Abstract
The house sparrow Passer domesticus has been declining in abundance in many localities, including Finland. We studied the genetic diversity and differentiation of the house sparrow populations across Finland in the 1980s, at the onset of the species' decline in abundance. We genotyped 472 adult males (the less dispersive sex) from 13 locations in Finland (covering a range of 400 × 800 km) and one in Sweden (Stockholm) for 13 polymorphic microsatellite markers. Our analysis of Finnish ringing records showed that natal dispersal distances are limited (90% <16 km), which confirmed earlier finding from other countries. The Finnish populations were panmictic, and genetically very homogeneous and the limited dispersal was sufficiently large to maintain their connectivity. However, all Finnish populations differed significantly from the Stockholm population, even though direct geographical distance to it was often smaller than among Finnish populations. Hence, the open sea between Finland and Sweden appears to form a dispersal barrier for this species, whereas dispersal is much less constrained across the Finnish mainland (which lacks geographical barriers). Our findings provide a benchmark for conservation biologists and emphasize the influence of landscape structure on gene flow.
Keywords: dispersal, DNA microsatellites, FST, gene flow, house sparrow, spatial population structure
Introduction
To make long-term conservation decisions, we need information on population structure and connectivity (Moritz, 1994). Dispersal ability, geographic variation, level of genetic differentiation and adaptation to local conditions are key aspects of a species' ecology (Frankham et al., 2002; Hartl and Clark, 2007), but gathering information on these aspects through traditional methods is often laborious and costly, or simply infeasible (Lowe et al., 2004). In many organisms, individuals can be marked and followed to determine population structure and track the movement of individuals, but this method cannot be used for all species for practical reasons. For example, dispersal of birds can be studied by monitoring individuals marked by ringing (that is, attaching an individually numbered metal or plastic ring on the leg), but not all species are equally well represented in ringing records (Baillie, 1995). When information based on traditional ecological techniques is patchy, a genetic approach can complement the picture because the data required can be gained by sampling a small part of a population in a short time.
Due to their ability to fly, birds can move over large geographical scales and their populations are therefore often spatially more homogeneous than in some other taxonomic groups (Barrowclough, 1983; Evans, 1987; Ward et al., 1992). However, there are some striking exceptions from this general pattern. Senar et al. (2006) found two populations of the highly mobile citril finch Serinus citrinella to be significantly differentiated (FST=0.094), even though their breeding areas were separated by only 5 km and the two populations mixed in a common overwintering area. Significant genetic differentiation (FST=0.043) was found also among migratory Swainson's warbler populations Limnothlypis swainsonii (Winker et al., 2000).
The medium-sized passerine, house sparrow Passer domesticus, is one of the avian species most associated with humans, and familiar to many. It is a sexually dimorphic, social and very site-tenacious bird (Summers-Smith, 1988). Its natural range includes Europe, North Africa and parts of Asia such as Middle East, Indian subcontinent and a narrowing band from northern Asia toward the Pacific coast. From these areas its large-scale spread has been enabled by humans who have introduced it either intentionally or by accident to almost every corner of the world, including North and South America, South Africa, Australia and New Zealand (Summers-Smith 1963, 1988; Anderson, 2006). The house sparrow has been very abundant all over the world, but major declines in its abundance have been reported in large parts of Europe during the last few decades (Summers-Smith, 1999; Crick and Siriwardena, 2002; Hole et al., 2002; Anderson, 2006; de Laet and Summers-Smith, 2007). Also non-native populations have been reported to decline in, for example, North America (Peterjohn et al., 1994). Reasons for these declines are not fully understood.
Conservation efforts should take into account knowledge on genetic population structure and the amount of gene flow between them, and focus management on units that are significantly genetically differentiated (Moritz, 1994). Field observations in the United Kingdom and on island populations in northern Norway indicate that the house sparrow is a resident species with limited dispersal undertaken mainly by juveniles, and that females are also more likely to disperse from their native areas than males (Summers-Smith, 1988; Skjelseth et al., 2007). Using ringing recovery data in the United Kingdom, Paradis et al. (1998) found that a mean natal dispersal distance was less than 2 km. Furthermore, long-distance recoveries from Britain and Ireland showed that only 3% of dispersal events extend further than 20 km (Siriwardena et al., 2002). In Norway only 10% of female and 6% of male fledglings that were later recruited to the breeding population had left their native islands (Altwegg et al., 2000). Furthermore, Tufto et al. (2005) found that most house sparrows did not disperse from their natal farms, and of the ones that did, 90% dispersed less than 36 km. Low dispersal suggests that populations may also be spatially structured. Nevertheless, the number of migrants per generation needed to homogenize populations is relatively low (Franklin, 1980; Frankel and Soulé, 1981; Allendorf, 1983), and low dispersal indicated by the field data need not translate into genetic differentiation between populations. Significant genetic differentiation has been found in Norway on the scale of less than 100 km (FST among mainland populations 0.018, FST among island populations 0.025; Jensen et al., in preparation), but earlier allozyme studies have not found significant genetic differentiation (Fleischer, 1983; Parkin and Cole, 1984, 1985). Thus, it is currently not clear whether low observed dispersal in the field indeed implies that house sparrow populations are genetically differentiated on a modest spatial scale or whether high-resolution DNA microsatellite markers are better able to capture genetic differentiation across house sparrow populations than allozymes.
In this work, we carry out a DNA microsatellite analysis of the house sparrow from an extensive material covering the species' entire distribution in Finland. Our aim is to gain insight into the spatial structure and connectivity of the populations and the levels of genetic diversity within them. We used historical house sparrow material collected in the mid-1980s, when the species was highly abundant in Finland (estimated at 400 000 pairs; Väisänen et al., 1998) but starting to decline. We use national ringing records to show that estimated field-based dispersal is low, but our genetic data indicate that, in absence of geographical barriers, house sparrows are panmictic over several hundreds of kilometers. We discuss our findings in relation to previous studies and provide a conservation genetic outlook.
Materials and methods
Ringing records
Individually marking animals (for example, by ringing them) is a traditional ecological method for estimating dispersal rates and distances. In the house sparrows, most of the species' dispersal is conducted by juvenile birds. Ringing records from various localities have indicated that house sparrow natal dispersal is limited. We analyzed Finnish ringing records (obtained from the Ringing Bureau, Finnish Museum of Natural History, Helsinki) to summarize the available information on natal dispersal in Finnish house sparrows from 1930 to 2004. We considered data on individuals ringed as a nestling and either found dead or caught alive on or after 1 March in the year after hatching. Despite their high abundance, house sparrows are rarely ringed, probably because they nest at sites that are difficult to access and are sensitive to nest disturbance. Due to the paucity of ringing records of natal dispersal (n=87), we simply pooled the entire data set and did not restrict analyses to any particular time period or sex.
In addition we looked at adult dispersal; that is, birds ringed as adults and caught the next calendar year or after that (n=1110). Sex-specific dispersal was determined from individuals with known sex (males, n=543; females, n=388).
Sampling
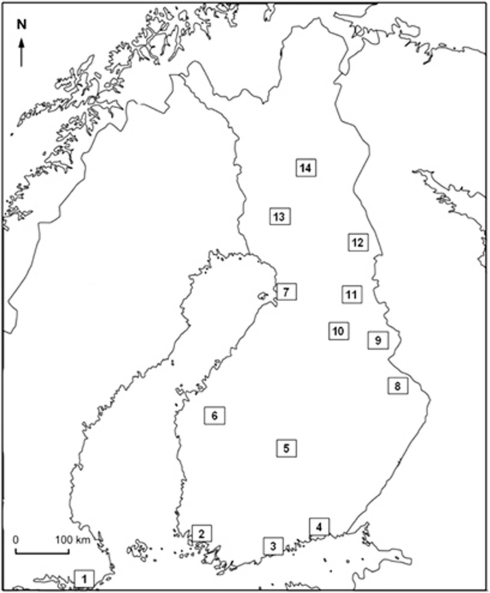
Tissue samples used in this study were collected between 1983 and 1986 from 13 localities in Finland (Figure 1, Table 1). Sampled locations included both urban and rural sites. Birds were sampled with permission by catching with mist nets and killed with carbon dioxide. Skins were dried and bones were cleared from tissue and preserved dry. Livers and kidneys were preserved at −18 °C. Samples were also collected in Stockholm (Sweden) for a comparison. For this study, we used only males, because these are supposedly the more sedentary sex (Summers-Smith 1988; Altwegg et al., 2000; Skjelseth et al., 2007). In addition, we selected only males that had survived their first winter (second calendar year or older, based on ossification of their skulls, Svensson, 1975) as they could be assumed to be resident individuals. Hence, the level of differentiation among the populations should be highest in this group.
Figure 1.
Map of Finland representing the sampling localities of house sparrows indicated by numbers from 1 to 14. Numbers refer to population names in Table 1. There are 13 sampling sites in Finland and 1 in Stockholm, Sweden. The edge of the map corresponds to north.
Table 1. Number of adult males sampled, by year, for the molecular analysis of this study.
| Population | 1983 | 1984 | 1985 | 1986 | Total | |
|---|---|---|---|---|---|---|
| 1 | Stockholm | 33 | 33 | |||
| 2 | Turku | 33 | 12 | 45 | ||
| 3 | Helsinki | 26 | 33 | 59 | ||
| 4 | Myllykoski | 7 | 22 | 29 | ||
| 5 | Jyväskylä | 23 | 12 | 35 | ||
| 6 | Seinäjoki | 30 | 30 | |||
| 7 | Oulu | 38 | 38 | |||
| 8 | Lieksa | 30 | 30 | |||
| 9 | Kuhmo | 36 | 36 | |||
| 10 | Kajaani | 25 | 25 | |||
| 11 | Ämmänsaari | 23 | 23 | |||
| 12 | Kuusamo | 14 | 7 | 21 | ||
| 13 | Rovaniemi | 29 | 19 | 48 | ||
| 14 | Sodankylä | 20 | 20 | |||
For these individuals, 11 or more loci were successfully genotyped.
Microsatellite genotyping
DNA was extracted from a small piece of liver tissue following the method described by Elphinstone et al. (2003), except that 70 μl of dH2O was used to elute DNA in the last step.
The samples were amplified in PCR for 14 microsatellite loci in two parallel panels. Panel 1 included primers Pdoμ16, Pdoμ19, Pdoμ22, Pdoμ27, Pdoμ32, Pdoμ44 and Pdoμ47 (Dawson et al., in preparation) and panel 2 primers Ase18 (Richardson et al., 2000), Pdoμ1 and Pdoμ3 (Neumann and Wetton, 1996), Pdoμ10 (Griffith et al., 2007), Pdoμ33 and Pdoμ40 (Dawson et al., in preparation) and Pdoμ5 (Griffith et al., 1999). PCR was conducted with Multiplex PCR kit (Qiagen), where one PCR reaction contained 5 μl of Qiagen Master mix-solution (Qiagen, Hilden, Germany), 3 μl of extracted DNA and 2 μl of primer mix (containing 0.28–0.94 μ of each primer). PCRs for both panels were completed using the following touchdown protocol on a Satellite Thermal Cycler MBS 0.2G (Thermo Fisher Scientific, Waltham, MA, USA): one denaturing step of 15 min at 94 °C followed by 12 cycles of 30 s at 94 °C, 90 s initially at a temperature of 62 °C (then decreasing by 1° every cycle), and 60 s at 72 °C. This was followed by 20 cycles of 30 s at 94 °C, 90 s at 50 °C and 60 s at 72 °C. Finally there was an additional 5 min at 60 °C and an indefinite hold at 4 °C.
PCR products were separated and visualized with MegaBACE 1000 (Amersham Biosciences), and genotypes were scored using the software package Fragment Profiler 1.2 (Amersham Biosciences, Buckinghamshire, UK).
Statistical analysis
The 14 microsatellite loci were checked for Hardy–Weinberg equilibrium with the program Cervus 3.0 (Marshall et al., 1998; Kalinowski et al., 2007) where Bonferroni correction was used to evaluate the significance of any deviation from equilibrium. Linkage disequilibrium, that is, whether the loci are independent from each other in each population, was tested with statistical software Genepop 3.4 (Raymond and Rousset, 1995) using the default dememorization number (1000), 100 batches and 1000 iterations per batch.
Genetic diversity within the populations was described with three statistics: allelic richness (AR), private allelic richness (APR) and expected heterozygosity according to Hardy–Weinberg equilibrium (HE). Allelic richness and private allelic richness (AR and APR) are genetic diversity measures that take into account uneven sample sizes by performing rarefaction, and calculate a statistic that is comparable across samples. These estimates were obtained using programs FSTAT 2.9.3 (Goudet, 2001) (for AR) and HP-RARE (Kalinowski, 2005) (for APR). Expected heterozygosity, as well as the inbreeding coefficient (FIS), was calculated for each population using FSTAT.
The level of genetic differentiation among the populations was assessed as FST (Wright, 1943) by using the θ estimator (Weir and Cockerham, 1984) as implemented in the software FSTAT 2.9.3 (Goudet, 2001). FST was estimated both globally and between all pairs of populations. To evaluate the significance of the pairwise tests, the program used randomizations (1820 times) for the genotypes. These P-values were then corrected with sequential Bonferroni correction that takes into account multiple tests instead of the standard Bonferroni correction that the program used. Mantel's test was used to test for isolation by distance; that is, correlation between the level of genetic differentiation and geographic distance. Only populations from Finland were used in this test.
The software Structure 2.2 (Pritchard et al., 2000; Falush et al., 2003a, 2007) was used to cluster populations into groups based on individual genetic data without any prior information on geographic sampling locality. Structure defines populations such that populations are more similar within the cluster than with populations in other clusters. Length of the burn-in period was set to 10 000 and the number of Markov chain Monte Carlo repetitions to 10 000. Simulation was ran 10 times for each values of K. Individuals were also assigned to populations using GeneClass 1.0.02 (Cornuet et al., 1999).
The program BOTTLENECK 1.2.02 (Cornuet and Luikart, 1996) was used to test whether study populations had gone through a decline in population size in recent history. In a bottlenecked population, allele numbers decline faster than heterozygosity, and the observed heterozygosity should be larger than the expected based on the allele count in the population. Here the expected heterozygosity was estimated by assuming the two-phase mutation model as recommended for microsatellite data. The variance of mutations was set to 30 and the proportion of mutations larger than one step to 30%. Significance of the mismatch between the observed and expected heterozygosities was tested by using the Wilcoxon test and the visual graphic test (Cornuet and Luikart, 1996).
Results
House sparrow dispersal in Finland based on ringing records
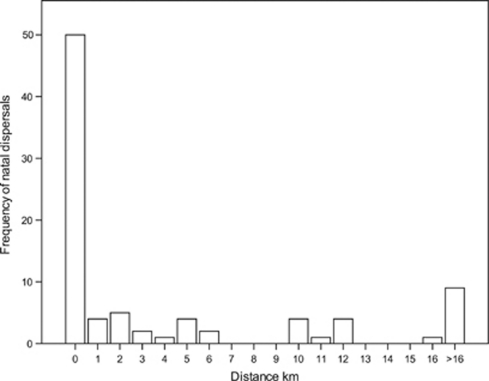
Median natal dispersal distance for house sparrows (n=87) was 0 km, and only 10% of individuals dispersed further than 16 km (Figure 2). Hence, the available ringing records suggest that natal dispersal of Finnish house sparrows is highly limited. Furthermore, adult dispersal (n=1110) was also expectedly low; that is, median was 0 km and only 10 % dispersed more than 3 km. When considering the sexes separately both had a median of 0 km, but from males 10% dispersed more than 2 km whereas from females 10% moved more than 5 km.
Figure 2.
Distribution of natal dispersal distances of Finnish house sparrows based on ringing data from 1930 to 2004.
Samples
For testing the quality of the data (that is, linkage equilibrium, Hardy–Weinberg equilibrium and sufficiency of the loci), we used only individuals that were successfully genotyped for all 14 loci (n=391). For all other analyses, we used individuals that had 11 or more loci successfully genotyped (n=472). In this case for each population, 20–59 adult males were included adding up to a total of 439 individuals from Finland and 33 from Stockholm (Table 1). Some populations were sampled in two consecutive years (Table 1), but because there were no genetic differences between years (data not shown), all the analyses were carried out on the pooled data.
Testing loci
All loci were independent from each other when testing for linkage disequilibrium. All loci except Pdoμ32 were also in Hardy–Weinberg equilibrium (Table 2). There was a large and highly significant deficiency of heterozygotes in Pdoμ32 (Table 2). This may be due to null alleles decreasing the number of observed heterozygotes, and in this case the estimated frequency of null alleles was 0.17 (estimated following methods of Pemberton et al. (1995)). Consequently, Pdoμ32 was not included in the subsequent analyses.
Table 2. Descriptive statistics of 14 microsatellite loci used.
| Locus | NA | N | HO | HE | HW |
|---|---|---|---|---|---|
| Ase18 | 21 | 391 | 0.852 | 0.880 | NS |
| Pdoμ1 | 20 | 391 | 0.854 | 0.869 | NS |
| Pdoμ3 | 18 | 391 | 0.864 | 0.897 | NS |
| Pdoμ5 | 19 | 390 | 0.854 | 0.874 | NS |
| Pdoμ10 | 15 | 391 | 0.867 | 0.870 | NS |
| Pdoμ16 | 14 | 391 | 0.877 | 0.889 | NS |
| Pdoμ19 | 7 | 391 | 0.565 | 0.568 | NS |
| Pdoμ22 | 16 | 391 | 0.655 | 0.733 | NS |
| Pdoμ27 | 13 | 391 | 0.785 | 0.822 | NS |
| Pdoμ32 | 18 | 391 | 0.575 | 0.814 | *** |
| Pdoμ33 | 23 | 391 | 0.928 | 0.915 | NS |
| Pdoμ40 | 18 | 391 | 0.895 | 0.909 | NS |
| Pdoμ44 | 18 | 391 | 0.905 | 0.888 | NS |
| Pdoμ47 | 18 | 391 | 0.821 | 0.890 | NS |
Number of alleles per locus (NA), number of individuals typed (N), observed and expected heterozygosities (HO and HE), deviance from Hardy–Weinberg equilibrium (HW significance indicated with stars, *** for P<0.001).
For testing the sufficiency of the number of loci used in this study, we calculated the change in FST and its standard error when loci were added one at a time (up to 13 loci). The standard error of FST was 0.008 with five loci and stabilized at 0.005 after the eighth locus was included. With 10 or more loci, the overall FST values did not change much. The quality and number of loci used were thus assumed to be sufficient for the analysis of genetic variability within and differentiation among the populations.
Genetic variability within populations
Basic population-level measures of genetic variability indicated that all populations were very similar with respect to intrapopulation levels of genetic variation (Table 3). Allelic richness ranged between 9.0 and 10.0 (on average 9.6) and private allelic richness between 0.02 and 0.29. Average expected heterozygosity, an unbiased estimate introduced by Nei (1987), was very similar in all populations (0.83–0.85). None of the inbreeding coefficients (FIS) of the populations differed significantly from zero.
Table 3. Basic population-level statistics of genetic variability.
| Population | N | AR | AP | APR | HE | FIS | |
|---|---|---|---|---|---|---|---|
| 1 | Stockholm | 33 | 9.59 | 3 | 0.208 | 0.849 | 0.013 |
| 2 | Turku | 45 | 9.76 | 1 | 0.088 | 0.844 | 0.025 |
| 3 | Helsinki | 59 | 9.58 | 3 | 0.161 | 0.842 | 0.000 |
| 4 | Myllykoski | 29 | 10.02 | 4 | 0.228 | 0.846 | 0.021 |
| 5 | Jyväskylä | 35 | 9.63 | 1 | 0.122 | 0.843 | 0.034 |
| 6 | Seinäjoki | 30 | 9.43 | 0 | 0.051 | 0.841 | 0.007 |
| 7 | Oulu | 38 | 9.47 | 2 | 0.096 | 0.834 | 0.062 |
| 8 | Lieksa | 30 | 9.57 | 0 | 0.023 | 0.845 | 0.011 |
| 9 | Kuhmo | 36 | 9.69 | 2 | 0.135 | 0.828 | 0.030 |
| 10 | Kajaani | 25 | 9.82 | 4 | 0.290 | 0.835 | −0.013 |
| 11 | Ämmänsaari | 23 | 9.51 | 2 | 0.261 | 0.835 | 0.022 |
| 12 | Kuusamo | 21 | 9.82 | 0 | 0.088 | 0.832 | 0.019 |
| 13 | Rovaniemi | 48 | 9.98 | 2 | 0.100 | 0.851 | 0.018 |
| 14 | Sodankylä | 20 | 9.01 | 0 | 0.053 | 0.833 | 0.026 |
Number of individuals (N), allelic richness (AR), number of private alleles (AP), private allelic richness (APR), expected heterozygosity (HE) and inbreeding coefficient (FIS).
Genetic differentiation among populations
The levels of genetic differentiation (measured by FST) were very low both globally (FST among Finnish populations=0.004±0.001 (s.e.)) and between all the population pairs (Table 4). Yet some of the pairwise FST estimates were significantly different from zero. Two rather centrally located populations, Myllykoski and Jyväskylä, did not differ from any other Finnish population, but the southern (Helsinki) and northernmost (Sodankylä) populations were significantly different from most other Finnish populations. The Stockholm population was significantly differentiated from all Finnish populations, with FST values ranging between 0.023 and 0.038.
Table 4. Genetic differentiation (FST estimates) among population pairs below the diagonal.
| Stockholm | Turku | Helsinki | Myllykoski | Jyväskylä | Seinäjoki | Oulu | Lieksa | Kuhmo | Kajaani | Ämmänsaari | Kuusamo | Rovaniemi | Sodankylä | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stockholm | — | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0006 |
| Turku | 0.025 | — | 0.0055 | 0.1154 | 0.3198 | 0.3093 | 0.3121 | 0.0006 | 0.0022 | 0.0017 | 0.0011 | 0.0071 | 0.2539 | 0.0006 |
| Helsinki | 0.027 | 0.001 | — | 0.0039 | 0.0110 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0082 | 0.0006 | 0.0006 | 0.0006 | 0.0006 |
| Myllykoski | 0.027 | 0.002 | 0.002 | — | 0.4670 | 0.0033 | 0.0140 | 0.1093 | 0.0824 | 0.0797 | 0.0039 | 0.1945 | 0.0747 | 0.0011 |
| Jyväskylä | 0.023 | −0.001 | 0.001 | −0.001 | — | 0.0121 | 0.0528 | 0.0791 | 0.0055 | 0.0137 | 0.0011 | 0.0082 | 0.0555 | 0.0022 |
| Seinäjoki | 0.033 | −0.003 | 0.002 | 0.003 | 0.000 | — | 0.0368 | 0.0006 | 0.0006 | 0.0006 | 0.0006 | 0.0593 | 0.0055 | 0.0006 |
| Oulu | 0.030 | 0.000 | 0.003 | 0.005 | 0.000 | 0.002 | — | 0.0006 | 0.0165 | 0.0017 | 0.0033 | 0.0270 | 0.1478 | 0.0077 |
| Lieksa | 0.024 | 0.003 | 0.007 | −0.001 | −0.000 | 0.005 | 0.005 | — | 0.0077 | 0.0033 | 0.0006 | 0.0022 | 0.0011 | 0.0006 |
| Kuhmo | 0.038 | 0.007 | 0.010 | 0.003 | 0.006 | 0.010 | 0.007 | 0.005 | — | 0.0011 | 0.0011 | 0.0017 | 0.0022 | 0.0006 |
| Kajaani | 0.031 | 0.007 | 0.002 | −0.000 | 0.002 | 0.007 | 0.006 | 0.005 | 0.010 | — | 0.0006 | 0.0187 | 0.0011 | 0.0006 |
| Ämmänsaari | 0.034 | 0.007 | 0.007 | 0.008 | 0.007 | 0.008 | 0.007 | 0.012 | 0.010 | 0.012 | — | 0.0061 | 0.0017 | 0.0011 |
| Kuusamo | 0.032 | 0.004 | 0.005 | 0.001 | 0.005 | 0.001 | 0.003 | 0.007 | 0.008 | 0.004 | 0.007 | — | 0.0022 | 0.0028 |
| Rovaniemi | 0.031 | 0.002 | 0.004 | 0.003 | 0.001 | 0.002 | 0.002 | 0.007 | 0.007 | 0.006 | 0.008 | 0.005 | — | 0.0028 |
| Sodankylä | 0.032 | 0.008 | 0.007 | 0.007 | 0.005 | 0.013 | 0.004 | 0.011 | 0.014 | 0.013 | 0.007 | 0.008 | 0.007 | — |
Uncorrected P-values (after 1820 randomizations) that program FSTAT provides given above the diagonal.
Significant FST values after sequential Bonferroni correction are in bold.
The low level of genetic differentiation among populations within Finland was corroborated by a number of additional findings. First, there was no isolation by distance among the Finnish populations, as shown by zero correlation between genetic differentiation and geographic distance (Mantel r=0.0000, P=0.941). Second, all the Finnish sampling localities formed one cluster in the clustering analyses (posterior probability for K=1 was Ln P(D)= −26 659.8, which was better than for K=2 for which Ln P(D) = −26 971.4). Finally, the lack of differentiation was further shown by the assignment test where the assignment of individuals to their population of origin was only 24.7%.
Bottlenecks
Using the two-phase mutation model, we found none of the study population to have gone through a drastic decline in numbers in the past. The graphic test gave normal L-shaped distribution and the Wilcoxon test a probability of P>0.85 for all populations.
Discussion
We studied the spatial structure of the house sparrow populations in Finland by using DNA microsatellite markers from the samples collected in mid-1980s, and found that populations were largely panmictic and homogenous over large areas at the time when the widespread decline in population sizes of this species had just started in Finland (Väisänen, 2003). We sampled and studied a large number of populations covering the whole country and a large number of individuals from each population. We focused on adult males, because they are the most sedentary sex and therefore the best indicator any differentiation. We also showed that the number of DNA microsatellite markers used was large enough to reach sufficient confidence for our genetic estimates.
Genetic diversity in the study populations
Genetic variability within house sparrow populations was very similar in all localities, including Stockholm. All measures of genetic diversity, allelic richness, number of private alleles and expected heterozygosity, were nearly equal (Table 3). None of the populations were inbred, which is not surprising given the large population sizes and strong gene flow between the populations (see below) indicated by this study. Also none of the populations had gone through a bottleneck in the recent past, which would have caused a reduction in genetic diversity. The measures of genetic diversity within house sparrow populations were in the same category as previously estimated for birds in general (Neff and Gross, 2001) as well for house sparrows (Neumann and Wetton, 1996; Griffith et al., 2007; Garnier et al., 2008). Because most populations were sampled only in 1 year, we could not estimate the effective population size formally from our data, but the equal genetic diversity across populations suggests that population sizes were of the same magnitude.
Spatial population structure
Despite the low dispersal indicated by the field data, the house sparrow populations showed hardly any spatial structuring across the Finnish range (about 400 × 800 km). We only found evidence of weak differentiation among some of the study populations, mainly concerning those at the southern (Helsinki) and northern (Sodankylä) edges of the region, but the global FST equaled zero showing no significant population structure overall (Table 4). Accordingly we could not differentiate any type of clusters of individuals within the data set by using the software Structure. Extensive homogeneity of populations was most likely due to populations being very large and possibly also better connected than field data suggested. At the time of sampling, the house sparrow was highly abundant in Finland, the census population size of the whole country being in the order of 106 individuals and distribution covering most human-inhabited areas from small farms to cities (Väisänen et al., 1998). Genetic drift has a small effect in such populations, and they diverge only slowly in the neutral markers, such as DNA microsatellites. Dispersal of the house sparrows may also have been more frequent and may have covered larger distances than previous ringing records have suggested. Conversely, only a small number of migrants is enough to homogenize populations (Franklin, 1980; Frankel and Soulé, 1981; Allendorf, 1983) and only few dispersal events may maintain connectivity in an unobstructed landscape.
Thus, Finland could be considered as a rather contiguous landscape for the house sparrow in the 1980s, and our findings suggest that it formed a large and genetically homogenous population in most of its range. Accordingly, previous allozyme studies on the house sparrows have also shown only slight differentiation among populations in contiguous landscapes in the lowlands of eastern England (FST=0.014) (Parkin and Cole, 1984), and among the introduced populations in New Zealand (FST=0.018) and Australia (FST=0.024) (Parkin and Cole, 1985). Also Fleischer (1983) found nonsignificant divergence (FST=0.076) among five farm populations in Kansas in a microgeographic scale (12 km2).
As adults rarely disperse, gene flow in the house sparrow is thought to be maintained by natal dispersal (Summers-Smith, 1988; Altwegg et al., 2000). Unfortunately, only few young birds are ringed in Finland and natal dispersal is poorly characterized. When looking at the limited data (n=87), most of the house sparrows had not dispersed from their natal sites. This low natal dispersal is in good agreement with studies based on ringing data in other European countries (Paradis et al., 1998; Siriwardena et al., 2002; Tufto et al., 2005). Adult dispersal was also very low even though there were more ringing records (n=1110). Nevertheless, dispersal distances based on recapture of marked individuals are underestimating connectivity (Koenig et al., 1996) and thus studies similar to this highlight the importance of genetic tools that can provide valuable information to be considered alongside with information gained by traditional ecological techniques. Finnish house sparrow may maintain gene flow over large geographical distances by a stepping-stone pattern of small migration distances in a fairly homogenously distributed population.
Another possible explanation for the low genetic differentiation between the house sparrow populations would be the colonization history of the house sparrow to Fennoscandia after the last ice age. Some species have shown less genetic variability as well as less genetic structuring in areas where they have expanded their ranges after the ice has retrieved (Hewitt, 2000). However, house sparrow populations in Norway have diverged from each other and because the ice seems to have retrieved approximately the same time from Norway and Finland (Kurtén, 1972; Koivisto, 2004), there should have been also time for the birds to colonize Finland and subsequently populations to diverge. We therefore believe that it is not so likely that a relatively recent postglacial recolonization explains the low genetic differentiation observed in Finland.
Despite the low differentiation on the scale of Finland (longest distance between sites 813 km), we also found that the distance of 250 km between Turku (Finland) and Stockholm (Sweden), including ca. 40 km of open sea, was sufficient to create significant genetic differentiation among the populations (FST=0.02–0.04). This is in line with two studies made on house sparrow populations on the coast of Norway, where populations are separated by fjords and mountains. Bjordal et al. (1986) showed that populations separated by 10–200 km differed from each other based on genetic identity calculated from allozyme frequencies. Later, a microsatellite analysis found that island populations separated by less than 100 km were more differentiated from each other than mainland populations (mainland FST=0.018, island FST=0.025, Jensen et al., in preparation). These results underline the strong effect that landscape composition can have on dispersal: a species that in a contiguous landscapes forms a homogenous population on the scale of hundreds of kilometers can have rather strong spatial structuring on a much smaller scale when barriers (such as open sea) limit natal dispersal over intermediate distances. This also supports the idea that the house sparrow gene flow in the contiguous landscape occurs in a stepwise manner among nearby populations.
Relatively large populations and low levels of genetic differentiation are found to be common in many temperate zone passerines (Barrowclough, 1983). Merilä et al. (1996) found only weak genetic differentiation among greenfinch Carduelis chloris populations on the scale of Europe. No population structuring was detected in the great tit Parus major among populations as distant as Spain and Lapland (Kvist et al., 1999). Kvist et al. (1998) also studied Finnish and Swedish willow tit Poecile montana populations and found no genetic differentiation despite interpopulation distances of up to 1000 km. Furthermore, when studying willow tits more extensively in their Palaearctic distribution range, Kvist et al. (2001) found almost no differentiation between subspecies or geographical localities. This was considered to be due to high gene flow across relatively homogenous landscape lacking geographical barriers. This scenario applies probably to Finnish house sparrows and their environment as well.
Perspectives
The house sparrow is a highly successful avian species that has used much of the habitats humans have provided. However, the situation has recently changed, as the house sparrow populations have declined in many localities throughout Europe (BirdLife International, 2004). For example, between 1976 and 2002, the Finnish house sparrow populations had declined by 63% (Väisänen, 2003). The decline has been particularly pronounced in certain European cities (De Laet and Summers-Smith, 2007; Brichetti et al., 2008) and it has been suggested that different habitats (urban, suburban and rural) present different subpopulations in need of specific conservation actions (De Laet and Summers-Smith, 2007). Our findings imply that small-scale spatial distance associated with an urban to rural gradient are unlikely to be associated with significant genetic structuring in Finland. ‘City sparrows' conceivably experience a different environment than their rural equivalents, but they all seem to form one population because no difference was found between the urban and rural sites in Finland. However, Finland does not have any real metropolis unlike some other countries where these man-made habitats may have a role in genetic differentiation even between nearby populations.
In view of the species' decline, a comparison of the results on the onset of, its decline with contemporary data from the same localities in Finland will be interesting. Results obtained from this study form an important backdrop against which the population genetic consequences of the species' rapid decline in abundance can be evaluated (cf. Wandeler et al., 2007). A population decline may result in an increase in differentiation among populations (Bouzat et al., 1998; Muños-Fuentes et al., 2005; Martínez-Cruz et al., 2007). Knowledge of the population structure before a decline is essential, as sometimes the pattern of low genetic diversity can be present already in historical samples (Matocq and Villablanca, 2001). Whether decline-derived loss of genetic diversity is found, possible conservation actions need to be considered more population-wise; for example, is it better to manage diverged and unique populations or the larger and more connected ones. As the house sparrow is dramatically declining, the need for understanding key population biological processes, for example, through population genetic research, is increasing.
Acknowledgments
This work was supported by the Academy of Finland (RAV (7183/303/82), JEB (1118484; 1131390)), the Nessling Foundation (to JEB) and the Norwegian Research Council (Project 191847/V40 to HJ). We thank those who helped in various phases of the sampling work. Ringing data from Finland was provided by Jari Valkama (Finnish Ringing Bureau). Author contributions: collection of material: RAV and IKH; analyses: JK, PS and HJ; writing the paper: JK and JEB.
The authors declare no conflict of interest.
References
- Allendorf FW.1983Isolation, gene flow, and genetic differentiation among populationsIn: Schonewald-Cox CM, Chambers SM, MacBryde B, Thomas WL (eds).Genetics and Conservation: A Reference for Managing Wild Animal and Plant Populations Benjamin/Cummings: Menlo Park, CA; 51–65. [Google Scholar]
- Altwegg RT, Ringsby TH, Saether B-E. Phenotypic correlates and consequences of dispersal in a metapopulation of house sparrows Passer domesticus. J Anim Ecol. 2000;69:762–770. doi: 10.1046/j.1365-2656.2000.00431.x. [DOI] [PubMed] [Google Scholar]
- Anderson TR. The Ubiquitous House Sparrow: From Genes to Populations. Oxford University Press: USA; 2006. [Google Scholar]
- Baillie SR. Uses of ringing data for the conservation and management of bird populations—a ringing scheme perspective. J Appl Stat. 1995;22:967–987. [Google Scholar]
- Barrowclough GF.1983Biochemical studies of microevolutionary processesIn: Brush AH, Clark GA (eds).Perspectives in Ornithology, Cambridge University Press, NY; 223–261. [Google Scholar]
- BirdLife International . Birds in the European Union: A Status Assessment. BirdLife International: Wageningen, The Netherlands; 2004. [Google Scholar]
- Bjordal H, Cole SR, Parkin DT. Genetic differentiation among some populations of the House Sparrow, Passer domesticus, from southwestern Norway. Hereditas. 1986;105:107–114. [Google Scholar]
- Bouzat JL, Levin HA, Paige KN. The ghost of genetic diversity past: historical DNA analysis of the greater prairie chicken. Am Nat. 1998;152:1–6. doi: 10.1086/286145. [DOI] [PubMed] [Google Scholar]
- Brichetti P, Rubolini D, Galeotti P, Fasola M. Recent declines in urban Italian Sparrow Passer (domesticus) italiae populations in northern Italy. Ibis. 2008;150:177–181. [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics. 1999;153:1989–2000. doi: 10.1093/genetics/153.4.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick H, Siriwardena G.2002National trends in the breeding performance of House Sparrows Passer domesticus Investigation into the Causes of the Decline of Starlings and House Sparrows in Great BritainCrick H, Robinson R, Appleton G, Clark NA, Rickard A (eds). BTO/DEFRA: Bristol. BTO Research Report Number 290.
- De Laet J, Summers-Smith JD. The status of the urban house sparrow Passer domesticus in north-western Europe: a review. J Ornithol. 2007;148 (Suppl. 2:275–278. [Google Scholar]
- Dawson DA, Horsburgh GJ, Krupa A, Stewart IRK, Skjelseth S, Jensen H, et al. A predicted microsatellite map of the house sparrow Passer domesticus genome (Re-submitted to Mol Ecol Res)(In preparation). [DOI] [PubMed]
- Elphinstone MS, Hinten GN, Anderson MJ, Nock CJ. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol Ecol Notes. 2003;3:317–320. [Google Scholar]
- Evans PGH.1987Electrophoretic variability of gene productsIn: Cooke F, Buckley PA (eds).Avian Genetics: A Population and Ecological Approach Academic Press: London; 105–162. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics. 2003a;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer RC. A comparison of theoretical and electrophoretic assessments of genetic structure in populations of the house sparrow (Passer domesticus) Evolution. 1983;37:1001–1009. doi: 10.1111/j.1558-5646.1983.tb05628.x. [DOI] [PubMed] [Google Scholar]
- Frankel OH, Soulé ME. Conservation and Evolution. Cambridge University Press: Cambridge; 1981. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA.2002Introduction to Conservation Genetics1st edn.Cambridge University Press: Cambridge [Google Scholar]
- Franklin IR.1980Evolutionary change in small populationsIn: Souĺe M, Wilcox B (eds).Conservation Biology: An Evolutionary-Ecological Perspective Sinauer Associates: Sunderland, MA; 135–149. [Google Scholar]
- Garnier S, Durand P, Arnathau C, Risterucci AM, Esparza-Salas R, Cellier-Holzem E, et al. New polymorphic microsatellite loci in th house sparrow Passer domesticus. Mol Ecol Resources. 2008;9:1063–1065. doi: 10.1111/j.1755-0998.2009.02552.x. [DOI] [PubMed] [Google Scholar]
- Goudet J.2001FSTAT: A Program to Estimate and Test Gene Diversities and Fixation IndicesVersion 2.9.3Lausanne University: Lausanne, Switzerland [Google Scholar]
- Griffith SC, Stewart IRK, Dawson DA, Owens IPF, Burke T. Contrasting levels of extra-pair paternity in mainland and island populations of the house sparrow (Passer domesticus): is there an ‘island effect'. Biol J Linn Soc. 1999;68:303–316. [Google Scholar]
- Griffith SC, Dawson DA, Jensen H, Ockendon N, Greig C, Neumann K, et al. Fourteen polymorphic microsatellite loci characterized in the house sparrows Passer domesticus (Passeridae, Aves) Mol Ecol Notes. 2007;7:333–336. [Google Scholar]
- Hartl DL, Clark AG.2007Principles of Population Genetics4th edn.Sinauer Associates: Sunderland, MA [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hole DG, Whittingham MJ, Bradbury RB, Anderson GQA, Lee PLM, Wilson JD, et al. Widespread local house sparrow extinctions. Nature. 2002;418:139–149. doi: 10.1038/418931a. [DOI] [PubMed] [Google Scholar]
- Jensen H, Moe R, Holand AM, Tufto J, Sæther B-E.Genetic variation and structure of house sparrow populations: is there an island effect(In preparation). [DOI] [PubMed]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005;5:187–189. [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Koivisto M. Jääkaudet (in Finnish) WSOY: Porvoo; 2004. [Google Scholar]
- Koenig WD, van Vuren D, Hooge PN. Detectability, philopatry, and the distribution of dispersal distances in vertebrates. Trends Ecol Evol. 1996;11:514–517. doi: 10.1016/s0169-5347(96)20074-6. [DOI] [PubMed] [Google Scholar]
- Kurtén B. The Ice Age. Rupert Hart-Davis Ltd: London; 1972. [Google Scholar]
- Kvist L, Ruokonen M, Thessing A, Lumme J, Orell M. Mitochondrial control region polymorphism reveal high amount of gene flow in Fennoscandian willow tit (Parus montanus borealis) Hereditas. 1998;128:133–143. doi: 10.1111/j.1601-5223.1998.00133.x. [DOI] [PubMed] [Google Scholar]
- Kvist L, Ruokonen M, Lumme J, Orell M. The colonization history and present-day population structure of the European great tit (Parus major major) Heredity. 1999;82:495–502. doi: 10.1038/sj.hdy.6885130. [DOI] [PubMed] [Google Scholar]
- Kvist L, Martens J, Ahola A, Orell M. Phylogeography of a Palaearctic sedentary passerine, the willow tit (Parus montanus) J Evol Biol. 2001;14:930–941. [Google Scholar]
- Lowe A, Harris S, Ashton P.2004Ecological Genetics: Desing, Analysis and Application1st edn.Blackwell Publishing: Oxford [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Martínez-Cruz B, Godoy JA, Negro JJ. Population fragmentation leads to spatial and temporal genetic structure in the endangered Spanish imperial eagle. Mol Ecol. 2007;16:477–486. doi: 10.1111/j.1365-294X.2007.03147.x. [DOI] [PubMed] [Google Scholar]
- Matocq MD, Villablanca FX. Low genetic diversity in an endangered species: recent or historic pattern. Biol Conserv. 2001;98:61–68. [Google Scholar]
- Merilä J, Bjorklund M, Baker AJ. Genetic population structure and gradual northward decline of genetic variability in the green finch (Carduelis chlorus) Evolution. 1996;50:2548–2557. doi: 10.1111/j.1558-5646.1996.tb03644.x. [DOI] [PubMed] [Google Scholar]
- Moritz C. Defining ‘evolutionary significant units' for conservation. Trends Ecol Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Muñoz-Fuentes V, Green AJ, Negro JJ, Sorenson MD. Population structure and loss of genetic diversity in the endangered white-headed duck, Oxyura leucocephala. Conserv Genet. 2005;6:999–1015. [Google Scholar]
- Neff BD, Gross MR. Microsatellite evolution in vertebrates: inference from ac dinucleotide repeats. Evolution. 2001;55:1717–1733. doi: 10.1111/j.0014-3820.2001.tb00822.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press: New York; 1987. [Google Scholar]
- Neumann K, Wetton K. Highly polymorphic microsatellites in the house sparrow Passer domesticus. Mol Ecol. 1996;5:307–309. [PubMed] [Google Scholar]
- Paradis E, Baillie SR, Sutherland WJ, Gregory RD. Patterns of natal and breeding dispersal in birds. J Anim Ecol. 1998;67:518–536. [Google Scholar]
- Parkin DT, Cole SR. Genetic variation in the house sparrow, Passer domesticus, in the East Midlands of England. Biol J Linn Soc. 1984;23:287–301. [Google Scholar]
- Parkin DT, Cole SR. Genetic differentiation and rates of evolution in some introduced populations of the house sparrow Passer domesticus in Australia and New Zealand. Heredity. 1985;54:15–23. [Google Scholar]
- Pemberton JM, Slate J, Bancroft DR, Barret JA. Nonamplifying alleles at microsatellite loci—a caution for parentage and population studies. Mol Ecol. 1995;4:249–252. doi: 10.1111/j.1365-294x.1995.tb00214.x. [DOI] [PubMed] [Google Scholar]
- Peterjohn BG, Sauer JR, Link WA. The 1992 and 1993 summary of the North American Breeding Bird Survey. Bird Populations. 1994;2:46–61. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Heredity. 1995;86:248–249. [Google Scholar]
- Richardson DS, Jury FL, Dawson DA, Salgueiro P, Komdeur J, Burke T. Fifty Seychelles warbler (Acrocephalus sechellensis) microsatellite loci polymorphic in Sylviidae species and their cross-species amplification in other passerine birds. Mol Ecol. 2000;9:2155–2234. doi: 10.1046/j.1365-294x.2000.105338.x. [DOI] [PubMed] [Google Scholar]
- Senar JC, Borras A, Cabrera J, Cabrera T, Bjorklund M. Local differentiation in the presence of gene flow in the citril finch Serinus citrinella. Biol Lett. 2006;2:85–87. doi: 10.1098/rsbl.2005.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena G, Robinson R, Crick H.2002Status and population trends of the House Sparrow Passer domesticus in Great Britain Investigation into the causes of the decline of Starlings and House Sparrows in Great BritainCrick H, Robinson R, Appleton G, Clark N, Rickard A (eds). BTO/DEFRA: Bristol. BTO Research Report Number 290.
- Skjelseth S, Ringsby TH, Tufto J, Jensen H, Sæther BE. Dispersal of introduced house sparrows Passer domesticus—an experiment. Proc R Soc B. 2007;274:1763–1771. doi: 10.1098/rspb.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers-Smith JD. The House Sparrow. Collins: London; 1963. [Google Scholar]
- Summers-Smith JD. The Sparrows. T. & A.D. Poyser: Staffordshire; 1988. [Google Scholar]
- Summers-Smith JD. Current status of the house sparrow in Great Britain. British Wildlife. 1999;10:381–386. [Google Scholar]
- Svensson L.1975Identification Guide to European Passerines2nd edn.Naturhistoriska Museet: Stockholm [Google Scholar]
- Tufto J, Ringsby TH, Dhondt AA, Adriaensen F, Matthysen E. A parametric model for estimation of dispersal patterns applied to five passerine spatially structured populations. Am Nat. 2005;165:13–26. doi: 10.1086/426698. [DOI] [PubMed] [Google Scholar]
- Väisänen RA, Lammi E, Koskimies P. Distribution, Numbers and Population Changes of Finnish Breeding Birds. Otava: Helsinki; 1998. [Google Scholar]
- Väisänen RA. Regional population trends of 33 common bird species in Finland during 27 winters. The Yearbook of the Linnut Magazine. 2003;2002:41–62. [Google Scholar]
- Wandeler P, Hoeck PEA, Keller LF. Back to the future: museum specimens in population genetics. Trends Ecol Evol. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Ward RD, Skibinski DOF, Woodwark M. Protein heterozygosity, protein structure, and taxonomic differentiation. Evol Biol. 1992;26:73–159. [Google Scholar]
- Weir B, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Winker K, Graves GR, Braun MJ. Genetic differentiation among populations of a migratory songbird. Limnothlypis swainsonii. J Avian Biol. 2000;31:319–328. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]