Abstract
Reliable population DNA molecular markers are difficult to develop for molluscs, the reasons for which are largely unknown. Identical protocols for microsatellite marker development were implemented in three gastropods. Success rates were lower for Gibbula cineraria compared to Littorina littorea and L. saxatilis. Comparative genomic analysis of 47.2 kb of microsatellite containing sequences (MCS) revealed a high incidence of cryptic repetitive DNA in their flanking regions. The majority of these were novel, and could be grouped into DNA families based upon sequence similarities. Significant inter-specific variation in abundance of cryptic repetitive DNA and DNA families was observed. Repbase scans show that a large proportion of cryptic repetitive DNA was identified as transposable elements (TEs). We argue that a large number of TEs and their transpositional activity may be linked to differential rates of DNA multiplication and recombination. This is likely to be an important factor explaining inter-specific variation in genome stability and hence microsatellite marker development success rates. Gastropods also differed significantly in the type of TEs classes (autonomous vs non-autonomous) observed. We propose that dissimilar transpositional mechanisms differentiate the TE classes in terms of their propensity for transposition, fixation and/or silencing. Consequently, the phylogenetic conservation of non-autonomous TEs, such as CvA, suggests that dispersal of these elements may have behaved as microsatellite-inducing elements. Results seem to indicate that, compared to autonomous, non-autonomous TEs maybe have a more active role in genome rearrangement processes. The implications of the findings for genomic rearrangement, stability and marker development are discussed.
Keywords: microsatellite containing sequences, transposable element, Mollusca, gastropod, genome, microsatellite isolation
Introduction
Microsatellites or tandem simple sequence repeats consist of highly variable repeated units of 2–6 bp. They have been widely and routinely used as molecular markers in diverse genetic studies (Selkoe and Toonen, 2006). Despite their usefulness, microsatellite molecular markers in certain taxa can sometimes be problematic to develop (Meglécz et al., 2004; Arthofer et al., 2007; Bailie et al., 2010). This is particularly true for molluscs, and the underlying causes of such observations have never been fully investigated or explained (Reece et al., 2004; Weetman et al., 2001, 2005). Preliminary findings from other taxa indicate that difficulties encountered with microsatellite development generally are not due to a paucity of microsatellites or a lack of polymorphic vs monomorphic microsatellites in the genome (Van't Hof et al., 2007). However, other studies rather suggest that methodological difficulties, at least in insects and crustaceans, may have been caused by genomic complexities contained within microsatellite flanking regions (Meglécz et al., 2004; Van't Hof et al., 2007; Bailie et al., 2010).
The two major genomic features thought to be responsible for PCR interference or inconsistencies are the following: (1) unstable flanking sequences and (2) occurrences of cryptic repetitive DNA (Meglécz et al., 2004). Thus, microsatellite flanking regions are either too similar or too variable in these taxa. Unstable flanking regions arise when indels or mutations occur at PCR primer binding sites, thereby causing PCR failure, commonly referred to as null alleles (Dakin and Avise, 2004). These can lead to underestimates in allele frequencies and heterozygosity. Consequently, demographic and biological inferences (for example, population size estimates, parentage) made from data sets affected by null alleles (without accounting for their bias) can be compromised (see Bonin et al. (2004) and DeWoody et al. (2006) for review). Second, the occurrence of cryptic repetitive DNA in microsatellite flanking regions overlapping with PCR binding sites can result in the amplification of products of unexpected sizes or in the difficulty in amplification of products representing a single locus (Zhang, 2004). Cryptic repetitive DNA and high similarity among microsatellite flanking regions, which are very commonly found in plants (Tero et al., 2006), have also been recently identified from a number of insects and crustaceans (Meglécz et al., 2004, 2007; Van't Hof et al., 2007; Bailie et al., 2010). These are thought to be primarily generated during DNA multiplication or duplication processes (Meglécz et al., 2004). Recombination-related events such as unequal crossing over and gene conversion may also be responsible. Evidence of genomic rearrangement processes can be inferred from proxies within microsatellite flanking regions (Meglécz et al., 2004; Van't Hof et al., 2007). For instance, although information is still limited, microsatellites can be found in close association with transposable elements (TEs) or as an integral component of the TE itself (Ramsay et al., 1999; Gaffney et al., 2003; Carreras-Carbonell et al., 2006). On the basis of these observations, Meglécz et al. (2004) suggest that genomic rearrangement processes could be mediated by TEs. TEs represent a class of repetitive DNA segments that can be extremely abundant and account for a large portion of a genome; for example, they comprise some 45% of the human genome and up to 80% of some grass genomes (see Feschotte et al. (2002a) for review). TEs can be classified as either autonomous or non-autonomous elements, both are mobile and can make duplicate copies of themselves, which are inserted into new genome locations. TE classes fundamentally differ in their transposition mechanism. Although autonomous TEs have the coding capacity for the production of all the enzymes required for their transposition, non-autonomous TEs do not and instead ‘hijack' the machinery of partner TEs to accomplish their transposition (Feschotte et al., 2002a, 2002b). It is still not known, however, whether autonomous or non-autonomous TEs contribute equally to genomic rearrangement processes.
A number of microsatellite marker systems were developed by the authors for three gastropod molluscs, Littorina saxatilis (Olivi, 1792), L. littorea (Linnaeus, 1758) (McInerney et al., 2009a, 2009b) and Gibbula cineraria (Linnaeus, 1758) (this study). In each case, identical protocols for microsatellite development based on enriched genomic libraries were implemented. Despite similar cloning efficiencies, success rates for viable marker development varied considerably among the species. To try to elucidate the reasons for such variation, we carried out a comparative genomic analysis of microsatellite containing sequences (MCS) from three gastropod molluscs in this study. More specifically, we tested the hypothesis that inter-specific variation in genomic complexity and stability of microsatellite flanking regions was responsible for the varied success rates observed.
Materials and methods
Microsatellite development, sequence data and PCR settings
The development and isolation of microsatellite markers for G. cineraria followed identical protocols used for both L. saxatilis and L. littorea as described by McInerney et al. (2009a, 2009b). All MCS isolated from the three independent enriched genomic libraries were edited to remove any vector contamination as identified with the UniVec vector base of NCBI (ftp://ftp.ncbi.nih.gov/pub/UniVec). Duplicate or redundant sequences (>98% identity) were identified using BLASTn (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1997) and excluded from further analyses. Microsatellite PCR primer sets were designed with PrimerSelect (DNASTAR, Inc., Madison, WI, USA) only from clones displaying suitable flanking regions. PCRs were performed with DNA template representing three to six individuals per species, isolated according to McInerney et al. (2009a). PCR primer sequences for markers amplifying single locus, cycling conditions and gastropod species tested are provided in Table 1. PCRs were undertaken in 12 μl reaction volumes containing 50 ng DNA, 100 μ dNTPs, 1 × PCR reaction buffer (Invitrogen Ltd, Paisley, UK), 0.5 U Taq DNA polymerase (Invitrogen), primer and MgCl2 concentrations as in Table 1. Fluorescently labelled PCR products were separated on a LI-COR 4200 automated system (LI-COR Inc., Lincoln, NE, USA). For further details of experimental conditions used, see McInerney et al. (2009a). Samples representing seven additional species were also used in PCR experiments to assess marker cross-species hybridization. These comprised the co-occurring intertidal gastropod molluscs of the family Littorinidae: L. fabalis (Turton, 1825), L. obtusata (Linnaeus, 1758), L. compressa (Jeffreys, 1865); and the family Trochidae: G. umbilicalis (da Costa, 1778), G. magus (Linnaeus, 1758), Calliostoma zizyphinum (Linnaeus, 1758) and Osilinus lineatus (da Costa, 1778).
Table 1. PCR primer sets and amplification conditions of loci used in Experiments I (only primer sets that amplified fragments representing single locus polymorphism are shown) and II.
| Experiment | Locus | Primer sequence (5′-3′)/Source | Tm (°C)a | PCR cycles | Primer (p)b | MgCl2 (m)c | Species testedd | GenBank accession no.e |
|---|---|---|---|---|---|---|---|---|
| I | Gcin1 | F: TCAAATTATGTATCACAAATCTCA R: AAAGTAACCAGTCTCGGGCTGTCG | 51 | 25 | 1 | 2.5 | GC, GU, GM, CZ, OL | GQ129680 |
| I | Gcin2f | F: CTTTTTGGGCCTGATGCTTCTG R: GATCCCGAGGTGCTTATGGTATG | 58 | 25 | 2 | 2.0 | GC, GU, GM, CZ, OL | GQ129681 |
| I | Gcin3f | F: TTGGGCATGATGCTTCTGGAG R: GATCCCGATGTGCTTATGGTATG | 58 | 25 | 1 | 2.0 | GC, GU, GM, CZ, OL | GQ129682 |
| I | Gcin4 | F: CTAGCATGCTATAGTTTAGGAGAT R: CGCCGAGCGTATTTTTA | 48 | 25 | 2 | 3.0 | GC, GU, GM, CZ, OL | GQ129683 |
| I | Llit52 | F: GTATTGTTGTCTTGCACCACGAA R: ATCCAACGAATTTAGACACAACCA | 56 | 25 | 1 | 2.5 | LS, LL, LF, LO, LC | FJ480222 |
| I | Lsax12 | F: AAAATGCCGAAGCGACAGT R: TGCCGAAAACCAAACCA | 53 | 25 | 1 | 2.5 | LS, LL, LF, LO, LC | FJ794786 |
| II | GcinCvA | F: GAGTCATTGGTTTCGGAGAAGAA R: GCTCGATCTCATCAGGTCTGAAA | 58 | 25 | 2 | 2.5 | GC, GU, GM, CZ, OL, LS, LL, LF, LO, LC | GQ129736 |
Annealing temperature.
Primer concentration pmol.
Magnesium chloride concentration.
Species tested: CZ, Calliostoma zizyphinum; GC, Gibbula cineraria; GM, G. magus; GU, G. umbilicalis; LS, Littorina saxatilis; LC, L. compressa; LL, L. littorea; LF, L. fabalis; LO, L. obtusata; OL, Osilinus lineatus.
GenBank accession numbers for the cloned sequences from which the markers were derived.
Instances where a touchdown PCR profile was used.
Inter-specific analysis of cryptic repetitive DNA and DNA families
To identify the possible existence of cryptic repetitive DNA among MCS flanking regions, we compared these sequences in an all-against-all BLASTn analysis (Altschul et al., 1997), with the option to mask for low-complexity repeat sequences following the approach of Meglécz et al. (2004). BLASTn alignments were initially screened by eye to ensure the exclusion of false alignments (relating to the tandem repeat regions) that had escaped the filtering process. Results were tabulated and limited to hits involving sequences larger than 40 bp in length with a >85% identity, occurring in the flanking regions as suggested by Meglécz et al. (2004). Multiple MCS containing similar cryptic repetitive DNA in the microsatellite flanking regions (as identified through BLASTn) were subsequently grouped into DNA families. For the purposes of this paper, we define DNA families as a group of MCS that have highly similar microsatellite flanking regions containing cryptic repetitive DNA. The proportion of grouped sequences into DNA families was calculated as grouped sequences over the sum of the total number of sequences examined per species.
Inter-specific analysis of TEs
To check for the possible presence of known TEs, we scanned all MCS against Repbase, a database of known TEs (Kohany et al., 2006) (http://www.girinst.org/censor/index.php) using default sensitivity parameters and the option to mask for low-complexity repeat sequences (that is, microsatellites). Hits below a similarity cut-off threshold of 0.65 were not considered, as these often showed little or no continuous similarity between the TE and microsatellite flanking regions. Treatment of raw results followed a parallel approach to the BLASTn analysis. Thus, Repbase hits were initially screened by eye to ensure the exclusion of false alignments (relating to the tandem repeat regions) that had escaped the filtering process. The proportion of sequences identified with TEs was calculated as sequences with TEs over the sum of the total number of sequences examined per species.
Results
Isolation of repetitive DNA and microsatellite characterization
A total of 180 MCS with a mean length of 260 bp, amounting to 47.2 kb, were isolated from three gastropod mollusc genomic libraries and deposited in GenBank (for details see Table 2). Of these, 17 have been already published as part of previous work (McInerney et al., 2009a, 2009b). Cloning efficiency (average 12.1%) and the numbers of unique MCS sequenced (N=58, 61 and 61 for L. saxatilis, L. littorea and G. cineraria, respectively) were similar for each of the genomic libraries. Despite this, PCR primer development success rate varied markedly among the species. Greater difficulty was encountered for marker development in G. cineraria in comparison to the two Littorina species. Despite a large number of G. cineraria PCR primer sets (N=150) developed on presumably unique microsatellite flanking regions, in a first instance no single locus amplification was achieved. In all cases, either multi-banding patterns or no amplification was the outcome.
Table 2. Summary of MCS examined in this study and the proportions (%) of MCS that grouped into DNA families and had significant homology to a known TE.
| Species | No. of MCSa | Total DNA (kb)b | Proportion (%) of MCS that Grouped into DNA families (N)c | TEsd | GenBank accession no. (Source)e |
|---|---|---|---|---|---|
| Gibbula cineraria (Linnaeus, 1758) | 61 | 14.7 | 74.6 (44) | 45.8 (27) | GQ129680–GQ129740 |
| Littorina littorea (Linnaeus, 1758) | 61 | 17.2 | 18.0 (11) | 19.7 (12) | FJ480214–FJ480222 (McInerney et al., 2009a), GQ129741–GQ129792 |
| Littorina saxatilis (Olivi, 1792) | 58 | 15.3 | 9.5 (6) | 3.2 (2) | FJ794781–FJ794788 (McInerney et al., 2009a), GQ129793–GQ129842 |
Number of MCS analysed.
Total length of the analysed genomic sequences in kilobases.
Proportion % (actual number of MCS in parenthesis) of MCS per species that grouped into DNA families, based on similarities in their flanking regions as identified by the all-against-all BLASTn analysis.
Proportion (actual number of identified TEs in parenthesis) of MCS per species with homology to TEs as identified in the scan against Repbase.
GenBank accession numbers, with the source of previously published MCS indicated.
Inter-specific variation in microsatellite flanking sequence similarities
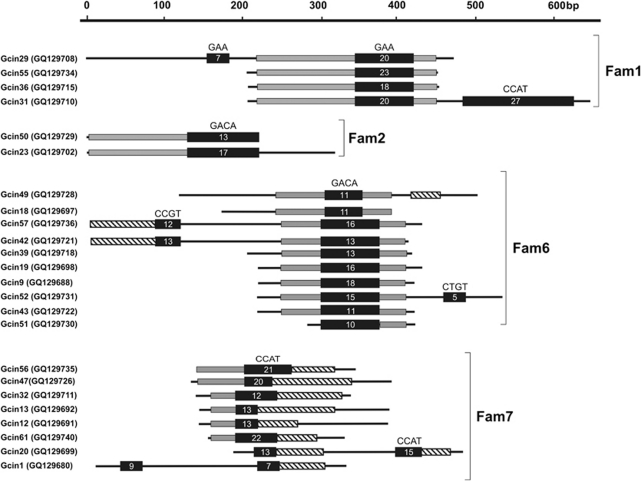
Numerous microsatellite flanking sequences contained cryptic repetitive DNA, the majority (83.3%) of these displayed a high level of intra-specific similarities (that is, similar to other cryptic repetitive DNA in the same species). A small (16.7%) number of inter-specific similarities were also identified between the littorinids. Of the 180 sequences examined, a total of 61 (33.9%) were grouped into 12 DNA families with the BLASTn analysis. A summary of the DNA families identified is provided in Table 3. Each of these comprised between 2 and 24 MCS. Five families comprised L. littorea and L. saxatilis MCS (families 8–12), each sharing a region within their flanking sequence of between ca. 40 and 80 bp. The remaining seven G. cineraria families (families 1–7) shared larger regions of similarity ranging between ca. 40 and 130 bp. The proportion of MCS per species that grouped into DNA families was far greater in G. cineraria (74.6%), compared to L. littorea (18%) and L. saxatilis (9.5%) (Table 2). A schematic representation of some of the DNA families identified with the all-against-all BLASTn analysis is presented in Figure 1. The cryptic repetitive DNA from the microsatellite flanking regions could be arranged both in symmetrical and asymmetrical orientations; that is, identical on both microsatellite flanking sequences or identical on one side only. Microsatellite repeats identified from the characterized DNA families were often imperfect. The two main motifs identified from the DNA families were GACA and CCAT with other repeat motifs occurring to a lesser extent (for example, GAA and GACG). These represent a fraction of the repeat motif types used in the enrichment procedure (McInerney et al., 2009a, 2009b).
Table 3. Summary of the DNA families identified with the BLASTn analysis and MCS with known TEs identified from their flanking regions (bold type, for details see Table 4).
| Species | DNA families |
|---|---|
| G. cineraria | |
| Family 1: Gcin29-Gcin31-Gcin36-Gcin55 | |
| Family 2: Gcin23-Gcin50 | |
| Family 3:Gcin20-Gcin30-Gcin57-Gcin61 | |
| Family 4: Gcin11-Gcin24 | |
| Family 5: Gcin4-Gcin46 | |
| Family 6: Gcin9-Gcin15-Gcin17-Gcin18-Gcin19- Gcin21-Gcin39-Gcin42-Gcin43-Gcin46-Gcin49-Gcin51- Gcin52-Gcin57-Gcin58 | |
| Family 7: Gcin1-Gcin2-Gcin3-Gcin8-Gcin12-Gcin13-Gcin16-Gcin20-Gcin24-Gcin26-Gcin30-Gcin32- Gcin35-Gcin39-Gcin40-Gcin42-Gcin44-Gcin47- Gcin48-Gcin49-Gcin53-Gcin56-Gcin57-Gcin59-Gcin61 | |
| L. saxatilis and L. littorea | |
| Family 8: Lsax12-Lsax47 | |
| Family 9: Llit95-Llit100-Llit101-Lsax43-Lsax39 | |
| Family 10: Llit52-Llit88 | |
| Family 11: Llit96-Llit104 | |
| Family 12: Llit80-Llit86-Llit113-Lsax54-Lsax65 |
Figure 1.
Schematic representation of DNA families identified with an all-against-all BLASTn analysis. MCS names are provided with GenBank accession numbers in parenthesis. Black boxes, microsatellite regions and number of repeat motif; grey boxes, cryptic repetitive DNA identified in flanking regions; black lines, unique DNA sequences (that is, no apparent similarity to any other regions). In families (Fam) 6 and 7, striped boxes indicate flanking regions of cryptic repetitive DNA identified as TEs (for details see Table 4). For simplicity, some sequences have been omitted from the schematic of DNA families 6 and 7.
It is clear that the presence of cryptic repetitive DNA in microsatellite flanking regions and the subsequent existence of DNA families with members scattered throughout the genome can hamper the development of PCR primer sets for single locus amplification. In an attempt to overcome this difficulty (that is, to amplify fragments representing single locus), we redesigned PCR primer sets (for primer sets and PCR conditions see Table 1 and Supplementary Table S1) in a way that their 3′ ends targeted unique nucleotide differences identifiable among DNA family members (Experiment I). Among the members belonging to the 12 identified DNA families, a total of 25 new PCR primer sets were tested for single locus amplification. Of these, only six primer sets amplified single microsatellite loci with a clear di-allelic pattern; four in G. cineraria (Gcin1, Gcin2, Gcin3, Gcin4) and one each for L. littorea and L. saxatilis (Lsax12, Llit52) (Table 1). In cross-species hybridization tests all redesigned microsatellite loci failed to amplify, except the locus Lsax12 (see McInerney et al. (2009b) for further details).
To facilitate future studies, we annotated the flanking region sequences characterizing the distinct DNA families (that is, cryptic repetitive DNA) that were not readily identified as TEs from Repbase scans with the identifier gastropod core sequences (GCS 1, 2 and so on). These core sequences are provided in Supplementary Table S2. Homologies between the GCS and sequences submitted to the GenBank database were surveyed in a BLASTn analysis. BLAST hits were limited to those with an E-value <0.025 and a BLAST score >40 (Van't Hof et al., 2007). No matches were observed for over half of the sequences tested suggesting they are novel. Five of the GCS (1, 4, 5, 10 and 11) produced matches with MCS of other molluscs albeit distantly related. These included bivalves and pelecypods in addition to other taxa (Supplementary Table S3).
Inter-specific variation in the occurrence of TEs
Identified TEs were classified following the universal classification system of Kapitonov and Jurka (2008) implemented in Repbase. Information regarding TE transpositional mechanism (autonomous/non-autonomous) was obtained from Repbase and Web of Science reports. TEs were found in 20.6% (N=37) of all the MCS examined (Tables 2 and 4). On average, regions displaying a high identity to a TE (ca.78%) were 74 bp in length. In most instances (89.2%), a single TE was identified from an MCS. In three instances, however, two different TEs were observed in the same MCS (Llit64, Llit66, Gcin26), and in one instance only, the same region of a TE was observed twice in the same MCS (Gcin20). Thus, the cryptic repetitive DNA identified from the DNA families, after Repbase scans, was sometimes shown to be composed of more than one TE (Table 4, Figure 1 (family 7)). L. saxatilis had the least proportion (3.2%, N=2) of MCS associated with TEs (Table 2). These were identified as MuDR (MULE) and Mu-like DNA transposons. L. littorea had a larger proportion of MCS with TEs (19.7%) and overall the highest number of different TEs (N=11), the majority of which were identified only once among MCS for this species. These included a variety of TEs divided in almost equal proportions between autonomous and non-autonomous as follows: DNA transposons (En/Spm (CACTA), Mariner, hAT, Arnold, MuDR (MULE)); LTR retrotransposon (Gypsy) and non-LTR retrotransposons (LINE and SINEs). Although G. cineraria had the greatest proportion of MCS with TEs (45.8%), these were represented by only six different types, all non-autonomous TEs, some of which were observed at high frequencies (Table 4). Among these were DNA transposons (MITE, helitrons) and LTR retrotransposons (endogenous retroviruses ERV1, ERV3).
Table 4. Summary of TEs identified in the scan of gastropod MCS against Repbase.
| Species | TE classa | TE descriptionb | Known TEc | A/NAd | MCS name | Similarity (%)e | Alignment length (bp)f | Alignment scoreg | GenBank accession no.h | Source |
|---|---|---|---|---|---|---|---|---|---|---|
| G. cineraria | ||||||||||
| DNA transposon | ||||||||||
| MITE | CvA | NA | Gcin1i | 81 | 63 | 345 | GQ129680 | This study | ||
| MITE | CvA | NA | Gcin2i | 66 | 70 | 222 | GQ129681 | This study | ||
| MITE | CvA | NA | Gcin8i | 71 | 63 | 217 | GQ129687 | This study | ||
| MITE | CvA | NA | Gcin11i | 77 | 71 | 377 | GQ129690 | This study | ||
| MITE | CvA | NA | Gcin12i | 66 | 70 | 246 | GQ129691 | This study | ||
| MITE | CvA | NA | Gcin16i | 73 | 63 | 275 | GQ129695 | This study | ||
| MITE | CvA | NA | Gcin20i | 72 | 60 | 258 | GQ129699 | This study | ||
| MITE | CvA | NA | Gcin20i | 84 | 66 | 424 | GQ129699 | This study | ||
| MITE | CvA | NA | Gcin24i | 80 | 63 | 375 | GQ129703 | This study | ||
| MITE | CvA | NA | Gcin26i | 77 | 63 | 315 | GQ129705 | This study | ||
| MITE | CvA | NA | Gcin30i | 78 | 66 | 362 | GQ129709 | This study | ||
| MITE | CvA | NA | Gcin35i | 73 | 63 | 279 | GQ129714 | This study | ||
| MITE | CvA | NA | Gcin40i | 67 | 125 | 346 | GQ129719 | This study | ||
| MITE | CvA | NA | Gcin44i | 86 | 56 | 374 | GQ129723 | This study | ||
| MITE | CvA | NA | Gcin48i | 75 | 63 | 286 | GQ129727 | This study | ||
| MITE | CvA | NA | Gcin49i | 100 | 24 | 234 | GQ129728 | This study | ||
| MITE | CvA | NA | Gcin56i | 77 | 63 | 312 | GQ129735 | This study | ||
| MITE | CvA | NA | Gcin57i | 85 | 66 | 453 | GQ129736 | This study | ||
| MITE | CvA | NA | Gcin61i | 75 | 66 | 305 | GQ129740 | This study | ||
| Helitron | DNAREP1_DYak | NA | Gcin13i | 71 | 123 | 436 | GQ129692 | This study | ||
| Helitron | DNAREP1_DYak | NA | Gcin32i | 74 | 101 | 414 | GQ129711 | This study | ||
| Helitron | DNAREP1_DYak | NA | Gcin42i | 80 | 101 | 471 | GQ129721 | This study | ||
| Helitron | DNAREP1_DYak | NA | Gcin47i | 66 | 122 | 376 | GQ129726 | This study | ||
| Helitron | Helitron-N1_SP | NA | Gcin46i | 73 | 87 | 359 | GQ129725 | This study | ||
| LTR retrotransposon | ||||||||||
| ERV1 | ERV46_MD_I | NA | Gcin53i | 74 | 150 | 489 | GQ129732 | This study | ||
| ERV1 | HERV17 | NA? | Gcin54 | 72 | 143 | 264 | GQ129733 | This study | ||
| ERV3 | MonoRep289C | NA? | Gcin26i | 80 | 78 | 212 | GQ129705 | This study | ||
| L. littorea | ||||||||||
| DNA transposon | ||||||||||
| En/Spm (CACTA) | EnSpm-1_HV | A? | Llit1 | 84 | 76 | 424 | FJ480214 | McInerney et al. (2009a) | ||
| En/Spm (CACTA) | EnSpm-1_HV | A? | Llit110 | 86 | 40 | 247 | GQ129786 | This study | ||
| En/Spm (CACTA) | ATENSPM5 | A | Llit65 | 74 | 85 | 247 | GQ129744 | This study | ||
| Mariner | Mariner-6_HM | NA? | Llit17 | 74 | 94 | 263 | FJ480220 | McInerney et al. (2009a) | ||
| Mariner | Mariner-4A_AF | NA | Llit64 | 88 | 33 | 231 | GQ129743 | This study | ||
| hAT | hAT-16_ZM | A | Llit64 | 73 | 60 | 222 | GQ129743 | This study | ||
| Arnold | ARNOLD1 | A | Llit66 | 89 | 34 | 222 | GQ129745 | This study | ||
| MuDR (MULE) | MuDR1_ZM | A | Llit66 | 88 | 32 | 216 | GQ129745 | This study | ||
| LTR retrotransposon | ||||||||||
| Gypsy | Gypsy41-LTR_DR | ? | Llit50 | 78 | 55 | 212 | FJ480221 | McInerney et al. (2009a) | ||
| Non-LTR retrotransposon | ||||||||||
| LINE | L2_Plat1h | A? | Llit55 | 68 | 135 | 301 | FJ480218 | McInerney et al. (2009a) | ||
| SINE | SINE2-4a1_SP | NA? | Llit62 | 91 | 53 | 251 | GQ129741 | This study | ||
| SINE | SINE2-1_SP | NA | Llit80i | 85 | 46 | 301 | GQ129759 | This study | ||
| L. saxatilis | ||||||||||
| DNA transposon | ||||||||||
| MuDR (MULE) | MDR2 | A? | Lsax20 | 80 | 83 | 322 | FJ794783 | McInerney et al. (2009b) | ||
| Mu-like | MERMITE-18 | NA? | Lsax66 | 80 | 82 | 262 | GQ129838 | This study | ||
Class of transposable element.
Transposable element description as defined by Repbase.
Name of known transposable element provided by Repbase.
Autonomous/non-autonomous/not specified transposition mechanism according to Repbase or Web of Science.
Value of similarity between two aligned sequences (for details see Kohany et al., 2006).
Alignment length between two aligned DNA sequences.
Sum of match/mismatch scores and penalties for gap opening/extension estimated from the alignment matrix (Kohany, personal communication).
GenBank accession numbers.
The MCS grouped into a DNA family based on sequence similarities among flanking regions (for more details see Table 3).
The most frequently identified TE in G. cineraria was the miniature inverted-repeat transposable element (MITE), CvA (Gaffney et al., 2003). CvA is part of a family of TEs known as pearl that was initially described in the oyster, Crassostrea virginica (Gaffney et al., 2003). A total of 19 copies of CvA were detected in 14.7 kb MCS from G. cineraria (1.29 copies per kb). Hits for CvA were on average 66 bp (range, 24–70 bp) in alignment length with varying degrees of homology (66–100%), and they all occurred between nucleotide positions 297–430 bp of the published sequence (∼600 bp). This region overlaps with the conserved terminal sequence and contains a proto-microsatellite (GACA)n and an RNA polymerase III promoter BoxA. To assess the relative abundance of CvA in molluscan genomes, we designed a PCR primer set (Table 1) to amplify the conserved terminal sequence of CvA in host genomic DNA and in the genomic DNA of phylogenetically close related species (Experiment II). Reliable amplification products were observed both from host genomic DNA (G. cineraria) and from the trochids G. umbilicalis, G. magus and O. lineatus. The presence of multiple amplified PCR fragments suggests that CvA are both abundant and phylogenetically conserved in close relatives from the family Trochidae. They are, however, absent in the genomes of C. zizyphinum and in the other littorinid species tested. The second most common TE identified from G. cineraria was the helitron DNAREP1_DYak (N=4). This TE is ∼793 bp in length and was initially identified from Drosophila yakuba (Kapitonov and Jurka, 2007). It is a deletion derivative of the autonomous Helitron-1_DYak, which is usually inserted in the ttw∣TTT target sites without the target site duplications (Kapitonov and Jurka, 2007).
Interestingly, although several different classes of TEs were identified from the MCS of six of the DNA families, in each case they consisted of non-autonomous TEs (families 3, 4, 5, 6, 7, 12). Specifically these included the DNA transposons CvA, the helitrons DNAREP1_DYak and Helitron-N1_SP, the LTR retrotransposon endogenous retroviruses MonoRep289C and ERV46_MD_I, and the Non-LTR retrotransposon SINE2-1_SP. These were predominantly found in association with the microsatellite repeat motifs CCAT.
Discussion
Inter-specific variation in cryptic repetitive DNA and DNA family abundance
Genomic complexities such as cryptic repetitive DNA and DNA family abundance identified in association with microsatellites have been commonly reported from plants, insects and crustaceans (Tero et al., 2006; Meglécz et al., 2004, 2007; Van't Hof et al., 2007; Bailie et al., 2010). This is the first study, however, to describe their occurrence and frequency in the largest class of molluscs, the gastropods. Previous reports of multiple-banding patterns observed during microsatellite development in other molluscs (Reece et al., 2004; Weetman et al., 2005) seem to suggest that this genomic idiosyncrasy may be far more widespread. Non-reporting of ‘negative' results in the published literature is likely to be responsible for their underestimation. Among distantly related molluscs, similar associations involving other classes of repetitive DNA have also been suggested in the scallop Pecten maximus, oyster C. virginica and clam Anadara trapezia (Gaffney et al., 2003; Biscotti et al., 2007).
In comparable studies that similarly attempted to quantify the frequency of cryptic repetitive DNA associated with MCS, the highest proportion of MCS that grouped into DNA families was just 55% (Meglécz et al., 2007). Thus, to the best of our knowledge, G. cineraria appears to have the greatest abundance of DNA families (74.6%), so far reported from invertebrates. The majority of cryptic repetitive DNA identified in this study, which grouped into distinct DNA families, was restricted to single species. This is congruent with reports from other studies involving insects and crustaceans (Meglécz et al., 2004, 2007; Van't Hof et al., 2007; Bailie et al., 2010). The occurrence of cryptic repetitive DNA grouped into DNA families can present a significant problem for the development of microsatellite markers. In this study, we described a possible solution to facilitate microsatellite marker development from genomic regions harbouring DNA families. This approach capitalized upon the existence of discrete nucleotide differences between DNA family members and could be implemented also for other taxa (molluscs, insects) where marker development has been otherwise unsuccessful.
The provision of the GCS database for the conserved mollusc cryptic repetitive DNA should prove useful for future studies. Shared similarities between GCS and MCS from bivalve and pelecypods suggest that the GCS are phylogenetically conserved among very distantly related molluscs. Interestingly, the majority of the cryptic repetitive DNA identified in this study were novel, thus suggesting that gastropod mollusc genomes harbour many as yet uncharacterized genomic elements.
TE abundance and DNA multiplication
TEs have been previously reported for the distantly related mollusc classes Pelecypoda and Bivalvia also in association with tandem repeated regions (Gaffney et al., 2003; Biscotti et al., 2007). This, however, is the first study to report the widespread occurrence of TEs in the largest mollusc class, the Gastropoda. The quantification of TEs in this study, however, is most likely an underestimate. This is because the identification of TEs is heavily reliant upon database entries, for which characterized molluscan TEs are still lacking. Nonetheless, we report greater abundance of TEs from microsatellite flanking regions, compared with all other previous similar studies (Meglécz et al., 2004; Van't Hof et al., 2007).
The comparative genomic analysis revealed that the three gastropods differed quite considerably in their TE abundance. The larger number of TEs in G. cineraria could explain a possible higher rate of genomic rearrangement processes occurring in this species, as a result of TE transpositional activity. In congruence with this hypothesis, we observed that G. cineraria had the highest amount of genomic complexities (cryptic repetitive DNA and DNA families) and increasingly unstable microsatellite flanking regions. Thus, it is reasonable to assume a possible link between DNA family frequencies and TEs in gastropods. This provides additional supporting evidence from a separate phylum, for the hypothesis that the creation of DNA families is mediated by TEs as suggested by Meglécz et al. (2004). Overall frequency of TEs and hence differential rates of DNA multiplication processes may not have been the only important factor to explain inter-specific variation in genome stability and marker development success rates.
Closer examination of the results revealed that the gastropods also differed significantly in their complement of TEs and the proportion of TEs that were identified as autonomous vs non-autonomous classes. One possibility is that dissimilar transpositional mechanisms differentiate the TE classes in terms of their propensity for fixation in the genome. As molluscs undergo high substitution rates (Davison, 2002), mutational changes in coding sequences necessary for transposition would lead to greater propensity for fixation of autonomous TEs. Conversely, non-autonomous TEs that ‘hijack' the machinery of partner TEs to accomplish their transposition (Feschotte et al., 2002a, 2002b) would probably be unaffected. Furthermore, as non-autonomous TEs continue to proliferate, their transposition would result in higher frequencies of mutational changes (Jiang et al., 2003; Nakazaki et al., 2003). This would further accelerate the process of fixation of autonomous TEs. Thus, as the combined result of these processes, compared to autonomous TEs, non-autonomous TEs may be more highly conserved and have higher transpositional activities. In this study, a number of different lines of evidence seem to support this new hypothesis.
First and foremost was the identification of a variety of exclusively non-autonomous TEs from the flanking regions of the DNA families. Second, the high number of non-autonomous TEs with highly conserved regions (for example, MITE, CvA) provides corroborating evidence that non-autonomous TEs in the G. cineraria genome have undergone recent transpositional activity (Ray, 2007; Kass et al., 2009). Thus, this indicates a lack of propensity for fixation for this TE class in the latter species. Non-autonomous TEs such as MITEs usually attain high copy numbers in the genome of many taxa (Feschotte et al., 2002b; Ray et al., 2005). Nonetheless it was interesting to note that MCS of G. cineraria contained a 10-fold greater copy number of CvA (1.29 copies per kb) compared to genomic DNA of C. virginica (0.19 copies per kb; Gaffney et al., 2003). A possible hypothesis is that non-autonomous TEs, such as CvA, tend to accumulate in genomic regions that are predominantly neutral, such as microsatellite regions. Remarkably, the identified regions, without exception, corresponded to an RNA polymerase III promoter BoxA. This promoter is involved in the transcription of TEs before the recruitment of enzymes from other TEs involved in their mobilization (Ray, 2007). Our results indicate that compared to autonomous TEs, non-autonomous TEs maybe have a more active role in the rearrangement processes occurring in mollusc (and possibly other) genomes. This mechanism may have been an important factor in determining the inter-specific variation observed among the three gastropods.
Alternatively, autonomous vs non-autonomous TEs may differ in their ability to successfully evade TE transcription silencing systems. These systems hamper the transcription and subsequent transposition of TEs through a process that involves RNA interference and sometimes DNA methylation (see Weil and Martienssen (2008) for a review). Interestingly, Weil and Martienssen (2008) suggest that TEs that are abundant, for instance non-autonomous MITEs, have successfully evaded TE transcription silencing systems. Although the authors have not suggested a direct link with non-autonomous TE transpositional mechanisms, we cannot rule out this hypothesis. The identification of active TE-transposase systems in other non-autonomous TEs, such as the mPing/Pong system in rice (see Feschotte and Pritham (2007) for review) should allow for the experimental investigation of these hypotheses in future investigations.
TE abundance and DNA recombination
Differential rates of DNA recombination processes may have also been an important factor to explain inter-specific variation in genome stability and marker development success rates.
In this investigation, we present additional evidence to support this by gene conversion and unequal crossing over. Gene conversion is the non-reciprocal exchange of genetic material among chromosomes and can be mediated by TE transposition (Meglécz et al., 2004). Specifically the non-autonomous TEs helitrons can capture and move gene fragments and are responsible for gene duplication and conversion (Morgante et al., 2005; Hollister and Gaut, 2007). This process can even lead to the formation of novel genes (Lockton and Gaut, 2009). Although Helitrons were not identified from the littorinids, the helitron DNAREP1_DYak was the second most highly abundant TE identified in G. cineraria. This TE is related to DNAREP1_DM, the most highly abundant TE in the Drosophila melanogaster genome (Kapitonov and Jurka, 2003). Thus, it may be possible that recombination rates are higher in G. cineraria due to the abundance of helitrons. The associated genomic rearrangements of helitrons transpositional activities can also explain genomic complexities and instability of microsatellite flanking regions in this species.
Unequal crossing over the other mechanism can result when ‘a chiasma occurs at two imperfectly aligned microsatellites with shared repeat units, leaving two new upstream–downstream combinations' (Van't Hof et al., 2007). The asymmetrical arrangement of similar microsatellite flanking regions from a DNA family is usually indicative of it having undergone unequal crossing over (Meglécz et al., 2004). Evidence for this type of arrangement was identified from all three species, thus it may not have been an important factor in differentiating the microsatellite flanking regions among species.
TE abundance and microsatellite proliferation
The genesis, behaviour and evolution of microsatellites within a genome are still a subject for ongoing discussions. One suggestion is that TEs that carry microsatellite repeated regions may have behaved as microsatellite-inducing elements in the host genomes of the plants pea and barley and some insects (Ramsay et al., 1999; Wilder and Hollocher, 2001; Coates et al., 2009; Smýkal et al., 2009). Likewise, Gaffney et al. (2003) proposed that CvA may be an ancient TE that has behaved as a source of satellite DNA in distantly related bivalves. This process would have involved the conversion of a proto-microsatellite (or imperfect microsatellite) contained within a TE, into heterochromatic satellite DNA (Wilder and Hollocher, 2001). Herein, we provide supporting evidence for the hypothesis that the non-autonomous TE, CvA, has behaved as a microsatellite dispersal agent in gastropod mollusc genomes. The phylogenetic conservation of CvA among the family Trochidae, in addition to the distantly related Bivalvia (Gaffney et al., 2003), confirms that CvA is an ancient molluscan TE. Furthermore, the establishment of the close association between this MITE and DNA families in G. cineraria observed in this study supports the idea that CvA is a dispersal agent for microsatellites, through hitchhiking within TEs during transposition (Coates et al., 2009; Smýkal et al., 2009). These processes can also explain the formation and account for the abundance of multilocus DNA families in G. cineraria. Finally, Wilder and Hollocher (2001) determined that the conversion of a proto-microsatellite (or imperfect microsatellite) contained within a TE into heterochromatic satellite DNA often leads to the existence of exclusively two tetra-nucleotide microsatellite repeat arrays. In our study, we likewise identified an abundance of imperfect tetra-nucleotide repeat motifs associated with the DNA families.
Conclusion
The study of MCS and neighbouring genomic regions is important to increase our knowledge about microsatellite genesis and evolution. Furthermore, it could also provide extra molecular tools available to resolve phylogenetic relationships and assist in the identification of population genetic structure. In this study, the comparative genomic analysis revealed considerable inter-specific variation in genomic complexity and stability of microsatellite flanking regions. We provide novel evidence regarding the differential importance of autonomous vs non-autonomous TEs in DNA multiplication and recombination rearrangement processes that explain genomic complexities. The discovery of many novel gastropod cryptic repetitive DNA associated with DNA families should provide a basis for further research into the description of new TEs. Molluscs, therefore, may prove useful as model genomes to investigate TE involvement in the behaviour and evolution of microsatellites and other genomic elements. Further research that involves whole-genome sequence data sets is required, however, to extend our understanding of the trends and observations outlined in this study to other genomic regions.
Acknowledgments
C McInerney was supported by a North-South strand 1 grant from the Higher Education Authority, Ireland awarded to MP Johnson, AL Allcock and PA Prodöhl. We are grateful to O Mulholland, M Jessopp, J Leal-Flórez, J Nunn and M McInerney for assistance with fieldwork. We thank P Watts, JM Pujolar, J Provan and H Fletcher for helpful discussions. We thank J Nunn (Ulster Museum) for providing us with Gibbula magus samples. Marine research in PA Prodöhl's laboratory is currently supported by the Beaufort Marine Research Award in Fish Population Genetics funded by the Irish Government under the Sea Change programme.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthofer W, Schlick-Steiner BC, Steiner FM, Avtzis DN, Crozier RH, Stauffer C. Lessons from a beetle and an ant: coping with taxon-dependent differences in microsatellite development success. J Mol Evol. 2007;65:304–307. doi: 10.1007/s00239-007-9012-1. [DOI] [PubMed] [Google Scholar]
- Bailie DA, Fletcher H, Prodöhl PA.2010High incidence of cryptic repeated elements in microsatellite flanking regions of galatheid genomes and its practical implications for molecular marker development J Crust Biol(in press).
- Biscotti MA, Canapa A, Olmo E, Barucca M, Teo CH, Schwarzacher T, et al. Repetitive DNA, molecular cytogenetics and genome organization in the King scallop (Pecten maximus) Gene. 2007;406:91–98. doi: 10.1016/j.gene.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetic studies. Mol Ecol. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Carreras-Carbonell J, Macpherson E, Pascual M. Population structure within and between species of the Mediterranean triplefin fish Tripterygion delaisi revealed by highly polymorphic microsatellite loci. Mol Ecol. 2006;15:3527–3539. doi: 10.1111/j.1365-294X.2006.03003.x. [DOI] [PubMed] [Google Scholar]
- Coates BS, Sumerford DV, Hellmich RL, Lewis LC. Repetitive genome elements in a European corn borer, Ostrinia nubilalis bacterial artificial chromosome library were indicated by bacterial artificial chromosome end sequencing and development of sequence tag site markers: implications for lepidopteran genomic research. Genome. 2009;52:57–67. doi: 10.1139/G08-104. [DOI] [PubMed] [Google Scholar]
- Dakin EE, Avise JC. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. [DOI] [PubMed] [Google Scholar]
- Davison A. Land snails as a model to understand the role of history and selection in the origins of biodiversity. Popul Ecol. 2002;44:129–136. [Google Scholar]
- DeWoody J, Nason JD, Hipkins VD. Mitigating scoring errors in microsatellite data from wild populations. Mol Ecol Notes. 2006;6:951–957. [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002a;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham E. DNA transposons and the evolution of eukaryotic genomes. Ann Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Zhang X, Wessler SR.2002bMiniature inverted-repeat transposable element in their relationship to established DNA transposonsIn: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds).Mobile DNA II American Society of Microbiology: Washington, DC; 1147–1158. [Google Scholar]
- Gaffney PM, Pierce JC, MacKinley AG, Titchen DA, Glenn WK. Pearl, a novel family of putative transposable elements in bivalve molluscs. J Mol Evol. 2003;56:308–316. doi: 10.1007/s00239-002-2402-5. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Population and evolutionary dynamics of helitron transposable elements in Arabidopsis thaliana. Mol Biol Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao ZR, Zhang XY, Hirochika H, Eddy SR, McCouch SR, et al. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. Helitrons in fruit flies. Repbase Rep. 2007;7:127. [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- Kass DH, Schaetz BA, Beitler L, Bonney KM, Jamison N, Wiesner C. Guinea pig ID-like families of SINEs. Gene. 2009;436:23–29. doi: 10.1016/j.gene.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;25:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Gaut BS. The contribution of transposable elements to expressed coding sequence in Arabidopsis thaliana. J Mol Evol. 2009;68:80–89. doi: 10.1007/s00239-008-9190-5. [DOI] [PubMed] [Google Scholar]
- McInerney CE, Allcock AL, Johnson MP, Prodöhl PA. Characterization of polymorphic microsatellite loci for the periwinkle intertidal gastropod Littorina littorea (Linneaus 1758) and their conservation in four congeners. Conserv Genet. 2009a;10:1417–1420. [Google Scholar]
- McInerney CE, Allcock AL, Johnson MP, Prodöhl PA. Characterization of polymorphic microsatellite loci for the rough periwinkle gastropod Littorina saxatilis (Olivi, 1972) and their conservation in four congeners. Conserv Genet. 2009b;10:1989–1992. [Google Scholar]
- Meglécz E, Anderson SJ, Bourgett X, Butcher R, Caldas A, Cassel-Lundhagen AC, et al. Microsatellite flanking region similarities among different loci within insect species. Insect Mol Biol. 2007;16:175–185. doi: 10.1111/j.1365-2583.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- Meglécz E, Petenian F, Danchin E, Coeur D'acier A, Rasplus J, Faure E. High similarity between flanking regions of different microsatellites detected within each of two species of Lepidoptera: Parnassius apollo and Euphydryas aurinia. Mol Ecol. 2004;13:1693–1700. doi: 10.1111/j.1365-294X.2004.02163.x. [DOI] [PubMed] [Google Scholar]
- Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, et al. Mobilization of a transposon in the rice genome. Nature. 2003;421:170–173. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, Cardle L, Morgante M, degli Ivanissevich S, Maestri E, et al. Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J. 1999;17:415–425. doi: 10.1046/j.1365-313x.1999.00392.x. [DOI] [PubMed] [Google Scholar]
- Ray DA. SINEs of progress: mobile element applications to molecular ecology. Mol Ecol. 2007;16:19–33. doi: 10.1111/j.1365-294X.2006.03104.x. [DOI] [PubMed] [Google Scholar]
- Ray DA, Hedges DJ, Herke SW, Fowlkes JD, Barnes EW, LaVie DK, et al. Chompy: an infestation of MITE-like repetitive elements in the crocodilian genome. Gene. 2005;362:1–10. doi: 10.1016/j.gene.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Reece KS, Ribeiro WL, Gaffney PM, Carnegie RB, Allen SK. Microsatellite marker development and analysis in the eastern oyster (Crassostrea virginica): confirmation of null alleles and non-mendelian segregation ratios. J Hered. 2004;95:346–352. doi: 10.1093/jhered/esh058. [DOI] [PubMed] [Google Scholar]
- Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- Smýkal P, Kalendar R, Ford R, Macas J, Griga M. Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity. 2009;103:157–167. doi: 10.1038/hdy.2009.45. [DOI] [PubMed] [Google Scholar]
- Tero N, Neumeier H, Gudavalli R, Schlotterer C. Silene tatarica microsatellites are frequently located in repetitive DNA. J Evol Biol. 2006;19:1612–1619. doi: 10.1111/j.1420-9101.2006.01118.x. [DOI] [PubMed] [Google Scholar]
- Van't Hof AE, Brakefield PM, Saccheri IJ, Zwann BJ. Evolutionary dynamics of multilocus arrangements in the genome of the butterfly Bicyclus anynana, with implications for Lepidoptera. Heredity. 2007;98:320–328. doi: 10.1038/sj.hdy.6800944. [DOI] [PubMed] [Google Scholar]
- Weetman D, Hauser L, Carvalho GR. Isolation and characterisation of di- and trinucleotide microsatellites in the freshwater snail Potamopyrgus antipodarum. Mol Ecol Notes. 2001;1:185–187. [Google Scholar]
- Weetman D, Hauser L, Shaw PW, Bayes MK. Microsatellite markers for the whelk Buccinum undatum. Mol Ecol Notes. 2005;5:361–362. [Google Scholar]
- Weil C, Martienssen R. Epigenetic interactions between transposons and genes: lessons from plants. Curr Opin Genet Dev. 2008;18:188–192. doi: 10.1016/j.gde.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Wilder J, Hollocher H. Mobile elements and the genesis of microsatellites in dipterans. Mol Biol Evol. 2001;18:384–392. doi: 10.1093/oxfordjournals.molbev.a003814. [DOI] [PubMed] [Google Scholar]
- Zhang D. Lepidopteran microsatellite DNA: redundant but prising. Trends Ecol Evol. 2004;19:507–509. doi: 10.1016/j.tree.2004.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.