Abstract
In many marine fish species, genetic population structure is typically weak because populations are large, evolutionarily young and have a high potential for gene flow. We tested whether genetic markers influenced by natural selection are more efficient than the presumed neutral genetic markers to detect population structure in Atlantic herring (Clupea harengus), a migratory pelagic species with large effective population sizes. We compared the spatial and temporal patterns of divergence and statistical power of three traditional genetic marker types, microsatellites, allozymes and mitochondrial DNA, with one microsatellite locus, Cpa112, previously shown to be influenced by divergent selection associated with salinity, and one locus located in the major histocompatibility complex class IIA (MHC-IIA) gene, using the same individuals across analyses. Samples were collected in 2002 and 2003 at two locations in the North Sea, one location in the Skagerrak and one location in the low-saline Baltic Sea. Levels of divergence for putatively neutral markers were generally low, with the exception of single outlier locus/sample combinations; microsatellites were the most statistically powerful markers under neutral expectations. We found no evidence of selection acting on the MHC locus. Cpa112, however, was highly divergent in the Baltic samples. Simulations addressing the statistical power for detecting population divergence showed that when using Cpa112 alone, compared with using eight presumed neutral microsatellite loci, sample sizes could be reduced by up to a tenth while still retaining high statistical power. Our results show that the loci influenced by selection can serve as powerful markers for detecting population structure in high gene-flow marine fish species.
Keywords: genetic markers, hitchhiking selection, statistical power, pelagic fish, MHC, Baltic
Introduction
Population genetic principles and methods are increasingly used in the management of harvested species and the conservation of natural populations (Allendorf and Luikart, 2007; Allendorf et al., 2008). Population subdivision results from genome-wide processes, such as reduced migration and genetic drift, that on an average affect all genes in the same way, as well as locus-specific processes such as recombination, selection and mutation that are limited to specific genomic regions. Population structure is typically assessed by estimating divergence in the allele frequencies at neutral marker loci, whereas coding gene regions are usually not used for testing demographic independence or for estimating migration rates because selection biases parameter estimation (for example, Whitlock and McCauley, 1999). However, in some cases, such as mixed stock analysis in harvested species, gene loci influenced by selection can be valuable as population markers on ecological time scales where isolated populations have not yet diverged at neutral loci (O'Malley et al., 2007; Westgaard and Fevolden, 2007; Hauser and Carvalho, 2008; Nielsen et al., 2007, 2009a). Moreover, coding loci will help in identifying locally adapted populations (Hemmer-Hansen et al., 2007; Andersen et al., 2009; Gebremedhin et al., 2009; Nielsen et al., 2009a; Árnason et al., 2009).
In marine fishes, population differentiation through drift is typically weak because populations have relatively shallow histories and are often very large (Hauser and Carvalho, 2008). Although some marine fish species show population structure even at small spatial scales (Bekkevold et al., 2005; Jorde et al., 2007; Galarza et al., 2009), a high number of neutral loci and large sample sizes are needed to resolve structure. Genetic differentiation at coding loci requires heterogeneous selection patterns and limited migration, and is expected to be more pronounced in large populations, in which even weak selection may override genetic drift. Temporal stability of divergence, whether selected or neutral, increases the likelihood that the detected structuring is biologically significant, and thus that the loci are useful as population markers (Waples, 1998; Nielsen et al., 2007).
In this study, we compare the performance of three putatively neutral genetic marker types (microsatellites, allozymes and mitochondrial DNA (mtDNA)) with two genetic markers allegedly influenced by selection (the major histocompatibility complex (MHC)-linked locus Clha-DAA-INTR3 and a microsatellite locus Cpa112) to detect structuring in Atlantic herring (Clupea harengus) populations within the North Sea/Baltic Sea region. Although there are several cases in which each of these three classes of neutral markers have shown to be under the influence of selection (Karl and Avise, 1992; Grant et al., 2006; Nielsen et al., 2006), the expectation is, in the absence of evidence to the contrary, that their evolutionary characteristics follow neutral expectations (Baer, 1999). It is, however, important to apply appropriate tests of neutrality beyond traditional measures of population structure (Rand, 1996).
Atlantic herring has traditionally been divided into different geographical ‘stocks' based on spawning behavior and meristic or morphological characters. However, there has been little concordance between these characters and genetic differentiation (Ryman et al., 1984; but see Clausen et al., 2007 and Jørgensen et al., 2008). Recent genetic investigations using microsatellite DNA and allozyme markers have found little differentiation among spatially discrete populations in the North Sea, but concordant divergence between the Baltic and North Sea populations (Bekkevold et al., 2005; Mariani et al., 2005; Larsson et al., 2007, 2010). Genetic mixed-stock analyses show that coastal Skagerrak is used as a nursery both for the Baltic and North Sea spawning populations (Ruzzante et al., 2006). Earlier studies of population structure in the NE Atlantic using mtDNA have generally indicated little divergence among the populations (Dahle and Eriksen, 1990).
Using two different methods, Larsson et al. (2007) and Gaggiotti et al. (2009) showed that a microsatellite locus, Cpa112, displayed highly divergent allele frequencies among populations, suggesting that this locus may be affected by hitchhiking selection. The identity and function of associated gene(s) is unknown, but the divergence pattern was stable over a 25-year time span (Larsson et al., 2010).
MHC genes encode proteins in the vertebrate immune response, and molecular variation among alleles has been linked directly to fitness and survival (Bos et al., 2008). For example, disease challenge trials on Atlantic salmon confirm that polymorphism at the MHC genes are associated with increased disease resistance (Grimholt et al., 2003). Wild populations show evidence for local adaptation (de Eyto et al., 2007), and several studies indicate that MHC genes, as well as a number of linked loci, can be more powerful than neutral microsatellites in detecting population differentiation in spatially structured fish species such as salmonids (Bernatchez and Landry, 2003; Beacham et al., 2005; Hansen et al., 2007). However, with the exception of a precursory characterization of the MHC class IIA gene and an embedded microsatellite locus in Atlantic herring (Stet et al., 2008), no studies exist to date that describe the MHC variation in an abundant and fully marine fish. It is worth noting, however, that divergence in genes under selection is not always expected; if fish populations share similar parasite environments, balancing selection can result in lower levels of differentiation in MHC-linked microsatellites than neutral microsatellites (Muirhead, 2001; Bernatchez and Landry, 2003).
Different genetic markers reflect different demographic histories (Avise, 2004) and also have varying statistical properties that affect the extent to which they can resolve the patterns of divergence (Ryman et al., 2006). For example, it has been shown that organelle markers such as mtDNA may be more powerful for detecting differentiation in recently diverged populations and especially when migration among populations is relatively high (Larsson et al., 2009).
In this study, we present an investigation of spatial and temporal patterns of divergence and statistical power obtained using three traditional genetic markers, allozymes, mtDNA and microsatellites, and two markers potentially influenced by selection, MHC and the microsatellite locus Cpa112. Using the same individuals for all analyses, we compare fish collected in 2002, and again in 2003, from four spawning grounds spanning the environmental gradient from the Baltic Sea to the North Sea.
Materials and methods
Sampling
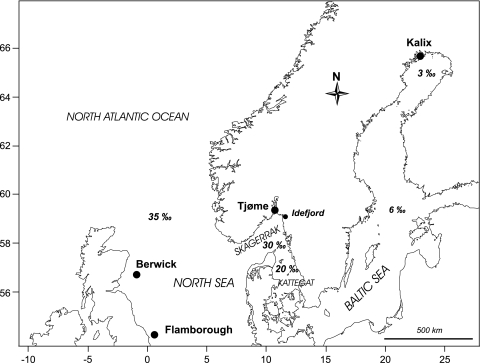
A total of eight samples of Atlantic herring were collected in 2002 and 2003 at four locations in the North Sea/Baltic Sea region: autumn-spawning herring from (1) Flamborough and (2) Berwick in the North Sea, (3) a spring-spawning population from Tjøme in the Skagerrak and (4) a second spring-spawning population from Kalix in the inner Baltic Sea (Figure 1; Table 1). These samples cover a salinity gradient from the fully marine environment in the North Sea (35‰) to almost fresh water in the Bothnian Bay of the Baltic Sea (3‰). Each sample consisted of ca 100 fish in full spawning condition. Otolith (sagitta) winter rings were counted as a proxy for age following standard procedures (ICES, 2003). The hatching season of individual fish (spring, autumn or winter) was estimated from otolith central area microstructure according to Clausen et al. (2007).
Figure 1.
Sampling locations for Atlantic herring collected in 2002 and 2003. Sea surface salinities are indicated.
Table 1. Sample descriptions including locations, coordinates, dates, spawning seasons, percentage of fish in spawning condition and sample sizes (n) of genetic markers used.
| Location | Sampling coordinates | Date | Spawning season | Spawning (%) | Fish (n) | Microsatellites (including Cpa112) (n) | MHC (n) | Allozymes (n) | mtDNA (n) | All markers in the same fish (n) |
|---|---|---|---|---|---|---|---|---|---|---|
| Flamborough | 54.60N/00.59W | 14-09-2002 | Autumn | 100 | 80 | 80a,b | 73 | 55 | 80 | 48 |
| Flamborough | 54.57N/00.45W | 17-09-2003 | Autumn | 99 | 93 | 93a,b,c,d | 80 | 93c | 91 | 79 |
| Berwick | 56.30N/00.97E | 20-08-2002 | Autumn | 98 | 90 | 90a,b | 83e | 85 | 85 | 73 |
| Berwick | 57.70N/01.45W | 22-08-2003 | Autumn | 88 | 96 | 96a,b,c | 90 | 96c | 80 | 72 |
| Tjøme | 59.35N/10.55E | 13-03-2002 | Spring | 98 | 95 | 95b,f | 95e | 95 | 84 | 84 |
| Tjøme | 59.35N/10.55E | 04-03-2003 | Spring | 100 | 104 | 104b,c,d,f | 94 | 104c | 86 | 75 |
| Kalix | 65.80N/22.72E | 12-06-2002 | Spring | 44 | 97 | 97g | 90 | 97g | 85 | 78 |
| Kalix | 65.80N/22.72E | 19-06-2003 | Spring | 94 | 88 | 88c,g | 79 | 88c,g | 69 | 64 |
| Sum | 743 | 743 | 684 | 713 | 660 | 573 |
Abbreviations: MHC, major histocompatibility complex; mtDNA, mitochondrial DNA.
Only fish that were successfully analyzed for at least three different genetic marker types were used in subsequent statistical analyses. Genotypes previously used by
Genetic analyses
The fish were genotyped using four genetic marker types: microsatellites, allozymes, mtDNA and MHC. One of the microsatellite loci, Cpa112, has previously been demonstrated to be influenced by selection (Gaggiotti et al., 2009; Larsson et al., 2007, 2010). Parts of the microsatellite, allozyme and MHC data have been presented previously (see Table 1), although here we provide full data sets to allow a direct comparison.
DNA for microsatellite, mitochondrial and MHC analyses was extracted from fin clip or muscle tissue using standard kits or the HotSHOT method (Truett et al., 2000). Nine microsatellite loci, Cha1017, Cha1020, Cha1027, Cha1202 (McPherson et al., 2001), Cpa101, Cpa111, Cpa113, Cpa114 and Cpa112 (Olsen et al., 2002), were amplified and genotyped using a BaseStation (MJ Research, MJ Research, Skovlunde, Denmark, Denmark) and Pharmacia ALF express (Amersham Pharmacia, Uppsala, Sweden) automated sequencers. Scoring consistency among runs and platforms was ensured by analyzing two heterozygote control individuals spanning the anticipated allelic ranges on all gels, in addition to internal and external size ladders.
MtDNA variation was examined by analysis of restriction fragment length polymorphism in the mitochondrial NADH dehydrogenase ND3/4 subunit. A 2.4 Kb sequence was PCR amplified using an existing universal primer (Hauser et al., 2001) and a new primer designed from a sardine (Sardinops melanostictus) mtDNA sequence (Inoue et al., 2000) published on GenBank (accession number NC 002616):
Universal vertebrate primer: 5′-TTTTGGTTCCTAAGACCAA(C/T)GGAT-3′
Newly designed primer: 5′-AAGACAGTACAGGTGGCTTCCAA-3′.
The PCR products were digested using six restriction enzymes: AluI, Hae III, Hinf I, Mbo I, Msp I and Rsa I, which produced 26 cut sites in Atlantic herring. Restricted fragments were separated on 6% polyacrylamide gels, together with a pGEM size marker (Promega, Southhampton, UK), and stained using a standard silver nitrate protocol. Fragments were sized using (http://tools.neb.com/NEBcutter2/index.php), and fragment patterns were expected to add up to a total of ∼2.4 Kb if the PCR product had been fully digested; partial digests were discarded.
MHC polymorphism was investigated using the linked microsatellite Clha-DAA-INTR3, located in the third intron of the MHC class II alpha gene Clha-DAA (Stet et al., 2008). PCR amplification of Clha-DAA was performed using standard cycling conditions with annealing at 50 °C. PCR products were analyzed with an ABI 377 automated sequencer and fragment sizes assessed by the software (Applied Biosystems, Foster City, CA, USA).
Allozyme starch gel electrophoresis was performed using procedures described in Andersson et al. (1981). The following eleven polymorphic loci were scored, (locus abbreviations used in early studies, enzyme names and EC numbers are given in parentheses): mAAT* (AAT-2, aspartate aminotransferase, EC 2.6.1.1), G3PDH* (GPD-1, glycerol-3-phosphate dehydrogenase, EC 1.1.1.8), GPI* (glucosephosphate isomerase, EC 5.3.1.9), mIDHP* (IDH-2, isocitrate dehydrogenase, EC 1.1.1.42), LDH-1*, LDH-2* (lactate dehydrogenase, EC 1.1.1.27), MDH-4* (malate dehydrogenase, EC 1.1.1.37), sMEP* (ME-1, malic enzyme, EC 1.1.1.40), mMEP* (ME-2, malic enzyme, EC 1.1.1.40), PGM-1* (phosphoglucomutase, EC 5.4.2.2, formerly EC 2.7.5.1) and SOD* (superoxide dismutase, EC 1.15.1.1). These loci have previously been shown to be variable in herring from this region (Andersson et al., 1981; Ryman et al., 1984; Larsson et al., 2007).
Statistical analyses
It was not possible to genotype all fish for all marker loci, and the original data set with ca 100 fish from each sample was cropped to include individual fish that were successfully analyzed for at least three of the four genetic marker types (microsatellites incl. Cpa112, the MHC locus Clha-DAA-INTR3, allozymes and mtDNA; Table 1). In total, 743 fish were analyzed, out of which 573 were genotyped for all marker types.
Amounts of genetic variation within herring samples were estimated as allelic richness (AR) and expected heterozygosity (gene diversity, HE). F-statistics were calculated using (Goudet, 2001). Statistical significance of FST for allozymes, MHC and microsatellites were examined using Fisher's exact test implemented in 3.4 (Raymond and Rousset, 1995), the χ2-test implemented in 1.3 (Ryman, 2006) and permutation tests using (Goudet, 2001); the results were very similar and only P-values obtained using (with 10 000 permutations and not assuming Hardy–Weinberg equilibrium) are presented here for brevity. Departure from Hardy–Weinberg equilibrium was tested using 10 000 permutations in . We used GDA version 1.1 (Lewis and Zaykin, 2001) to examine linkage disequilibria. 3.1 (Excoffier et al., 2005) was used to estimate gene diversity and pairwise FST between samples for the mtDNA haplotype data; statistical significance was obtained from 10 000 permutations. The presence of outlier loci was investigated with the test of Beaumont and Nichols (1996), in which the null distribution was created using (Beaumont, 2002). 6.0 (StatSoft Inc., Uppsala, Sweden) was used to visualize genetic relationships among samples for the different marker types with multidimensional scaling using pairwise FST estimates. Concordance in FST estimates between markers was investigated using correlation of FST and Mantel tests between all sample pairs.
Temporal stability in differentiation for the different marker types was estimated using hierarchical analysis of molecular variance, with sampling years nested within locations. We used the locus-by-locus option in statistical significance was obtained from 10 000 permutations. Further, we divided all fish into year classes, based on otolith ageing, and tested for genetic heterogeneity among cohorts with n⩾20, within and among locations.
The statistical power for detecting genetic differentiation at various levels of FST was evaluated for the separate markers using the software (Ryman and Palm, 2006; Ryman et al., 2006). This analysis simulates sampling from a specified number of populations that have diverged to predefined levels of divergence. We used sample sizes, number of loci and allele frequencies obtained from the eight samples in this study. Finally, we used a modified version of to obtain sampling distributions of FST for the different markers; using the observed allele frequencies over all samples, we simulated a drift process in which two large populations diverge until an expected FST of 0.002 is reached (see Table 2). We then sampled these simulated populations at n=100 and calculated FST. This procedure was repeated 10 000 times.
Table 2. F-statistics (FIS, FST, FIT) for allozyme, microsatellite, MHC and mtDNA loci for unpooled (s=8) and pooled (s=4) temporal herring samples.
| Locus |
s=8 |
s=4, temporal samples pooled |
||||
|---|---|---|---|---|---|---|
| FIS | FST | FIT | FIS | FST | FIT | |
| Allozymes | ||||||
| mAAT* | −0.0115 | 0.0020 | −0.0095 | −0.0113 | 0.0020 | −0.0092 |
| G3PDH* | −0.0032 | −0.0008 | −0.0040 | −0.0041 | 0.0003 | −0.0038 |
| GPI* | 0.0100 | −0.0006 | 0.0095 | 0.0104 | −0.0011 | 0.0092 |
| mIDHP | −0.0155 | 0.0011 | −0.0143 | −0.0164 | 0.0025 | −0.0139 |
| LDH-1* | 0.0092 | 0.0031* | 0.0123 | 0.0125 | −0.0007 | 0.0118 |
| LDH-2* | −0.0317 | 0.0029* | −0.0287 | −0.0322 | 0.0041* | −0.0280 |
| MDH-4* | 0.0422 | 0.0016 | 0.0438 | 0.0440 | −0.0007 | 0.0434 |
| sMEP* | 0.0000 | −0.0003 | −0.0003 | −0.0007 | 0.0008 | 0.0001 |
| mMEP* | −0.0072 | −0.0026 | −0.0098 | −0.0084 | −0.0014 | −0.0098 |
| PGM-1* | −0.0101 | 0.0020 | −0.0080 | −0.0123 | 0.0052 | −0.0069 |
| SOD* | 0.0754 | 0.0162** | 0.0904** | 0.0781 | 0.0150** | 0.0919** |
| Allozymes 11 loci | 0.0184 | 0.0011* | 0.0195 | 0.0192 | 0.0002** | 0.0194 |
| Microsatellites | ||||||
| Cha1017 | 0.0679*** | −0.0012 | 0.0668*** | 0.0669*** | 0.0001 | 0.0669*** |
| Cha1020 | 0.0020 | 0.0056*** | 0.0075 | 0.0026 | 0.0057*** | 0.0083 |
| Cha1027 | 0.0364** | 0.0004* | 0.0368*** | 0.0365*** | 0.0002* | 0.0368*** |
| Cha1202 | 0.0355* | 0.0051*** | 0.0405** | 0.0385* | 0.0019*** | 0.0403** |
| Cpa101 | 0.0109 | 0.0008 | 0.0117 | 0.0109 | 0.0010 | 0.0118 |
| Cpa111 | −0.0008 | 0.0062*** | 0.0054 | −0.0007 | 0.0071*** | 0.0064 |
| Cpa113 | 0.0172 | 0.0025** | 0.0197* | 0.0169 | 0.0034*** | 0.0202* |
| Cpa114 | 0.0334** | 0.0017* | 0.0350** | 0.0348** | 0.0000 | 0.0348** |
| Microsatellites 8 loci | 0.0268*** | 0.0024*** | 0.0291*** | 0.0273*** | 0.0021*** | 0.0294*** |
| Cpa112 | 0.0132 | 0.0360*** | 0.0487*** | 0.0135 | 0.0415*** | 0.0544*** |
| MHC, Clha-DAA-INTR3 | 0.0020 | 0.0027 | 0.0047 | 0.0012 | 0.0043 | 0.0055 |
| MtDNA | — | 0.0145*** | — | — | 0.0065*** | — |
| All loci, excluding mtDNA | 0.0225*** | 0.0052*** | 0.0276*** | 0.0230*** | 0.0055*** | 0.0283*** |
| All, excluding mtDNA, Cpa112 | 0.0234*** | 0.0022*** | 0.0256*** | 0.0238*** | 0.0020*** | 0.0258*** |
| All, excluding mtDNA Cpa112, SOD | 0.0233*** | 0.0021*** | 0.0254*** | 0.0237*** | 0.0019*** | 0.0256*** |
Abbreviations: MHC, major histocompatibility complex; mtDNA, mitochondrial DNA.
*P<0.05; **P<0.01; ***P<0.001.
Sample-specific heterozygosities and FIS estimates for microsatellite loci and MHC are presented in Supplementary Appendix 1.
In all statistical analyses that involve multiple tests, we provide both uncorrected and sequential Bonferroni-corrected significance levels for comparison.
Results
Sampling
The hatching date determined from otolith microstructure verified that the collected samples represented the presumed seasonal spawning stocks: autumn spawners in the North Sea and spring spawners in the Skagerrak and the Baltic. Between 48 and 84 individuals from each sample were genotyped for all five genetic markers: the three putatively neutral genetic markers (8 microsatellite loci, 11 allozyme loci, 1 mtDNA locus) and the two genetic markers allegedly influenced by selection (the microsatellite Cpa112 and the MHC-embedded Clha-DAA-INTR3).
Within-population descriptive analyses
Allelic richness and expected heterozygosity, HE, for the MHC-embedded microsatellite locus ranged between 8.5–11.6 and 0.70–0.80 among samples, respectively (Supplementary Appendix 1a). These values are similar to those obtained for the other microsatellite loci, including Cpa112, in the same populations. For allozymes the average number of alleles per locus and HE were considerably lower, ranging between 2.1–2.7 and 0.12–0.14 among samples (Supplementary Appendix 1b). In total, we found 237 mtDNA haplotypes among the 660 analyzed fish. Accordingly, gene (haplotype) diversity within samples was high, ranging between 0.93 and 0.98 among samples (Supplementary Appendix 1a).
Deficiency of heterozygotes relative to Hardy–Weinberg expectations was found in one locus-sample combination for the allozymes, and in 12 out of 64 for the presumed neutral microsatellites; there was no sample- or locus-specific pattern, and only one persisted after Bonferroni correction. One sample showed heterozygote deficiency at Cpa112, whereas no deviations were found at the MHC locus (Supplementary Appendix 1a). Combining the information from all localities and/or samples show that microsatellites displayed significantly high overall FIS values at four loci (Table 2); however, separate statistically significant locus-sample combinations appeared evenly distributed throughout the locus-sample table (Supplementary Appendix 1a). There was little indication of linkage disequilibria between loci. Out of a total of 1848 pairwise locus combinations (22 loci) within samples, 57 showed significant deviation from linkage equilibrium (ca 3% of the tests). Only one significance remained after Bonferroni correction.
Outlier test
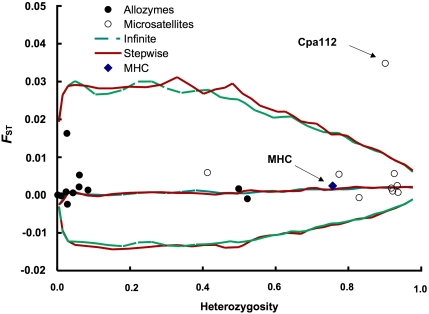
The microsatellite locus Cpa112 was identified as an outlier (Figure 2), indicating that this locus is influenced by directional selection. While within the 95% confidence limits of the simulated FST distribution, the allozyme locus SOD* had a conspicuously large FST value compared with the other allozyme loci (Table 2; Figure 2). There was no indication of selection affecting the MHC locus.
Figure 2.
FST for individual microsatellite, allozyme and MHC loci plotted against heterozygosity. Lines denote the 2.5, 50 and 97.5 percentiles of simulation-based expected distributions of FST assuming an infinite allele or stepwise mutation model, respectively. The simulations were based on the combined overall FST=0.002 for all three marker types.
Statistical power and expected FST
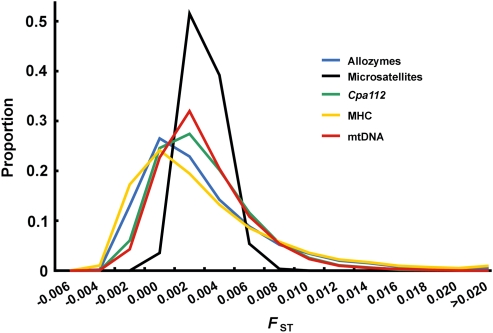
Before comparing statistical power, we investigated the expected distribution of FST for all markers in this case under the assumption of selective neutrality. Two simulated populations differentiated at FST=0.002 were sampled repeatedly at n=100. The various markers had similar mean FST values, close to 0.002, but the medians and variances differed (Figure 3). The FST distributions were markedly skewed to the right, except for the eight microsatellites, in which the distribution was more symmetric and had a smaller variance (Figure 3). This pattern is also seen in our empirical data in which the average and variance were 0.0022 and 24.5 × 10−6 for the 11 allozyme loci and 0.0026 and 7.4 × 10−6 for the eight microsatellite loci (see Table 2; also see Larsson et al., 2007).
Figure 3.
Distributions of FST for various marker types based on 10 000 simulated samples of 100 individuals from two populations with a true FST of 0.002. The sample parameters mean, median and variance ( × 10−6) are for microsatellites (eight loci): 0.0020, 0.0019, 1.4; allozymes: 0.0019, 0.0009, 19; mtDNA: 0.0020, 0.0014, 9.3; MHC: 0.0021, 0.0008, 25; Cpa112: 0.0020, 0.0014, 10.
Analysis of statistical power indicated that all marker sets will detect FST=0.0050 or larger with a probability close to one, for present sample sizes (Table 3). At lower levels of divergence, eight microsatellites are more powerful for detecting structuring than eleven allozymes, mtDNA, MHC and Cpa112. The individual microsatellite loci have, however, statistical power comparable to the latter markers (Table 3).
Table 3. Statistical power of different marker types for detecting various true levels of population differentiation (FST) by means of Fisher's exact test when using present sample sizes and allele frequencies obtained from the eight samples in this study.
| FST | Micro- satellites 8 loci | Allozymes 11 loci | mtDNA | MHC | Cpa112 | Median of individual microsatellite loci |
|---|---|---|---|---|---|---|
| 0.0000 | 0.063 | 0.057 | 0.063 | 0.064 | 0.049 | 0.054 |
| 0.0001 | 0.125 | 0.056 | 0.072 | 0.068 | 0.070 | 0.065 |
| 0.0010 | 0.958 | 0.342 | 0.455 | 0.240 | 0.386 | 0.366 |
| 0.0020 | 1.000 | 0.774 | 0.866 | 0.572 | 0.799 | 0.770 |
| 0.0050 | 1.000 | 0.998 | 1.000 | 0.982 | 0.998 | 1.000 |
| 0.0100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
Abbreviations: MHC, major histocompatibility complex; mtDNA, mitochondrial DNA.
The locus Cpa112 showed strong divergence and had an FST more than ten times higher than the other loci (Table 2). This raises the question: How much smaller could sample sizes be using Cpa112 and still have the same power as several neutral loci to detect divergence? Using , we estimated power at Cpa112 for different sample sizes, and we show that a sample size reduced to n≈20 would be sufficient to detect FST=0.025 (Table 4).
Table 4. Statistical power of Cpa112 for detecting various true levels of population differentiation (FST) by means of Fisher's exact test when using allele frequencies obtained from the eight samples in this study and different sample sizes (n).
| FST | n=100 | n=50 | n=30 | n=20 | n=10 |
|---|---|---|---|---|---|
| 0.000 | 0.056 | 0.049 | 0.049 | 0.057 | 0.051 |
| 0.001 | 0.512 | 0.216 | 0.145 | 0.088 | 0.064 |
| 0.002 | 0.898 | 0.462 | 0.220 | 0.163 | 0.083 |
| 0.005 | 1.000 | 0.956 | 0.669 | 0.397 | 0.171 |
| 0.010 | 1.000 | 1.000 | 0.971 | 0.778 | 0.325 |
| 0.025 | 1.000 | 1.000 | 1.000 | 1.000 | 0.844 |
| 0.050 | 1.000 | 1.000 | 1.000 | 1.000 | 0.996 |
Population differentiation
The patterns of differentiation among the herring populations using the different marker types were explored in three different ways: using individual samples (s=8), pooling temporal samples from the same locality (s=4), and using year-class cohorts at sampling locations as the basic unit (s=10; Supplementary Appendix 3). Overall FST (s=8) was 0.0024 for microsatellites, 0.0011 for allozymes, 0.0145 for mtDNA, 0.0027 for MHC and 0.0360 for Cpa112 (Table 2). FST over all nuclear marker types (that is, excluding mtDNA) was 0.0052. Pooling the temporal samples within locations gave similar estimate of FST for microsatellites, whereas the estimates decreased for mtDNA and allozymes, and increased for MHC and Cpa112 (Table 2).
Spatial and temporal patterns
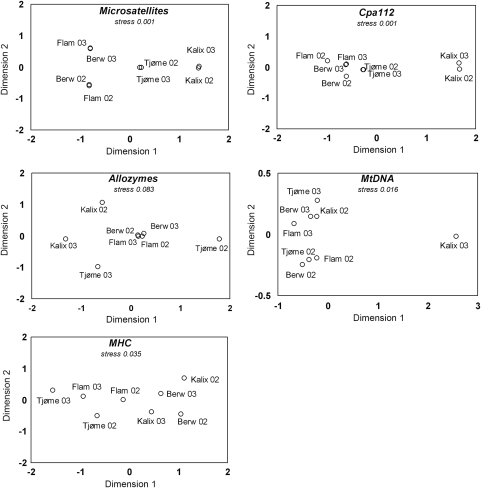
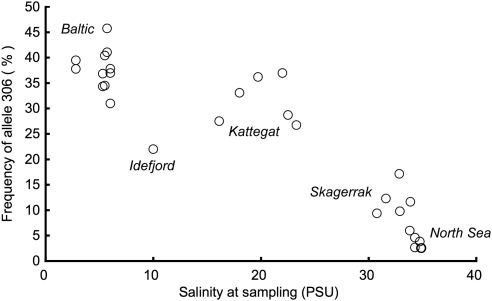
The microsatellite markers showed significant differentiation between the three regions, the North Sea, the Skagerrak and the Baltic (Figure 4, Table 2, Supplementary Appendix 2). At both Kalix and Tjøme, samples were genetically similar between years. In contrast, the four North Sea samples clustered by year of sampling. The cohort analysis indicates that this could be a year-class effect; younger fish from Berwick and Flamborough are more similar genetically than they are to older fish from the same locations (Supplementary Appendices 3, 4). The locus Cpa112 showed a similar pattern, but with a more pronounced divergence for the Baltic samples. Pairwise comparisons involving the Baltic had one magnitude higher FST values than comparisons outside the Baltic. Larsson et al. (2007) showed that the differentiation in Cpa112 is due to frequency differences especially for one allele, Cpa112(306). In this study, this allele had average frequencies of 38.6% at Kalix, 10.5% at Tjøme and 3.4% among the North Sea samples (see Figure 5). This pattern corroborates that Cpa112 is likely influenced by selection. The allozymes showed little divergence: 4 out of 11 loci had negative overall FST, and 16 out of 28 pairwise comparisons also had a negative FST estimate. As an exception, the herring sample collected in Tjøme in 2002 was highly divergent from other samples at the SOD* locus (Figure 4, Supplementary Appendix 2). Owing to this rare pattern, we repeated the genotyping of this specific sample, with an identical result. Notwithstanding this specific sample, and despite the many negative FST values, the North Sea samples clearly grouped together (Figure 4). Similar to the allozymes, the mtDNA data included a single highly divergent sample (Figure 4, Supplementary Appendix 2): the Kalix 2003 sample had one haplotype occurring at a frequency of 20% compared with around 2% in all other samples. In addition, mtDNA displayed a number of statistically significant pairwise differences. Finally, MHC showed a more complex pattern. Some pairwise comparisons were negative and some had relatively high point estimates of FST of around 1%, without any obvious geographical pattern (Figure 4, Supplementary Appendix 2).
Figure 4.
Multidimensional scaling plots of herring samples based on pairwise FST estimated from different genetic markers.
Figure 5.
Frequency of the allele 306 in the locus Cpa112 in samples of herring collected in 2002 and 2003 along the salinity gradient from the North Sea to the inner part of the Baltic Sea. Genetic data are compiled from Mariani et al., 2005; Bekkevold et al., 2005, Larsson et al., 2010 and André C (unpublished) (sample collected in 2002 in Idefjord, a sheltered Skagerrak fjord with low salinity, see Figure 1). Salinities were obtained as in Bekkevold et al., 2005 or by the Swedish Meteorological and Hydrological Institute.
The hierarchical analysis of molecular variance analysis (Table 5) indicated spatial differentiation among basins for microsatellites and Cpa112. MtDNA, on the other hand, showed a strong temporal variation, presumably masking any spatial pattern (see Table 2, s=4).
Table 5. Hierarchical analysis of genetic differences (AMOVA) among four sampling locations (see Figure 1) and between temporal samples (2002 and 2003) of Atlantic herring for different genetic markers.
| Source of variation | d.f. |
Microsatellites |
Allozymes |
MtDNA |
MHC |
Cpa112 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F-value | P-value | F-value | P-value | F-value | P-value | F-value | P-value | F-value | P-value | ||
| Among locations | 3 | 0.0016 | 0.005 | −0.0007 | 0.78 | −0.0013 | 0.63 | 0.0052 | 0.067 | 0.0410 | 0.010 |
| Between yrs within locations | 4 | 0.0011 | 0.037 | 0.0018 | 0.25 | 0.016 | 0.001 | −0.0017 | 0.81 | 0.0007 | 0.32 |
Abbreviations: AMOVA, analysis of molecular variance; MHC, major histocompatibility complex; mtDNA, mitochondrial DNA.
Marker correlations
Across samples, there was strikingly little concordance in sample pairwise FST between marker types (R=0.000–0.005; P>0.05), except between Cpa112 and microsatellites (R=0.70; P<0.001), and Cpa112 and mtDNA (R=0.44; P=0.021).
Discussion
Using the same fish for all analyses, thus excluding year-class effects, we show that levels of population differentiation in herring were low for the presumed neutral genetic markers (allozymes, mtDNA and microsatellites), in agreement with earlier studies from the same region (Ryman et al., 1984; Hauser et al., 2001; Bekkevold et al 2005; Jørgensen et al., 2005; Mariani et al., 2005; Larsson et al., 2007). For the putatively selected markers, the microsatellite locus Clha-DAA-INTR3 embedded in the MHC gene showed characteristics similar to those of the neutral microsatellite loci, and there was no indication of selection influencing this locus. Cpa112 on the other hand was highly divergent for Baltic samples, indicating strong influence by selection as reported in previous studies (Gaggiotti et al., 2009; Larsson et al., 2007, 2010). The higher level of divergence in Cpa112 makes this locus efficient at detecting even weak overall population structure in Atlantic herring.
Statistical power of neutral vs selected markers
All genetic markers, including Cpa112, displayed high statistical power for detecting low levels of genetic differentiation in simulations; the applied set of eight neutral microsatellites could even detect FST as low as 0.0010 with a power of 95% at n≈100. Statistical power is related to the distribution of expected FST under neutrality, and the smaller variance of the eight microsatellites explains their higher power (Figure 3). In turn, this is likely an effect of the larger total number of alleles (Kalinowski, 2002; Ryman et al., 2006; Larsson et al., 2007). The eight microsatellite loci had 192 alleles in total, whereas the eleven allozyme loci had 38 alleles. Interestingly, the median microsatellite locus harboring 26 alleles had a power comparable to the allozymes (Table 3).
These evaluations were made under the assumption of selective neutrality; if selection is taken into account, expectations change. For Baltic vs North Sea herring, Cpa112 displayed a 10–20 times higher overall FST compared with the eight neutral microsatellite loci (Table 2), and this higher level of differentiation increase the statistical power. Consequently, a substantial reduction in sample size is possible when using a selected locus vs neutral markers (Table 4), in agreement with predictions by Schmidt et al. (2008). However, it should be noted that these estimates relate to FST over all eight samples, and that power varied with specific comparisons, that is, North Sea vs Skagerrak comparisons were less powerful, and Baltic vs Skagerrak or North Sea comparisons were even more powerful than the estimates above (see Supplementary Appendix 2 for detailed pairwise comparisons of FST).
Several pairwise comparisons using mtDNA were statistically significant (Supplementary Appendix 2), indicating that mtDNA can be a powerful marker, even at low levels of nuclear divergence (Hoarau et al., 2004; Larsson et al., 2009).
Spatial and temporal patterns of differentiation
Neutral markers
The microsatellite results support the existence of a barrier to gene flow between Baltic and Skagerrak/North Sea populations, which has been shown earlier for herring (Bekkevold et al., 2005; Larsson et al., 2007), as well as for several other species (Johannesson and André, 2006; Limborg et al., 2009). Using the microsatellite data for the same samples collected at Berwick and Tjøme as in this study, as well as several other samples, Ruzzante et al. (2006) showed that herring collected at Rügen in the southwestern Baltic is clearly distinct from the populations in the North Sea and Skagerrak. In this study, the differentiation estimated with neutral microsatellites was stable over two subsequent years (Figure 4). The allozyme data for the 2003 herring showed a concordant spatial pattern of differentiation, as was also reported by Larsson et al. (2007). In 2002, however, the sample collected at Tjøme was highly divergent at the SOD* locus. Owing to the low overall level of divergence, a single odd genotype at one locus had a disproportional impact on the FST estimate. A similar case is shown in the mtDNA data in which one specific haplotype had an elevated frequency in the Kalix 2003 sample (Figure 4). Owing to the low levels of differentiation typical for marine fish, a slightly different set of samples, or loci, may yield different patterns of population structure (cf. Waples, 1998; Allendorf and Seeb, 2000; Hauser and Carvalho, 2008). Such variance is particularly pronounced in genetic markers with low heterozygosities and a high expected range of FST, such as allozymes (Figure 3; also refer Fauvelot et al., 2007). Therefore, careful sampling including temporal replication is warranted, especially in species that undertake ontogenetic shifts in habitat use, such as herring and cod, in which the inclusion of juvenile fish may bias patterns of spatial genetic differentiation (Nielsen et al., 2009b).
Markers influenced by selection
The MHC-embedded microsatellite locus, Clha-DAA-INTR3, showed similar patterns of allelic richness and divergence as the presumed neutral microsatellite loci (Table 2; Supplementary Appendix 1a), and there was no indication of either directional or balancing selection acting on Clha-DAA-INTR3 (Figure 2). The locus is situated in the third intron of the MH IIA gene, and the extent to which the variability in the microsatellite reflects exonic variability is unknown (Stet et al., 2008). Previous attempts to estimate the variability in MHC genes using embedded micro- or minisatellites in salmonids have yielded varying results (Stet et al., 2002; Grimholt et al., 2002, 2003; Hansen et al., 2007; de Eyto et al., 2007). de Eyto et al. (2007) found a one-to-one relationship between minisatellite and MH class II allelic variability; however, for MH class I, a specific microsatellite allele could be found in several different MH I alleles. Wynne et al. (2007) showed that, although alleles of a microsatellite located in the untranslated region of the salmon MH II gene were linked to several different MH II sequences, the microsatellite diversity was correlated to disease susceptibility, and thus acted as a good proxy for actual MH class II polymorphism. In a recent study, Tonteri et al. (2010) demonstrated that 18 microsatellite loci linked to immune-relevant genes show stronger signal of selection than 76 loci without such association.
Baltic herring are highly divergent at the microsatellite locus Cpa112. Larsson et al. (2010) showed this for herring collected in the 1980s, and it is also evident for herring collected in 2002 and 2003 (Figure 4; Supplementary Appendix 2). Skagerrak and North Sea samples are also divergent at this locus, but to a substantially lesser degree. Presently, there is no information on any gene associated with Cpa112, but the lower salinity of the Baltic Sea is an obvious candidate as selective agent (Figure 5), although other environmental factors that covary with salinity may also have a role. This is also supported by preliminary data indicating a high frequency of the Cpa112(306) allele in the low-saline Idefjord in NE Skagerrak (Figure 5).
Conclusion
Neutral markers have traditionally been used to estimate population structure and also migration rates. A decade ago, Whitlock and McCauley (1999) pointed out that indirect estimates of migration rates using gene frequency data often tend to be flawed because underlying assumptions are violated. Recently, new ways of estimating migration, m, independently of FST that do not rely on equilibrium assumptions have been developed (Beerli and Felsenstein, 2001), but most investigations of population structure per se still use presumed neutral markers, most likely because it is believed that selection may bias FST. However, as long as selection pressure is temporally stable (decades–centuries), genetic markers influenced by selection, such as Cpa112, can be of use in detecting population structure on ecological time scales, where neutral loci have not yet diverged substantially. Selected loci may be particularly useful in genetic stock identification based on individual assignment and as markers for population origin in mixed-stock analysis (see Nielsen et al., 2009a, p. 3136), for example, MHC in Pacific sockeye salmon (Beacham et al., 2005), the pantophysin locus in mixed stocks of coastal and oceanic cod in Lofoten, northern Norway (Wennevik et al., 2008) and Cpa112 in mixed-stock analysis of herring in the Skagerrak (Ruzzante et al., 2006). It may be preferable to use selection-influenced genetic markers with known function (Hoffmann and Willi, 2008), or with at least some knowledge of the nature of the selection pressure, for example, hemoglobin (Andersen et al., 2009), genes involved in osmoregulation (Larsen et al., 2008) and depth adaptation (Árnason et al., 2009). With recent advances in molecular technologies, it is anticipated that such information will become increasingly available. Moreover, loci under selection may allow for an estimation of the adaptive value of subpopulations, useful for conservation priorities (Bonin et al., 2007; Gebremedhin et al., 2009).
Acknowledgments
This paper is dedicated to Rene Stet who completed the initial research on the herring MHC. We thank two anonymous reviewers for very valuable comments and suggestions. Sampling and fish ageing were undertaken by the Marine Scotland Marine Laboratory in the United Kingdom, at IMR in Norway, and by the Swedish National Board of Fisheries. This work is part of the research project HERGEN (http://www.hull.ac.uk/hergen/) funded by the 5th EU Frame work program, and the efforts of the consortium are acknowledged. Financial support was provided by the Swedish Research Council for Environment FORMAS (CA, LL, NR), the Swedish Research Council VR (CA, LL, NR), and Sven and Lilly Lawski's Foundation (LCL). Parts of the analyses were conducted within the framework of the BaltGene project funded by BONUS Baltic Organisations' Network for Funding Science EEIG, and the Linnaeus Centre for Marine Evolutionary Biology at University of Gothenburg (http://www.cemeb.science.gu.se).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allendorf FW, Seeb LW. Concordance of genetic divergence among sockeye salmon populations at allozyme, nuclear DNA, and mitochondrial DNA markers. Evolution. 2000;54:640–651. doi: 10.1111/j.0014-3820.2000.tb00065.x. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Blackwell Publishing: Malden, MA; 2007. [Google Scholar]
- Allendorf FW, England PR, Luikart G, Ritchie PA, Ryman N. Genetic effects of harvest on wild animal populations. Trends Ecol Evol. 2008;23:327–337. doi: 10.1016/j.tree.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Andersen Ø, Wetten OF, De Rosa MC, André C, Alinovi CC, Colafranceschi M, et al. Haemoglobin polymorphisms affect the oxygen-binding properties in Atlantic cod populations. Proc R Soc B. 2009;276:833–841. doi: 10.1098/rspb.2008.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Ryman N, Rosenberg R, Ståhl G. Genetic variability in Atlantic herring (Clupea harengus harengus): description of protein loci and population data. Hereditas. 1981;95:69–78. [Google Scholar]
- Árnason E, Hernandez UB, Kristinsson K. Intense habitat-specific fisheries-induced selection at the molecular Pan I locus predicts imminent collapse of a major cod fishery. PLoS ONE. 2009;4:e5529. doi: 10.1371/journal.pone.0005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC.2004Molecular Markers, Natural History, and Evolution2 edn,Sinauer: Sunderland, MA [Google Scholar]
- Baer CF. Among-locus variation in FST: fish, allozymes and the Lewontin-Krakauer test revisited. Genetics. 1999;152:653–659. doi: 10.1093/genetics/152.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham TD, Candy JR, McIntosh B, MacConnachie C, Tabata A, Kaukinen K, et al. Estimation of stock composition and individual identification of sockeye salmon on a Pacific rim basis using microsatellite and major histocompatibility complex variation. Trans Am Fish Soc. 2005;134:1146. [Google Scholar]
- Beaumont MA.2002FDIST2Available from http://rubic.rdg.ac.uk/∼mab/software.html .
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc B. 1996;263:1619–1626. [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations using a coalescent approach. Proc Natl Acad Sci USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkevold D, André C, Dahlgren TG, Clausen LAW, Torstensen E, Mosegaard H, et al. Environmental correlates of population differentiation in Atlantic herring. Evolution. 2005;59:2656–2668. [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years. J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Bonin A, Nicole F, Pompanon F, Miaud C, Taberlet F. Population adaptive index: a new method to help measure intraspecific genetic diversity and prioritize populations for conservation. Conserv Biol. 2007;21:697–708. doi: 10.1111/j.1523-1739.2007.00685.x. [DOI] [PubMed] [Google Scholar]
- Bos DH, Gopurenko D, Williams RN, DeWoody JA. Inferring population history and demography using microsatellites, mitochondrial DNA, and major histocompatibility complex (MHC) genes. Evolution. 2008;62:1458–1468. doi: 10.1111/j.1558-5646.2008.00364.x. [DOI] [PubMed] [Google Scholar]
- Clausen LAW, Bekkevold D, Hatfield EMC, Mosegaard H. Application and validation of otolith microstructure as a stock identification method in mixed Atlantic herring (Clupea harengus) stocks in the North Sea and western Baltic. ICES J Mar Sci. 2007;64:377–385. [Google Scholar]
- Dahle G, Eriksen AG. Spring and autumn spawners of herring (Clupea harengus) in the North Sea, Skagerrak and Kattegat; population genetic analysis. Fisheries Res. 1990;9:131–141. [Google Scholar]
- de Eyto E, McGinnity P, Consuegra S, Coughlan J, Tufto J, Farrell K, et al. Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc R Soc B. 2007;274:861–869. doi: 10.1098/rspb.2006.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fauvelot C, Lemaire C, Planes S, Bonhomme F. Inferring gene flow in coral reef fishes from different molecular markers: which loci to trust. Heredity. 2007;99:331–339. doi: 10.1038/sj.hdy.6801005. [DOI] [PubMed] [Google Scholar]
- Gaggiotti OE, Bekkevold D, Jørgensen HBH, Foll M, Carvalho GR, André C, et al. Disentangling the effects of evolutionary, demographic, and environmental factors influencing genetic structure of natural populations: Atlantic herring as a case study. Evolution. 2009;63:2939–2951. doi: 10.1111/j.1558-5646.2009.00779.x. [DOI] [PubMed] [Google Scholar]
- Galarza JA, Carreras-Carbonell J, Macpherson E, Pascual M, Roques S, Turner GF, Rico C. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA. 2009;106:1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebremedhin B, Ficetola GF, Naderi S, Rezaei H-R, Maudet C, Rioux D, et al. Combining genetic and ecological data to assess the conservation status of the endangered Ethiopian walia ibex. Anim Conserv. 2009;12:89–100. [Google Scholar]
- Goudet J.2001FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3)Available from http://www.unil.ch/izea/softwares/fstat.html .
- Grant WS, Spies IB, Canino MF. Biogeographic evidence for selection on mitochondrial DNA in North Pacific walleye pollock Theragra chalcogramma. J Hered. 2006;97:571–580. doi: 10.1093/jhered/esl033. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Drablos F, Jorgensen SM, Hoyheim B, Stet RJM. The major histocompatibility class I locus in Atlantic salmon (Salmo salar L. polymorphism, linkage analysis and protein modelling. Immunogenetics. 2002;54:570–581. doi: 10.1007/s00251-002-0499-8. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, et al. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Hansen MM, Skaala O, Jensen LF, Bekkevold D, Mensberg KLD. Gene flow, effective population size and selection at major histocompatibility complex genes: brown trout in the Hardanger Fjord, Norway. Mol Ecol. 2007;16:1413–1425. doi: 10.1111/j.1365-294X.2007.03255.x. [DOI] [PubMed] [Google Scholar]
- Hauser L, Turan C, Carvalho GR. Haplotype frequency distribution and discriminatory power of two mtDNA fragments in a marine pelagic teleost (Atlantic herring, Clupea harengus) Heredity. 2001;87:621–630. doi: 10.1046/j.1365-2540.2001.00956.x. [DOI] [PubMed] [Google Scholar]
- Hauser L, Carvalho GR. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. Fish Fish. 2008;9:333–362. [Google Scholar]
- Hemmer-Hansen J, Nielsen EE, Frydenberg J, Loeschcke V. Adaptive divergence in a high gene flow environment: Hsc70 variation in European flounder (Platichthys flesus L.) Heredity. 2007;99:592–600. doi: 10.1038/sj.hdy.6801055. [DOI] [PubMed] [Google Scholar]
- Hoarau G, Piquet AMT, Van der Veer HW, Rijnsdorp AD, Stam WT, Olsen JL. Population structure of plaice (Pleuronectes platessa L.) in northern Europe: a comparison of resolving power between microsatellites and mitochondrial DNA data. J Sea Res. 2004;51:183–190. [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- ICES 2003Report of the Herring Assessment Working Group for the area South of 62°NICES CM 2003 /ACFM: 17.
- Inoue JG, Miya M, Tsukamoto K, Nishida M. Complete mitochondrial DNA sequence of the Japanese sardine Sardinops melanostictus. Fish Sci. 2000;66:924–932. [Google Scholar]
- Johannesson K, André C. Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol. 2006;15:2013–2029. doi: 10.1111/j.1365-294X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- Jorde PE, Knutsen H, Espeland SH, Stenseth NC. Spatial scale of genetic structuring in coastal cod Gadus morhua and the geographic extent of local populations. Mar Ecol Prog Ser. 2007;343:229–237. [Google Scholar]
- Jørgensen HBH, Hansen MM, Bekkevold D, Ruzzante DE, Loeschcke V. Marine landscapes and population genetic structure of herring (Clupea harengus L.) in the Baltic Sea. Mol Ecol. 2005;14:3219–3234. doi: 10.1111/j.1365-294X.2005.02658.x. [DOI] [PubMed] [Google Scholar]
- Jørgensen HBH, Pertoldi C, Hansen MM, Ruzzante DE, Loeschcke V. Genetic and environmental correlates of morphological variation in a marine fish: the case of Baltic Sea herring (Clupea harengus) Can J Fish Aquat Sci. 2008;65:389–400. [Google Scholar]
- Kalinowski ST. How many alleles per locus should be used to estimate genetic distances. Heredity. 2002;88:62–65. doi: 10.1038/sj.hdy.6800009. [DOI] [PubMed] [Google Scholar]
- Karl SA, Avise JC. Balancing selection at allozyme loci in oysters: implications from nuclear RFLPs. Science. 1992;256:100–102. doi: 10.1126/science.1348870. [DOI] [PubMed] [Google Scholar]
- Larsen PF, Nielsen EE, Koed A, Thomsen DS, Olsvik PA, Loeschcke V. Interpopulation differences in expression of candidate genes for salinity tolerance in winter migrating anadromous brown trout (Salmo trutta L.) BMC Genetics. 2008;9:12. doi: 10.1186/1471-2156-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson LC, Laikre L, Palm S, André C, Carvalho GR, Ryman N. Concordance of allozyme and microsatellite differentiation in a marine fish, but evidence of selection at a microsatellite locus. Mol Ecol. 2007;16:1135–1147. doi: 10.1111/j.1365-294X.2006.03217.x. [DOI] [PubMed] [Google Scholar]
- Larsson LC, Charlier J, Laikre L, Ryman N. Statistical power for detecting genetic divergence–organelle versus nuclear markers. Conserv Genet. 2009;10:1255–1264. [Google Scholar]
- Larsson LC, Laikre L, André C, Dahlgren TG, Ryman N. Temporally stable genetic structure of heavily exploited Atlantic herring (Clupea harengus) in Swedish waters. Heredity. 2010;104:40–51. doi: 10.1038/hdy.2009.98. [DOI] [PubMed] [Google Scholar]
- Lewis PO, Zaykin D.2001Genetic Data Analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c)Free program distributed over the internet from http://lewis.eeb.uconn.edu/lewishome/software.html .
- Limborg MT, Pedersen JS, Hemmer-Hansen J, Tomkiewicz J, Bekkevold D. Genetic population structure of European sprat, Sprattus sprattus: differentiation across a steep environmental gradient in a small pelagic fish. Mar Ecol Prog Ser. 2009;379:213–224. [Google Scholar]
- Mariani S, Hutchinson WF, Hatfield EMC, Ruzzante DE, Simmonds EJ, Dahlgren TG, et al. North Sea herring population structure revealed by microsatellite analysis. Mar Ecol Prog Ser. 2005;303:245–257. [Google Scholar]
- McPherson AA, O′Reilly PT, McParland TL, Jones MW, Bentzen P. Isolation of nine novel tetranucleotide microsatellites in Atlantic herring (Clupea harengus) Mol Ecol Notes. 2001;1:31–32. [Google Scholar]
- Muirhead CA. Consequences of population structure on genes under balancing selection. Evolution. 2001;55:1532–1541. doi: 10.1111/j.0014-3820.2001.tb00673.x. [DOI] [PubMed] [Google Scholar]
- Nielsen EE, Hansen MM, Meldrup D. Evidence of microsatellite hitch-hiking selection in Atlantic cod (Gadus morhua L.) implications for inferring population structure in nonmodel organisms. Mol Ecol. 2006;15:3219–3229. doi: 10.1111/j.1365-294X.2006.03025.x. [DOI] [PubMed] [Google Scholar]
- Nielsen EE, MacKenzie BR, Magnussen E, Meldrup D. Historical analysis of Pan I in Atlantic cod (Gadus morhua): temporal stability of allele frequencies in the southeastern part of the species distribution. Can J Fish Aquat Sci. 2007;64:1448–1455. [Google Scholar]
- Nielsen EE, Hemmer-Hansen J, Larsen PF, Bekkevold D. Population genomics of marine fishes: identifying adaptive variation in space and time. Mol Ecol. 2009a;18:3128–3150. doi: 10.1111/j.1365-294X.2009.04272.x. [DOI] [PubMed] [Google Scholar]
- Nielsen EE, Wright PJ, Hemmer-Hansen J, Poulsen NA, Gibb IM, Meldrup D. Microgeographical population structure of cod Gadus morhua in the North Sea and west of Scotland: the role of sampling loci and individuals. Mar Ecol Prog Ser. 2009b;376:213–225. [Google Scholar]
- O'Malley KG, Camara MD, Banks MA. Candidate loci reveal genetic differentiation between temporally divergent migratory runs of Chinook salmon (Oncorhynchus tshawytscha) Mol Ecol. 2007;16:4930–4941. doi: 10.1111/j.1365-294X.2007.03565.x. [DOI] [PubMed] [Google Scholar]
- Olsen JB, Lewis CJ, Kretschmer EJ, Wilson SL, Seeb JE. Characterization of 14 tetranucleotide microsatellite loci derived from Pacific herring. Mol Ecol Notes. 2002;2:101–103. doi: 10.1046/j.1365-294x.2000.105317.x. [DOI] [PubMed] [Google Scholar]
- Rand DM. Neutrality tests of molecular markers and the connection between DNA polymorphism, demography and conservation biology. Conserv Biol. 1996;10:665–671. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Ruzzante DE, Mariani S, Bekkevold D, André C, Mosegaard H, Clausen LAW, et al. Biocomplexity in a highly migratory pelagic marine fish, Atlantic herring. Proc R Soc B. 2006;273:1459–1464. doi: 10.1098/rspb.2005.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N, Lagercrantz U, Andersson L, Chakraborty R, Rosenberg R. Lack of correspondence between genetic and morphologic variability patterns in Atlantic herring. Heredity. 1984;53:687–704. [Google Scholar]
- Ryman N. CHIFISH: a computer program testing for genetic heterogeneity at multiple loci using chi-square and Fisher's exact test. Mol Ecol Notes. 2006;6:285–287. [Google Scholar]
- Ryman N, Palm S. POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes. 2006;6:600–602. doi: 10.1046/j.0962-1083.2001.01345.x. [DOI] [PubMed] [Google Scholar]
- Ryman N, Palm S, André C, Carvalho GR, Dahlgren TG, Jorde PE, et al. Power for detecting genetic divergence: differences between statistical methods and marker loci. Mol Ecol. 2006;15:2031–2045. doi: 10.1111/j.1365-294X.2006.02839.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Serrão EA, Pearson GA, Riginos C, Rawson PD, Hilbish TJ, et al. Ecological genetics in the North Atlantic: environmental gradients and adaptation at specific loci. Ecology. 2008;89:S91–S107. doi: 10.1890/07-1162.1. [DOI] [PubMed] [Google Scholar]
- Stet RJM, de Vries B, Mudde K, Hermsen T, van Heerwaarden J, Shum BP, et al. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics. 2002;54:320–331. doi: 10.1007/s00251-002-0477-1. [DOI] [PubMed] [Google Scholar]
- Stet RJM, Mudde K, Wynne JW, Nooijen A, Dahlgren TG, Ruzzante DE, et al. Characterization of a major histocompatibility class II A gene (Clha-DAA) with an embedded microsatellite marker in Atlantic herring (Clupea harengus L.) J Fish Biol. 2008;73:367–381. [Google Scholar]
- Tonteri A, Vasemägi A, Lumme J, Primmer CR. Beyond MHC: signals of elevated selection pressure on Atlantic salmon (Salmo salar) immune-relevant loci. Mol Ecol. 2010;19:1273–1282. doi: 10.1111/j.1365-294X.2010.04573.x. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Waples RS. Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered. 1998;89:438–450. [Google Scholar]
- Wennevik V, Jørstad KE, Dahle G, Fevolden S-E. Mixed stock analysis and the power of different classes of molecular markers in discriminating coastal and oceanic Atlantic cod (Gadus morhua L.) on the Lofoten spawning grounds, Northern Norway. Hydrobiologia. 2008;606:7–25. [Google Scholar]
- Westgaard J-I, Fevolden S-E. Atlantic cod (Gadus morhua L.) in inner and outer coastal zones of northern Norway display divergent genetic signature at non-neutral loci. Fisheries Res. 2007;85:306–315. [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST≠1/(4 Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Wynne JW, Cook MT, Nowak BE, Elliott NG. Major histocompatibility polymorphism associated with resistance towards amoebic gill disease in Atlantic salmon (Salmo salar L.) Fish Shellfish Immunol. 2007;22:707–717. doi: 10.1016/j.fsi.2006.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.