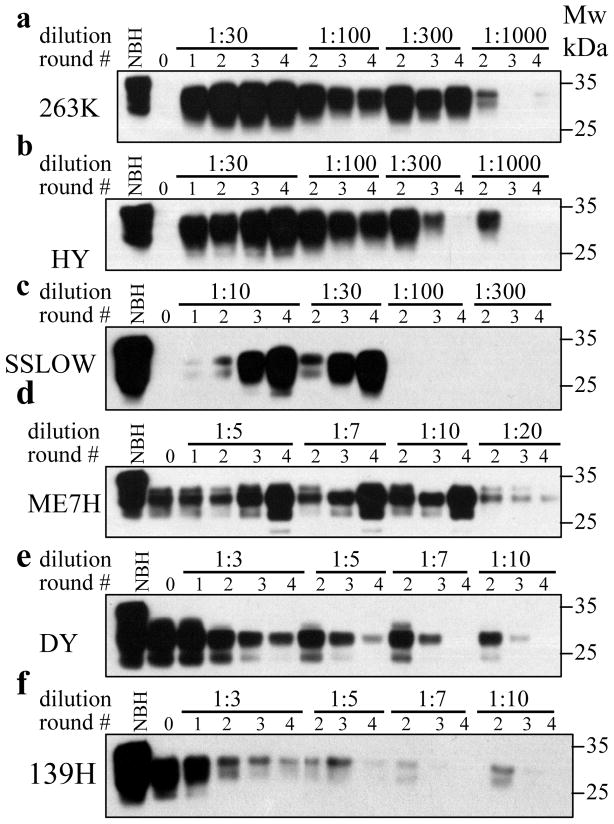
To establish strain-specific amplification rates, a set of serial PMCAb (sPMCAb) reactions were conducted for each strain with the dilution factors between serial rounds ranging from 1:3 to 1:1000 (Fig. 3). The amplification rate is defined operationally as the highest dilution between serial PMCAb rounds at which amplification is still capable of compensating the effect of dilution, i.e. the highest dilution at which a strain can be maintained steadily. For instance, the amount of 263K PK-resistant material remained stable in sPMCAb at a 1:300 dilution but decayed at a 1:1000 dilution (Fig. 3a); therefore, the amplification rate of 263K was determined to be 300. This experiment established the following rank order with respect to PMCAb amplification rates (starting from a strain with the highest rate): 263K>HY>SSLOW>ME7H>DY>139H (Fig. 3a,b,c,d,e,f). This experiment also revealed a relatively good correlation between strain-specific amplification rates and their conformational stability. The strains with the lowest stability (DY and 139H) displayed the poorest amplification rate, while the strains with high amplification rates (263K, HY and SSLOW) were the most stable.
FIGURE 3. Analysis of PrPSc amplification fold.
Scrapie brain materials were diluted 104-fold for 263K (panel a), HY (panel b), or SSLOW (panel c), 103- fold for ME7H (panel d), or 102-fold for DY (panel e) or 139H (panel f) into 10% NBH and subjected to four serial PMCAb rounds. The material amplified in each round was diluted 3-, 5-, 7-, 10-, 20-, 30-, 100-, 300-, or 1000-fold into 10% NBH for the next PMCA round, as indicated. Prior to electrophoresis, samples were digested with PK. Undigested 10% NBH is provided as a reference.
A synergy between effects of beads and RNA
To test whether the effect of beads is mediated by mechanisms independent of PrPSc fragmentation, normal brain homogenate (NBH) was subjected to 48 cycles of sonication (an equivalent of one PMCA round) in the presence or absence of beads and used as a source of PrPC in PMCA (Fig. 4). PMCAs with pre-sonicated NBH were conducted in the presence or absence of beads. Sonication of NBH prior to PMCA reduced the conversion yield (compare lanes 3 and 4 to 11 and 12, respectively, in Fig. 4). While the presence of beads during pre-sonication of NBH partially restored the yield (compare lanes 7 and 8 to 3 and 4, respectively, in Fig. 4), the yield did not reach the levels observed with fresh NBH (compare lanes 7 and 8 to 11 and 12, respectively, in Fig. 4). Therefore, (i) NBH was no longer suitable for PMCA, after it was subjected to sonication cycles for 24 h; and (ii) the presence of beads during sonication of NBH prior to PMCA had minimal if any effects.
FIGURE 4. Synergy between effects of beads and RNA.
263K scrapie brain material was diluted 105-fold (lane 2) into NBH pre-sonicated without beads (lanes 3–6), NBH pre-sonicated with three beads (lanes 7–10), or fresh NBH (lanes 11–14) and subjected to a single PMCA round in the absence or presence of beads as indicated. Total mouse liver RNA was added to PMCA reactions as indicated. Prior to electrophoresis, samples were digested with PK. Undigested 10% NBH was provided as a reference (lane 1).
To test whether the negative effect of NBH pre-sonication could be eliminated by adding extra RNA, total liver RNA was added to PMCA reactions. In the absence of beads in PMCA reactions, supplementing RNA showed no notable improvements (compare lane 5 to 3, lane 9 to 7, or line 13 to 11 in Fig. 4). However, adding RNA to the reactions conducted with beads improved the conversion yield (compare lane 6 to 4, lane 10 to 8, or lane 14 to 12 in Fig. 4). Similarly, addition of extra RNA to fresh NBH showed no notable improvement (compare lane 13 to 11 in Fig. 4) unless beads were present during PMCA (compare lane 14 to lanes 13 and 11 in Fig. 4). Thus, in the presence of extra RNA the improvements in yield were observed regardless of whether pre-sonicated or fresh NBH was used. While in the absence of extra RNA beads improved the efficiency of conversion too, (compare lane 12 to 11, lane 8 to 7 and lane 4 to 3 in Fig. 4), these improvements were less substantial than those observed in the presence of beads and extra RNA. These modest improvements could be due to the effect of beads on the RNA present in NBH.
Taken together, these results pointed to a synergy between RNA and beads and illustrated that both beads and RNA must be present during PMCA to have the optimal effect. To explore the synergetic effects of RNA and beads further, PMCA reactions were conducted using RNA-depleted NBH in the absence or presence of beads (Fig. 5). Treatment of NBH with RNase for 1 h was found to effectively remove RNAs (Fig. S1). As predicted, neither the presence of RNA (lanes 9 and 10 in Fig. 5)) nor beads (lanes 5 and 6 in Fig. 5)) could guarantee an efficient amplification. However, in the presence of both RNA and beads, the yield of amplification increased significantly (lanes 11 and 12 in Fig. 5). This experiment provided further support that beads and RNA had a synergistic effect on amplification.
FIGURE 5. Effect of RNA-depletion on RNA-amplification.
263K scrapie brain material was diluted 105-fold (lanes 2 or 8) into RNA-depleted NBH (lanes 3-6) or NBH (lanes 9–12) and subjected to a single PMCA round in the absence or presence of beads as indicated. Prior to electrophoresis, samples were digested with PK. Undigested 10% NBH were provided as a reference (lanes 1 or 7). Amplification in two independent PMCA reactions is shown for each condition.
RNA-dependency of amplification of hamster strains
Considering the results from the previous experiments, one can presume that strain-specific differences in the effect of beads could be attributed in part to the differences in RNA-dependency in strain amplification. Toward this question, we tested whether amplification of six hamster strains depended on RNA. Serial PMCAb reactions were conducted in NBH or in RNA-depleted NBH (Fig. 6). Four PMCAb rounds were performed for each condition, to ensure that small amounts of RNA in the seed material did not affect the results in RNA-depleted NBH. Because of strain-specific differences in amplification rates, the dilution factors between serial rounds were adjusted individually for each strain based on the results of previous experiment. Higher dilutions were used for strains with higher amplification rates.
FIGURE 6. Analysis of RNA-dependency of prion amplification.
Scrapie brain materials were diluted 104-fold for 263K (panel a), HY (panel b), or SSLOW (panel c), 103- fold for ME7H (panel d), and 102-fold for DY (panel e) or 139H (panel f) into 10% NBH or RNA-ablated NBH (referred to as NBH(-RNA)) as indicated and subjected to four serial PMCAb rounds. The material amplified in each round was diluted 300-fold for 263K, 100-fold for HY, 10-fold for SSLOW or ME7H, and 5-fold for DY or139H for the next PMCAb round. Prior to electrophoresis, samples were digested with PK. Undigested 10% NBH is provided as a reference. The lower panels in e and f represent the results of serial 1:5 dilution of DY and 139H seeds, respectively, in the absence of amplification.
In the absence of RNA, the amplification was suppressed for all strains (Fig. 6a,b,c,d,e,f). However, in RNA-depleted NBH, DY and 139H showed positive signals up to the third PMCAb round (Fig. 6e,f). High amounts of seeds and a very low 1:5 dilution factor between serial PMCAb rounds could account for the signal in the 2nd and 3rd rounds in these two strains. The control experiments with a simple 1:5 serial dilution of seeds in the absence of a substrate showed the same decay rates as those observed during serial PMCA in RNA-depleted NBH (Fig. 6e,f). This result confirmed that the signal observed in the 2nd and 3d rounds for DY or 139H was simply due to seed dilution but not amplification. Therefore, in agreement with the previous studies (3;9–11;20), this experiment revealed that amplification of all hamster strains tested (263K, HY, SSLOW, ME7H, DY and 139H) was dependent on RNA. Unfortunately, due to intrinsically low amplification rates for the strains with low conformational stability, establishing the ranking order of strain amplification with respect to their RNA-dependency is not feasible.
Sonication fragments RNA
Previous studies illustrated that RNAs of sizes above 200 base pairs were the most effective in stimulating prion conversion in PMCA (11;20). To explore the mechanisms behind RNA-stimulated prion conversion further, we tested whether sonication changes the RNA size distribution during PMCA. Total liver RNA was subjected to 48 sonication cycles and the size distribution was analyzed as a function of sonication cycle using agarose gel electrophoresis or microfluidics-based technology using an Agilent 2100 Bioanalyzer. Both approaches revealed a gradual decrease in size of RNA as a function of sonication cycle number (Fig. 7a,b). Before sonication, the profile of total RNA was dominated by two major bands at ~1.8 and 3.6 kbases that correspond to 18S and 28S ribosomal RNAs, respectively. By the 8th cycle, substantial amounts of RNAs were observed within 200–1000 base range (Fig. 7b). At the 48th cycle, most of RNA molecules were found within the 50–200 base range (Fig. 7b). The experiment was repeated several times and produced consistent results. No notable differences with respect to fragmentation kinetics or fragment size distribution were observed between RNA samples sonicated in the presence or absence of beads. While this experiment failed to explain the synergy between beads and RNA, it revealed that under the sonication conditions employed for PMCA, by the 8th cycle large RNA molecules were degraded into smaller fragments within a size range that was previously shown to be the most effective in stimulating prion conversion (11;20). The same experiment showed that by the 48th cycle, the RNA was largely degraded into a size range previously shown to be too small to be effective in facilitating prion conversion (11;20).
FIGURE 7. Analysis of RNA fragmentation.
Total liver RNA was subjected to a sonication procedure identical to the one employed in PMCA (30 sec sonication pulses applied every 30 min during a 24 hour period) in the presence (blue lines) or absence of beads (red lines) as indicated. RNA size distribution was analyzed as a function of sonication cycle number by agarose gel (a) or microfluidic technology (b).
DISCUSSION
Amplification of prions in PMCA is believed to involve two main steps: fragmentation of PrPSc particles and their growth. To date, the mechanism of amplification remains hypothetical. It is not known whether the amplification rate is limited by PrPSc fragmentation or binding and conversion of PrPC into PrPSc. Furthermore, the extent to which degradation of PrPC and/or PrPSc limits the amplification rate and the extent to which the amounts and availability of cellular cofactors impact the reaction also remain to be established. For a number of technical reasons, direct measurement of PrPSc size distribution during PMCA sonication cycles is very challenging and might not be informative. For instance, assuming that a PMCA round which consisted of 48 cycles amplifies PrPSc by ~10 fold, simple mathematical calculation shows that on average, only 5 out of 100 PrPSc particles fragment in each sonication cycle producing two particles from one.
The current work revealed several important findings that provide new insight into prion replication mechanisms. The largest improvements in the amplification with beads were observed for strains with the highest conformational stability. This result supports the notion that conformational stability controls the PrPSc fragmentation rate and that beads assist in fragmentation (18). Alternatively, this result is also consistent with the view that conformationally stable strains are prone to form large PrPSc aggregates and, therefore, require bead assistance to break up aggregates during amplification. In previous studies, a correlation between conformational stability and incubation time to disease was observed for hamster and mouse synthetic prions generated in animals by inoculating recombinant PrP fibrils (19;21;22). Shortening of the incubation time to disease observed during serial transmission of synthetic strains was found to coincide with a decrease in their conformational stability (19;22). Furthermore, a strong correlation between the incubation time to disease and PrPSc conformational stability was also observed for a broad range of synthetic strains produced by inoculating recombinant PrP fibrils of various stability (21). In vitro studies with recombinant PrP fibrils showed that amyloid structures with lower conformational stability produce smaller particles when fragmented by sonication than the amyloid fibrils of higher stability (23). Taken together, these results support the hypothesis that PrPSc conformational stability is one of the factors that controls the incubation time to disease and that the PrPSc fragmentation rate presumably links the stability to the incubation time. Notably, previous studies on cross-species transmission of hamster-adapted strains suggested that PrPSc conformational stability also influences the transmission efficiency of prions between species (24).
The linear relationship between conformational stability and incubation time did not hold if a broad group of hamster-adapted strains with diverse biological properties was considered (25). The study by Ayers et al. suggested that other features such as strain-specific differences in neuro- and cell tropism as well as involvement of astrocytes and microglia impact the rate of disease progression to a larger extent than PrPSc conformational stability (25). It is possible that the incubation time to disease does not depend on the PrPSc accumulation rate as much as it reflects the strain-specific intrinsic neurotoxicity of PrPSc. For instance, some PrPSc strains or amyloid structures might be intrinsically more neurotoxic than other structures (26;27). The precise relationship between strain-specific PrPSc structures and their neurotoxicity has yet to be established.
Consistent with the previous studies (25), the strains with long incubation time to disease (139H, DY and ME7H), with an exception of SSLOW, had very low amplification rates in PMCAb. Considering that SSLOW is characterized by a very slow progression of clinical disease in addition to long incubation time, the unique biological properties of this synthetic strain might account for its exceptional position. One can speculate that low amplification rates for 139H, DY and ME7H could be due to their fast degradation rate rather than slow amplification per se, a hypothesis that needs to be tested in future studies. Low conformational stability of 139H, DY and ME7H is consistent with this hypothesis. Taking into account all data available to date, the relationship between the incubation time to disease and conformational stability could be described by a biphasic rather than a linear curve. According to this model, the strains that have the shortest incubation time (263K or HY) are those for which the conformational properties are optimized to avoid rapid cell clearance while providing fast amplification. The strains with conformational stabilities lower than optimal (139H, DY and ME7H) or higher than optimal (synthetic strains) exhibit long incubation times because of their high clearance rates or lower than optimal amplification rates, respectively.
Consistent with the previous studies (9–11), amplification of all hamster strains tested here was found to be RNA-dependent. Whether or not RNA serves as a co-factor for prion replication in a cell is currently not known (9;10;28). One could envision that ribophagy (authophagy of ribosomal RNAs (29)) supplies RNA fragments for prion conversion in the cellular compartments (autophagosomes or lysosomes) where PrPSc replication might take place. Unexpectedly, under PMCA sonication conditions, large RNA molecules were found to degrade into smaller fragments producing a size distribution that was previously shown to be the most efficient in facilitating prion conversion (11;20). Considering that multiple sonication cycles were required for generating RNAs of the most active sizes, one can speculate that sonication-induced fragmentation of RNA might represent one of the rate-limiting steps in prion amplification in vitro. However, by the 48th sonication cycle, most of the RNA was found to be degraded to fragments within the 50–200 bases range. Because RNAs of this size were previously shown to be not very effective in facilitating amplification (11), further increase in the number of PMCAb cycles might not be productive for amplification of strains that rely on RNA. Taken together, these data suggest that the most productive time-window for amplification of RNA-dependent strains in PMCAb could be only between cycle # 8 and 24.
The current study revealed a synergy between the effects of beads and RNA on PrPSc amplification. The mechanism responsible for this synergistic effect has yet to be explored. The kinetics of sonication-induced RNA fragmentation was not affected by beads (Figure 7). We do not know whether beads alter accessibility of RNAs and/or change its secondary structure. Nevertheless, when all experiments on beads and RNA are considered together, the strongest effect of RNA in stimulating amplification was observed when beads were present in PMCA reactions and when RNA fragmentation occurred in parallel but not prior to the conversion. Because polymeric cofactors of different biochemical natures might be involved in amplification of strains from different species (10;16;30), the effect of sonication and beads on size distribution of these polymers have to be considered. Future optimization of PMCA conditions for slow-replicating strains might involve finding the right balance for effective fragmentation of PrPSc and a cofactor, while avoiding degradation of PrPSc.
Supplementary Material
Acknowledgments
We thank Pamela Wright for editing the manuscript.
This work was supported by NIH grant NS045585 to (I.V.B.) and by a Beatriu de Pinós Fellowship with the support of the Commission for Universities and Research of the Department of Innovation, Universities and Enterprise of the Government of Catalonia to (N.G.M).
Abbreviations
- PMCA
protein misfolding cyclic amplification
- PMCAb
PMCA with Teflon beads
- sPMCAb
serial PMCAb
- PrPC
normal cellular isoform of the prion protein
- PrPSc
infectious, disease-related isoform of the prion protein
- NBH
normal brain homogenate
- HY
Hyper strain
- DY
Drowsy strain
- PK
proteinase K
Footnotes
Supplemental Information. Analysis of RNA content in RNA-depleted NBH (Figure S1).This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 2.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Deleault NR, Harris BT, Rees JR, Supattapone S. Formation of native prions from minimal components in vitro. Proc Acad Natl Sci U S A. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Wang X, Yuan CG, Ma J. Generating a Prion Bacterially Expressed Recombinant Prion Protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C. Cell-free propagation of prion strains. EMBO J. 2008;27:2557–2566. doi: 10.1038/emboj.2008.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, Telling GC. Accelerated High Fidelity Prion Amplification Within and Across Prion Species Barriers. PLOS Pathog. 2008;4:e1000139. doi: 10.1371/journal.ppat.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castilla J, Gonzalez-Romero D, Saa P, Morales R, De Castro J, Soto C. Crossing the Species Barrier by PrPSc Replication In Vitro Generates Unique Infectious Prions. Cell. 2008;134:757–768. doi: 10.1016/j.cell.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol. 2010;84:5706–5714. doi: 10.1128/JVI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 10.Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry. 2010;49:3928–3934. doi: 10.1021/bi100370b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson RA, Supattapone S. Protease-resistant Prion Protein Amplification Reconstituted with Partially Purified substrates and Synthetic Polyanions. J Biol Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 12.Mays CE, Ryou C. Plasminogen stimulates propagation of protease-resistant prion protein in vitro. Faseb J. 2010;24:5102–5112. doi: 10.1096/fj.10-163600. [DOI] [PubMed] [Google Scholar]
- 13.Nishina K, Deleault NR, Mahal S, Baskakov I, Luhrs T, Riek R, Supattapone S. The Stoichiometry of Host PrPC Glycoforms Modulates the Efficiency of PrPSc formation in vitro. Biochemistry. 2006;45:14129–14139. doi: 10.1021/bi061526k. [DOI] [PubMed] [Google Scholar]
- 14.Saa P, Castilla J, Soto C. Presymptomatic Detection of Prions in Blood. Science. 2006;313:92–94. doi: 10.1126/science.1129051. [DOI] [PubMed] [Google Scholar]
- 15.Tattum MH, Jones S, Pal S, Collinge J, Jackson GS. Discrimination between prion-infected and normal blood samples by protein misfolding cyclic amplification. Transfusion. 2010;50:2619–2627. doi: 10.1111/j.1537-2995.2010.02595.x. [DOI] [PubMed] [Google Scholar]
- 16.Murayama Y, Yoshioka M, Masujin K, Okada H, Iwamaru Y, Imamura M, Matsuura Y, Fukuda S, Onoe S, Yokoyama T, Mohri S. Sulfated dextrans enhance in vitro amplification of bovine spongiform encephalopathy PrP(Sc) and enable ultrasensitive detection of bovine PrP(Sc) Plos ONE. 2010;5:e13152. doi: 10.1371/journal.pone.0013152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritzkow S, Wagenfuhr K, Daus ML, Boerner S, Lemmer K, Thomzig A, Mielke M, Beekes M. Quantitative detection and biological propagation of scrapie seeding activity in vitro facilitate use of prions as model pathogens for disinfection. Plos ONE. 2011;6:e20384. doi: 10.1371/journal.pone.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Montalban N, Makarava N, Ostapchenko VG, Savtchenko R, Alexeeva I, Rohwer RG, Baskakov IV. Highly Efficient Protein Misfolding Cyclic Amplification. PLoS Pathogen. 2011;7:e1001277. doi: 10.1371/journal.ppat.1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarava N, Kovacs GG, Bocharova OV, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild type animals. Acta Neuropathol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. Selective incorporation of polyanionic molecules into hamster prions. J Biol Chem. 2007;282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Acad Natl Sci U S A. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legname G, Nguyen HOB, Baskakov IV, Cohen FE, DeArmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Aca Sci USA. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Makarava N, Lee CI, Laksanalamai P, Robb FT, Baskakov IV. Conformational stability of PrP amyloid firbils controls their smallest possible fragment size. J Mol Biol. 2008;376:1155–1167. doi: 10.1016/j.jmb.2007.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton D, DeArmond S, Prusiner S, Scott MR. A Change in the Conformation of Prions Accompanies the Emergence of a New Prion Strain. Neuron. 2002;34:921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 25.Ayers JL, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLOS Pathog. 2011;7:e1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petkova AT, Leapman RD, Gua Z, Yau WM, Mattson MP, Tycko R. Self-Propagating, Molecular-Level Polymorphism in Alzheimer's b-Amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 27.Lee YJ, Savtchenko R, Ostapchenko VG, Makarava N, Baskakov IV. Molecular Structure of Amyloid Fibrils Controls the Relationship between Fibrillar Size and Toxicity. Plos ONE. 2011;6:e20244. doi: 10.1371/journal.pone.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abid K, Morales R, Soto C. Cellular factors implicated in prion replication. FEBS Lett. 2010;584:2409–2314. doi: 10.1016/j.febslet.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama T, Takeuchi A, Yamamoto M, Kitamoto T, Ironside JW, Morita M. Heparin enhances the cell-protein misfolding cyclic amplification efficiency of variant Creutzfeldt-Jakob disease. Neroscience Lett. 2011 doi: 10.1016/j.neulet.2011.04.072. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.