Abstract
Recently developed Bayesian genotypic clustering methods for analysing genetic data offer a powerful tool to evaluate the genetic structure of domestic farm animal breeds. The unit of study with these approaches is the individual instead of the population. We aimed to empirically evaluate various individual-based population genetic statistical methods for characterization of genetic diversity and structure of livestock breeds. Eighteen British pig populations, comprising 819 individuals, were genotyped at 46 microsatellite markers. Three Bayesian genotypic clustering approaches, principle component analysis (PCA) and phylogenetic reconstruction were applied to individual multilocus genotypes to infer the genetic structure and diversity of the British pig breeds. Comparisons of the three Bayesian genotypic clustering methods (, and ) revealed some broad similarities but also some notable differences. Overall, the methods agreed that majority of the British pig breeds are independent genetic units with little evidence of admixture. The three Bayesian genotypic clustering methods provided complementary, biologically credible clustering solutions but at different levels of resolution. detected finer genetic differentiation and in some cases, populations within breeds. Consequently, it estimated a greater number of underlying genetic populations (K, in the notation of Bayesian clustering methods). Two of the Bayesian methods ( and ) and phylogenetic reconstruction provided similar success in assignment of individuals, supporting the use of these methods for breed assignment.
Keywords: Bayesian, domestic pig, genetic structure, livestock breeds, microsatellites
Introduction
Farm animal domestic breeds are recognized for their economic, social and cultural value (DEFRA, 2006). Traditional characterization of breeds by phenotypic attributes can now be complemented with information on genetic diversity and structure. Adequate understanding of the genetic structure and diversity of breeds is useful to provide an objective evaluation for future breed preservation (DEFRA, 2006). Population genetic statistics, developed to infer the population structure of naturally occurring populations, are now commonly used to describe the genetic diversity and structure of livestock breeds (for example, MacHugh et al., 1998; SanCristobal et al., 2006). The genetic structure within and between populations have traditionally been measured using population genetic measures such as Wright's F-statistics or their derivatives, and population genetic distances typified by Nei's D (Hartl and Clark, 1989). However, analysis at the population level may be restrictive and, increasingly, these measures have received scrutiny and criticism (Pearse and Crandall, 2004). First, a priori assignment of individuals to specific populations, usually based on sampling locations or phenotypes, might introduce bias. It imposes a subjective pre-existing structure that may not reflect reality. Second, statistical estimates are averaged across individuals within populations. As a result, processes such as migration, genetic introgression and cross-breeding are blurred. Traditional statistical population genetic approaches can miss admixed or hybrid individuals, and related biological processes, leading to inaccurate representations of breed diversity and structure.
Pritchard et al. (2000) recognized the need for a procedure to identify genetically differentiated populations directly from individual genetic polymorphism data, instead of relying on a priori population information. Several genotypic clustering models have since been developed in a Bayesian statistical framework and are available in special purpose software packages: (Pritchard et al., 2000; Falush et al., 2003), (Guillot et al., 2005), (Bayesian Analysis of Population Structure; Corander et al., 2006) and (Huelsenbeck and Andolfatto, 2007). The methods operate by creating clusters in which the assumptions of Hardy–Weinberg and linkage equilibria are met, and simultaneously each individual is assigned to a cluster based on a probabilistic model. Each method has slightly different underlying assumptions and different methods of searching the parameter space. Genotypic clustering techniques are now prominent computational tools as they have proved very useful in the field of population genetics and the range of practical applications is broad (Beaumont and Rannala, 2004; Pearse and Crandall, 2004).
In addition to Bayesian genotypic clustering approaches, two other individual-based methods are also available, principal component analysis (PCA; Menozzi et al., 1978) and phylogenetic reconstruction (Bowcock et al., 1994). Both techniques have been used for several decades to study genetic structure and diversity, but are most often used on population-averaged data where populations are defined a priori. Population-based phylogenetic reconstruction may be an unreliable approach (Toro and Caballero, 2005). First, as mentioned above, within population genetic variation is ignored as estimates are averaged across individuals. Second, the principles of phylogenetic reconstruction are not upheld when applied to admixed populations, because one assumption is that there is no genetic exchange between populations (Toro and Caballero, 2005). Adopting an individual-based approach to livestock genetic diversity analyses may be more appropriate for considering past cross-breeding. Like Bayesian genotypic clustering, individual-based PCA and phylogenetic reconstruction make no assumptions about the number or identity of separate populations from which individuals are drawn.
All the approaches described above are potentially useful for the elucidation of livestock breed diversity and structure. However, it is still not clear how the methods differ in their power and appropriateness for particular data and questions. In addition, individual-based PCA and phylogenetic reconstruction approaches are rarely used in conjunction with Bayesian genotypic clustering methods. The objective of this study was to empirically assess various individual-based population genetic statistical methods for inference of genetic diversity and structure of livestock breeds. Using microsatellite data from British pig breeds, the genetic structure was inferred with various ‘individual-based' approaches: three Bayesian genotypic clustering techniques, PCA and phylogenetic reconstruction. The applicability, efficacy and complementarity of the chosen methods were considered.
Materials and methods
The genotypic data of British pig breeds used in this study were a subset from an extensive European project on pig breed biodiversity (PigBioDiv http://www.projects.roslin.ac.uk/pigbiodiv/). The microsatellites recommended for the European pig biodiversity had high polymorphism and good typing performance and were well spaced across the genome (SanCristobal et al., 2006). Forty-six microsatellites were selected for this study as they were genotyped across all of the selected populations. The proportion of missing data was 6.8%. Additional information on the chromosomal location and polymorphism level of each marker is provided in Supplementary Materials 1. Deviation from Hardy–Weinberg equilibrium within loci and the presence of genotypic linkage disequilibrium between pairs of loci were tested using version 4.0.7 (Rousset, 2008). No markers showed consistent evidence of linkage disequilibrium or deviations from Hardy–Weinberg equilibrium (Supplementary Materials 2).
In brief, there were a total of 18 populations. Twelve British pig breeds were included with two breeds represented by more than one line (Table 1). The term ‘population' was used to represent commercial lines sampled within a breed. An Asian breed (Meishan) was chosen as an outgroup, composed of two populations, one from Great Britain (sampled from both the Roslin Institute and from PIC, a UK-based pig breeding company) and another from France. The number of individuals sampled per population ranged from 25 to 53, giving a total of 819 individuals (Table 1). Additional information on the sampling and genotyping of the microsatellite markers can be found at http://www.projects.roslin.ac.uk/pigbiodiv/ and in SanCristobal et al. (2006).
Table 1. Pig breeds.
| Population | Breed | Categorya | Classificationb | Sample size |
|---|---|---|---|---|
| 1 | Berkshire | T | V | 50 |
| 2 | British Lop | T | E | 35 |
| 3 | British Saddleback | T | AR | 42 |
| 4 | Duroc (PIC) | C | 50 | |
| 5 | Gloucester Old Spots | T | M | 53 |
| 6 | Hampshire (PIC) | C | 50 | |
| 7 | British Landrace (PIC1) | C | 50 | |
| 8 | British Landrace (PIC2) | C | 50 | |
| 9 | British Landrace (PIC3) | C | 48 | |
| 10 | Large Black | T | V | 52 |
| 11 | Large White (PIC1) | C | 50 | |
| 12 | Large White (PIC2) | C | 50 | |
| 13 | Large White (PIC3) | C | 50 | |
| 14 | Middle White | T | E | 38 |
| 15 | Pietrain | C | 50 | |
| 16 | Tamworth | T | V | 42 |
| 17 | Meishan (France) | A | 25 | |
| 18 | Meishan (UK) | A | 36 |
Traditional breed (T), Commercial breed (C), Asian import (A).
Endangered (E), vulnerable (V), at risk (AR), minority (M): according to the Rare Livestock Breed Trust (RBST).
Bayesian genotypic clustering techniques
In this study, we evaluated the utility of two widely used Bayesian methods ( and ) and a newer method (). The clustering methods perform a Bayesian analysis, using the multilocus genotypes, to probabilistically assign individuals to clusters and infer the number of genetically distinguishable populations (K). The three methods assume that all markers are in Hardy–Weinberg Equilibrium and genotypic linkage equilibrium. Both and allow individuals to be of mixed ancestry, proportionally assigning an individual genome to clusters (estimated individual coefficients of ancestry, ‘q'), but differ in their approach to estimating admixture.
uses a Monte Carlo Markov Chain method and estimates the natural logarithm of the probability (Pr) of the observed genotypic array (X), given a pre-defined number of clusters (parameter K) in the data set (ln Pr(X∣K)) (Pritchard et al., 2000). In a Bayesian context the estimate of ln Pr(X∣K) is a direct indicator of the posterior probability of having K clusters, given the observed genotypic array. The model with correlated allele frequencies was implemented, assuming admixture. This model assumes that frequencies from different populations are likely to be similar due to either migration or shared ancestry. The Markov Chain was run for 1 000 000 iterations, after a burn-in of 500 000 iterations, for values of K from 1 to 20, with five replicates for each K value. From each Monte Carlo Markov Chain chain, simultaneously infers the posterior probability of K and membership probabilities (q) for each individual. Individuals may be assigned probabilistically to more than one cluster, reflecting admixture. To help identify the optimal K value, an ad hoc statistic, ΔK, was calculated (Evanno et al., 2005). It is based on the second order rate of change of Pr(X∣K) with respect to K, where the magnitude of the estimated values indicate the strength of the population subdivision.
The most recent version of (v 5.2) uses a ‘greedy stochastic optimization algorithm' to directly estimate the most likely K and assign individuals to clusters (Corander et al., 2008). In the value of K can be either pre-defined to investigate the clustering solutions of populations with successive K values or left undefined so that the algorithm searches for the most likely K value. For each K value searches for the optimal partitions, stores them internally and, after all K value have been processed, it merges the stored results according to log-likelihood values. Five independent replicate runs for every level of K from 1 to 20 were conducted. In , estimating individual admixture is a two-tiered approach. First the clustering solutions of populations are determined and then the admixture of genotypes is quantified by establishing the ancestral sources of alleles for each individual with respect to the determined clusters. In , evidence for admixture was considered significant for individuals with P-values <0.05 (Corander and Marttinen, 2006).
The third Bayesian clustering approach was performed using the program (Huelsenbeck and Andolfatto, 2007). The method implemented is similar to that in except that treats K as a random variable. A prior distribution is placed on K such that the data determines the most appropriate value. The number of clusters and the assignment of genotypes to those clusters were estimated simultaneously. A Markov chain of 100 000 iterations following a burn-in of 50 000 iterations was sufficient for convergence and production of consistent results. A partition was sampled from the Markov chain every 100 iterations and the mean partition, which minimizes the squared distance to the sampled partitions, was calculated to make assignments. This process was independently replicated five times.
Multivariate analysis
The second approach was a multivariate PCA (Menozzi et al., 1978) performed using the statistical package R (Team RDC, 2009). PCA is a technique that reduces multi-dimensional data to a few principal components (PCs) that explain the most variability in the data set. PCs that explain the most genetic variation are identified and individuals can then be clustered along axes based on their allele distribution. The data for individual genotypes were prepared by scoring a ‘0' if a particular allele was not present, a ‘1' if it was present and ‘2' if two copies were present in the homozygous state (MacHugh et al., 1998; Patterson et al., 2006). Two statistical tests were conducted to determine the number of PCs to retain. Horn's parallel analysis is a simulation procedure where a random correlation matrix, of the same dimensions as the empirical data, is generated and subjected to PCA. The components to retain from the PCA on the empirical data are those that account for more variance than the components derived from random simulation. The parallel analysis was conducted in the R package paran (Dinno, 2009). The second test is Velicer's minimum average partial test (O'Connor, 2000), which involves a PCA on the empirical data followed by an examination of a series of matrices of partial correlations. Partial correlation analysis measures the degree of association between two random variables after removing the effects of other variables. The average of the squared correlations of the off-diagonal partial correlation matrix is then computed. This average should decrease as long as shared variance is being extracted, but begin to increase when error variance predominates and extraction of components should stop. A script was written and executed in R.
Phylogenetic reconstruction
The final approach was an unrooted phylogenetic analysis implemented in MICROSAT (Minch et al., 1997). Pairwise individual genetic distances (shared allele distance, DSA) were estimated using the proportion of shared alleles (Bowcock et al., 1994). At any locus, two individuals' genotypes share either 0, 1 or 2 alleles; across large numbers of loci, the proportion of shared alleles becomes almost continuously distributed and is an index of genetic similarity. An unrooted neighbour-joining cladogram was constructed from the distance matrix of proportion of shared alleles values for all pairs of individuals using the R package APE (Paradis et al., 2004). Bootstrapping of 1000 replicates was performed across loci in MICROSAT and the consensus cladogram was calculated using CONSENSE (Phylip v 3.67; Felsenstein, 1989).
Results
Bayesian genotypic clustering
Number of populations (K) and clustering solutions
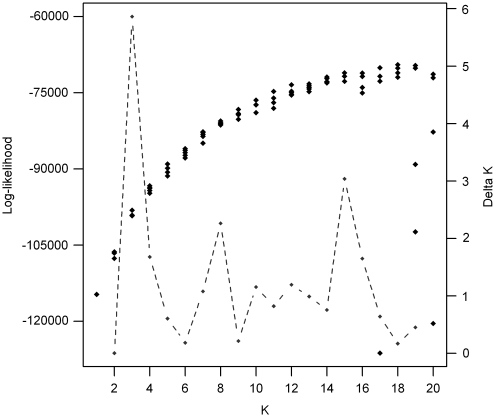
Results from the analysis showed steadily increasing values of log likelihoods from K=1 to 20 subpopulations (Figure 1). The largest ΔK value, ΔK=5, was at K3–K2, followed by second and third modes at K8–K7 and K15–K14, respectively (Figure 1). These were not strong or conclusive as compared with, for example, ΔK=130 reported by Frantz et al. (2006). Variance in the log likelihoods for a given K increased at high values of K (K>16) as has been reported with other data sets (Rosenberg et al., 2001; Evanno et al., 2005).
Figure 1.
Likelihood plot of results. The black points are the likelihood values and the grey points are the estimated delta values. The plot illustrates the difficulty in deciding the most likely number of subpopulations in the data set.
At K=2 there was inconsistent clustering solutions between runs. At K=3 the Asian lines either clustered with the British Landrace line or independently. Regarding the clustering patterns, until K=4 the Asian lines clustered with the British populations. The British Lop and the British Landrace lines consistently clustered together (Figure 2a). At K=12, in three out of five replicates British Saddleback individuals split over two clusters and the Berkshire and Gloucestershire Old Spots clustered together (Figure 2a, Table 2). The British Lop split from the British Landrace lines at K=13 to form its own cluster. At values greater than K=16 the confidence of assignments fell dramatically, with ‘ghost' or empty clusters observed. For the remaining breeds the results were inconsistent between the runs such that a large number of clustering solutions were depicted. The clustering solutions reproduced in Figure 2a are a consensus of 5 replicate runs. As can be seen, there is partial hierarchical splitting of clusters at each stage (Figure 2a), but also some inconclusive splitting: for instance, Gloucestershire Old Spots splitting (K=10) and then rejoining Berkshire (K=12). A sensitivity analysis was conducted where various starting parameters including ALPHA (the degree of admixture), ALPHAPROPSD (standard deviation for ALPHA that allows for better mixing in the Metropolis-Hasting chain) and LAMBDA (distribution of allelic frequencies) varied from the default values in an attempt to produce more repeatable results. This did not decrease the variation in log likelihood estimates at high K values, nor alter the log likelihood curve or the inconsistent clustering solutions.
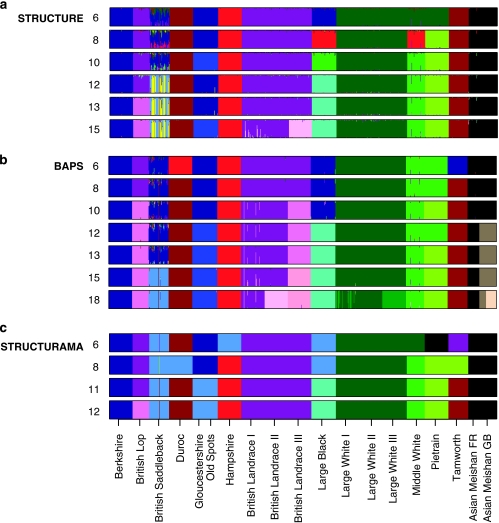
Figure 2.
Individual assignment based on Bayesian genotypic cluster analysis, at various values of K. Histograms demonstrate the proportion of each individual's genome that originated from each of 18 populations. Each individual is represented by a vertical line corresponding to its membership coefficient (q). (a) Histograms constructed from results. Histograms are a consensus view across five replicates. (b) Histograms constructed from results. (c) Histograms constructed from results. could not converge on a solution for a fixed value of K=10.
Table 2. Comparison of breed allocations to groups using different methods.
| Breed | DSA | |||
|---|---|---|---|---|
| Berkshire | Aa | Aa | A | A |
| British Lop | Ba | Ba | B | Ba |
| British Saddleback | C,D | Aa | Ca | C,D |
| Duroc | E | C | D | E |
| Gloucestershire Old Spots | Aa | D | Ca | F |
| Hampshire | F | E | E | G |
| British Landrace | Ba | F | Ba | |
| Line 1 | F | |||
| Line 2 | F | |||
| Line 3 | Ba | |||
| Large black | G | G | G | H |
| Large White | H | H | H | I |
| Line 1 | ||||
| Line 2 | ||||
| Line 3 | ||||
| Middle White | I | Ia | I | J |
| Pietrain | J | Ia | J | K |
| Tamworth | K | J | K | L |
| Meishan | L | L | M | |
| Line 1 | K | |||
| Line 2 | L |
Unlike , provided a probabilistic approximation of the number of clusters when K was left undefined and the optimal partition was identified at K=18 (Pr (K=18∣X)=1.0). At K=18, all populations formed their own independent cluster except: (i) the two Large White lines formed one population and, (ii) the Asian Meishan GB line split over two populations. When K was predefined the clustering solutions were identical between replicate runs at a given K value. At K=2, the Asian populations formed one cluster separate from the British populations. As K increased, the commercial breeds first split away to form their own clusters: the Large White (K=4), British Lop-Landrace lines (K=6), Hampshire (K=7), Duroc (K=8) and Pietrain (K=9). The Middle White and Tamworth were the only two traditional British breeds that split at lower K values to occupy independent clusters (K=9 and K=8, respectively). This left the remaining four traditional British breeds Berkshire, British Saddleback, Gloucestershire Old Spots and Large Black as a single cluster from K=7 to 10 (Figure 2b). Once the commercial breeds inhabited independent clusters, at K=9, the group of four indigenous breeds began to split. Berkshire and British Saddleback formed a single cluster from K=11–14. At K=12, the two Asian Meishan lines split to occupy independent clusters (Figure 2b, Table 2). The grouping of British Lop-Landrace into a single cluster was observed until K=9. At K=19 the first ‘ghost' population was observed. In the analysis the British Saddleback did not split into two clusters at any point; instead all individuals formed a single genetic population from K=14–18 (Figure 2b).
The final Bayesian implementation was performed using , which like , allows K to be a random variable and thus estimates K. The estimated number of populations was 11 across the five independent runs (Pr (K=11∣X)=0.99). The clustering of British Lop-Landrace lines in a single cluster was again observed and British Saddleback and Gloucestershire Old Spots were placed in one cluster. All other breeds formed independent clusters. Clustering solutions are given in Figure 2c for various fixed K values. Hierarchical splitting of clusters at lower K values was not observed (Figure 2c). At K=12 British Lop split from the British Landrace (Figure 2c, Table 2). could not converge on a clustering solution for a fixed value of K=10 and from K=13 upwards.
Assignment of individuals and admixture
The majority of individuals clustered to pre-labelled population origin (proportion of genome assignments, q>0.9, Figure 2). At lower K values (—K⩽11 and —K⩽15), British Saddleback individuals appeared to be admixed, probably a reflection of the inability of the algorithms to resolve the clustering of this breed. At higher K values split the British Saddleback into two separate clusters with four individuals being admixed, whereas retained the breed as a single genetic unit with admixed individuals present (Figures 2a and b). At higher K values, (both and at K⩾16) the Large White individuals split into two clusters, but not strictly according to population identities. Some individuals from Large White Line I clustered with individuals from Line III. Both and identified the same individual-labelled British Saddleback as being of Tamworth origin (q>0.9) and individuals of Middle White with a proportion of DNA from other breeds (q>0.15). identified, from K⩾5, one individual from Gloucestershire Old Spots with a substantial proportion of British Saddleback DNA (q>0.25) and one individual from the French Meishan population with a proportion of Large Black DNA (q>0.15). identified five individuals of British Landrace line I with proportions of DNA from British Landrace III and British Lop.
Principle component analysis
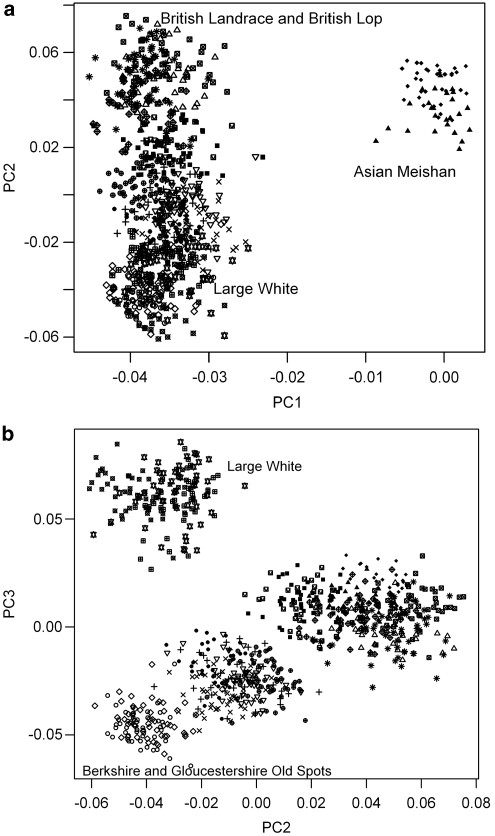
The first two principle components are shown plotted in Figure 3. The first PC accounted for 29.3% of the underlying variation and the second PC accounted for 4.3%. The first PC clearly split the British from the Asian populations. The second PC gives a coarse separation of the British breed individuals: British Landrace lines and British Lop group clustering at the top of the quadrant with Large White lines grouping at the bottom (Figure 3a). The third PC, which accounted for 4.1% of the variation, showed additional structuring amongst the British breeds (Figure 3b). The Large White lines were clearly separated from the other breeds at the top left of the plot and the Berkshire and Gloucestershire Old Spots clustered together at the bottom left. With increasing dimensions there was further breed partitioning: PC 5 (3.3%) separated the Hampshire and Tamworth breeds, PC 6 (2.4%) separated the Duroc breed and PC 7 (2.1%) separated the Pietrain, Middle White and Large Black breeds. When PCA was conducted on just the British populations, congruent results were produced. The first PC (30%) spread out individuals within the breeds and the second PC (4.6%) gave a coarse separation of the populations. The PCA projection was similar to that shown in Figure 3a, save for the Asian populations. According to Velicer's minimum average partial test over 50 principle components should be retained whereas the Parallel Test indicated 28. However, components were noisy and non-informative by the 20th PC as there visually appeared to be no structure present.
Figure 3.
Principle component analysis projections. (a) scatterplot diagram showing the first and second PCs and allele distribution from all individuals. (b) scatterplot diagram showing the second and third PCs and allele distribution from all individuals.
Phylogenetic reconstruction
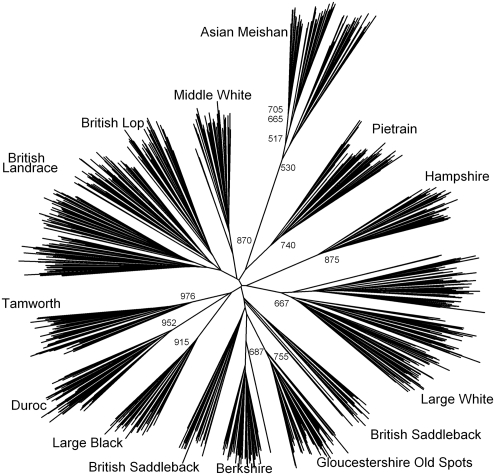
The phylogenetic reconstruction based on the proportion of shared alleles distance measure is presented in Figure 4. All individuals clustered to their designated breed origin except for the British Saddleback, in which individuals split into two clusters (Figure 4, Table 2). One British Saddleback individual fell within the Tamworth clade (the same individual identified in the Bayesian genotypic clustering analyses). There was high bootstrap support for individuals belonging to their breed of origin, except for the British Landrace-Lop and the British Saddleback groupings. The longest branches separated individuals within breeds implying that there is greater variation within than between breeds. There was no bootstrap support for genetic relationships between the pig breeds.
Figure 4.
A neighbour-joining tree constructed from allele-sharing distances among all individuals. Bootstrap values greater than 500 are shown.
Discussion
Bayesian genotypic clustering tools
Bayesian genotypic clustering methods offer the prospect of inferring the number of underlying populations, K, present in an empirical data set. However, obtaining a definitive value of K for the British pig breeds was a challenge as the three Bayesian methods, , and , yielded slightly different answers. The algorithm does not provide a statistical indication of the most likely K. Instead, K is identified at a point of inflection on the log-likelihood curve that leads to a plateau or by the maximum value (Pritchard and Wen, 2004). However, when there is a continual increase in the log likelihood (Figure 1) choosing K may be problematic and is often a subjective task (Frantz et al., 2006). The Evanno et al. (2005) ΔK method did not clarify the best value of K. Through visual observation of the log-likelihood curve, it is probable that the value of K lies between 10 and 15 (Figure 1). This is also supported by the fact that ‘ghost' populations started to appear from K=16 in the analysis.
Other studies on both domestic and wild species have similarly experienced difficulty in identifying K using (cat breeds (Menotti-Raymond et al., 2008) and red deer populations (Frantz et al., 2006)). In analysis the very gradual increase in log-likelihood values up to an asymptote may be indicative of the presence of genetic continuity across breeds. Such a pattern could be a possible consequence of limited breed barriers due to a short history, out-crossing and a common ancestry (Menotti-Raymond et al., 2008). In wild populations, similar results have been attributed to an isolation-by-distance relationship (Frantz et al., 2006). In that situation, individuals are spatially distributed across space and allelic frequencies of sampled populations vary gradually across the region. The underlying model is not well suited to data from this kind of scenario and defining distinct genetic units can be challenging (Pritchard and Wen, 2004).
implements a simpler version of the model but it also allows K to be a random variable. As the manual selection of the number genetic populations may be considered a drawback, this is a useful improvement. gave a value of 11 for the number of underlying populations. At this value of K, also produced a biologically credible clustering solution, in which all breeds were independent units except for British Lop-British Landrace and British Saddleback-Gloucestershire Old Spots.
estimated a higher value of K (18) than and . The clustering result at this value of K did not entirely correlate with the 18 identified populations (Table 1) in that the Asian Meishan GB line was split into two clusters, the Large White lines I and II clustered together and a few individuals of Large White I clustered with Large White III (Figure 2b). Rowe and Beebee (2007) concluded that overestimated K in British natterjack toad populations, generating more genetically distinct groups than did . However, tests using simulated data showed that was reliable at estimating the true number of populations, with a performance comparable to that of (Waples and Gaggiotti, 2006). Weak random fluctuations in allele frequencies detected by the algorithm may be considered as evidence of genetic differentiation among sub-populations (Corander et al., 2008).
It is difficult to compare the clustering solutions of versus at higher values of K as the former was not consistent between runs and also produced one or more ‘ghost' populations. Comparison of returned clustering solutions at lower K values between the Bayesian genotypic clustering approaches showed that results were not entirely complementary. The methods concurred that certain breeds became distinct genetic units at low K values (for example, Duroc, Tamworth, Large White; Figure 2). Yet, the clustering solutions of other breeds (namely Berkshire, British Saddleback and Gloucestershire Old Spots) were largely unresolved due to inconsistent results between the methods. At a specific value of K, different pairings of breeds were observed (for example, K=12, Table 2). In addition, sometimes the methods returned the same clustering observations but at different levels of K. For instance, observed subdivision between the lines of British Landrace from K=10 (Figure 2b), whereas produced this result at a higher value of K=15 (Figure 2a).
Another incongruency between the Bayesian genotypic clustering methods was in the detection of substructure within breeds (Figure 2). The British Saddleback did not form a single cluster according to the (Figure 2a) and phylogenetic analysis (although the bootstrap support for this was low, ∼30% Figure 4), whereas did not provide any evidence of substructuring within the British Saddleback breed (Figure 2b). In contrast, separated the French (FR) Meishan from the British (GB) Meishan, and furthermore, detected substructure with the GB Meishan (K=18, Figure 2b). Phylogenetic reconstruction of only the British Saddleback individuals reproduced the same result of the full data set (result not shown), although again there was very low bootstrap support for the separation of the two groups. In contrast, there was high bootstrap support (>50%) for the division of GB Meishan into two genetic clusters (Figure 4). The GB Meishan was known to be composed of individuals from two separate populations, thus these findings reflect true differentiation. The British Saddleback, on the other hand, was considered a panmictic population.
did not reveal definitive substructure beyond the level of breed in those composed of separate lines. At high values of K there was evidence for further substructure in the British Landrace and Large White (Figure 2a). However, ‘ghost' populations, inconsistent clustering solutions and a large variation in Ln [(Pr (X∣K)] were evident at these high K values (Figure 1). This indicates that the Monte Carlo Markov Chain had not converged, which could suggest that genetic differentiation was weak (Latch et al., 2006; Waples and Gaggiotti, 2006). However, the levels of genetic differentiation in this analysis should have been sufficient (FST>0.05, suggested by Latch et al., 2006) for the detection of the substructure in this data set (British Landrace lines, FST=0.156 (mean FST between the three lines); the two identified genetic clusters of British Saddleback, FST=0.077; the two identified genetic clusters of GB Meishan, FST=0.178).
Overall, detected genetic structure at a finer scale than the other Bayesian clustering methods. This resulted in a higher estimated value of K, but the genetic groups detected were all biologically credible (Figure 2b). Incongruent results between different Bayesian clustering methods have been encountered in previous studies (Frantz et al., 2006; Latch et al., 2006; Rowe and Beebee, 2007). It is not apparent what causes these inconsistencies. They may arise from differences in the underlying models, the statistical estimators or the algorithms used (Guillot et al., 2009). Alternatively, there may not be sufficient genetic information to differentiate groups within breeds (for example, British Saddleback) and, consequently, the methods may be operating at their limits. Thus, it remains uncertain whether and phylogenetic reconstruction have uncovered real genetic structure within the British Saddleback.
Assignment of individuals to origin and genetic diversity
The majority of the individuals were successfully assigned to their pre-designated breed origin using both the Bayesian genotypic clustering methods and phylogenetic reconstruction. This is reflected by the estimation of high membership proportions (Figure 2, q>0.9, clustering methods) and high bootstrap values following resampling of loci (phylogenetic reconstruction). The lack of evidence of admixture indicates that the majority of the British pig breeds are distinct genetic units and that there is little hidden substructure within breeds. In a study on dog breeds, Koskinen (2003) also reported that Bayesian genotypic clustering and phylogenetic reconstruction performed similarly in terms of assigning individuals to their breed origin.
Phylogenetic reconstruction also indicated that more genetic variation lies within than between the British pig breeds. A cladogram was produced, where the longest branches separated individuals within breeds (Figure 4). This is substantiated by multivariate analysis of the microsatellite allele distribution. The PCA projection shows individuals are not tightly clustered, and are instead spread out and populations overlapping (Figures 3a and b). An analysis of molecular variance (Excoffier et al., 1992), a method that partitions variance among groups, also revealed that most of the variation was explained within the pig breeds (∼72%, P<0.0001). Greater genetic variation within, than between breeds, is a common observation in domestic animal breeds (MacHugh and Bradley, 2001; Bruford et al., 2003).
Defining the genetic boundaries of breeds
In PCA and Bayesian genotypic clustering, data can be examined at a number of dimensions, where populations may separate to form their own independent genetic unit with each increase in principal component or value of K. This likely indicates distinctive multilocus genetic combinations for these particular populations (Rosenberg et al., 2001). In both PCA and , the Large White was the first British breed to form a distinct cluster, with the other commercial breeds following with increasing dimensions.
Some credible genetic grouping of breed pairs was observed. The first was a clustering of the British Lop breed with the British Landrace lines, a breed of European origin. Megens et al. (2008) showed, using phylogenetic reconstruction, a genetic affinity of British Lop with other European pig breeds. This suggests that British Lop may either be a breed of European origin or has experienced substantial genetic introgression from British Landrace (Hall and Clutton-Brock, 1988). The second was the pairing of Berkshire and Gloucestershire Old Spots breeds. These observations were consistent across all the statistical approaches (Figures 2a, 3a and b and 4; Table 2). In a third case, some of the methods revealed that the British Saddleback breed shared a genetic affinity with the Berkshire-Gloucestershire Old Spots grouping. clustered the breed with Gloucestershire Old Spots and placed British Saddleback with the Berkshire-Gloucestershire Old Spots cluster in the majority of the replicates for low K values. In addition, the three breeds shared an internal branch on the phylogenetic topology (Figure 4). The observed genetic similarities between these pig breeds are supported by historical information. First, the three breeds are indigenous to Great Britain and share a common geographic origin: the counties of the south of England (Porter, 1993; BPA, 2002). Second, the Berkshire was once a popular and prevalent pig used to improve other breeds (Porter, 1993; BPA, 2002). Historic genetic introgression of the Berkshire could have augmented or maintained the genetic affinities between these indigenous breeds.
Beyond pairs of breeds, the genetic structure among the British pig breeds could not be discerned (Figure 4). There are likely to be a limited number of microsatellite loci that differentiate individuals between the breeds. This is reflected by low bootstrap support for relationships between the breeds, a star-shaped topology and the short internal branches that separate individuals from those of other breeds. The lack of a robust and coherent evolutionary tree has also been seen for a larger group of European pig breeds (SanCristobal et al., 2006; Megens et al., 2008). One common assumption in tree-building methods is bifurcation, which is likely to be violated in livestock as breeds most likely did not follow a strict dichotomous development (Rosenberg et al., 2001). Cross-breeding during the history of these breeds could lead to smaller estimated distances between populations and lower bootstrap support (Eding and Bennewitz, 2007). The inconsistent clustering solutions between replicate runs found using , as have previously been reported in chicken (Rosenberg et al., 2001) and cattle breeds (Li et al., 2007), could reflect the same phenomenon. Historical cross-breeding for improvement may have created a genetic structure such that the individuals of the British pig breeds are genetically similar, leading to multiple possible clustering solutions.
Conclusions
In this paper we compared the performance of three Bayesian genotypic clustering methods, PCA and phylogenetic reconstruction for inferring population structure in a livestock breed. Except for PCA, which was only able to separate breeds into related groups and could not identify the individual breeds, the methods were similarly effective in delineating breeds and assigning individuals to breed of origin. However, there were incongruent results between the different Bayesian genotypic clustering techniques with respect to the determination of K, clustering solutions and the detection of substructure within breeds. Of the Bayesian clustering methods, detected finer genetic differentiation within the breeds with known substructure.
Acknowledgments
This work received support from the European Union (PigBioDiv, http://www.projects.roslin.ac.uk/pigbiodiv/), which is gratefully acknowledged. S Wilkinson acknowledges funding from BBSRC and Rare Breeds Survival Trust. Thanks also to Andy Law (Roslin) for maintenance of the ‘PigBioDiv' database.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Beaumont MA, Rannala B. The Bayesian revolution in genetics. Nat Rev Genet. 2004;5:251–261. doi: 10.1038/nrg1318. [DOI] [PubMed] [Google Scholar]
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. High resolution of human evolutionary trees with polymorphic microsatellites. Nature. 1994;368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- BPA . British Pig Breeds. British Pig Association, Cambridge; 2002. [Google Scholar]
- Bruford MW, Bradley DG, Luikart G. DNA markers reveal the complexity of livestock domestication. Nat Rev Genet. 2003;4:900–910. doi: 10.1038/nrg1203. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multi-locus molecular markers. Mol Ecol. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P, Mantuniemi S. A Bayesian method for identificaiton of stock mixtures from molecular marker data. Fish Bull. 2006;104:9. [Google Scholar]
- Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539–552. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEFRA . UK National Action Plan on Farm Animal Genetic Resources. DEFRA; 2006. [Google Scholar]
- Dinno A.2009pp paran—Horn's Parallel Analysis of Components/Factors . http://cran.r-project.org/web/packages/paran/index.html .
- Eding H, Bennewitz J.2007Measuring genetic diversity in farm animalsIn: Oldenbroek K (ed).Utilisation and Conservation of Farm Animal Genetic Resources Wageningen Academic Publishers [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software : a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Frantz A, Pourtois JT, Heuertz M, Flamand M, Krier A, Bertouille S, et al. Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol. 2006;15:3191–3203. doi: 10.1111/j.1365-294X.2006.03022.x. [DOI] [PubMed] [Google Scholar]
- Guillot G, Estoup A, Mortier F, Cosson JF. A spatial statistical model for landscape genetics. Genetics. 2005;170:1261–1280. doi: 10.1534/genetics.104.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot G, Leblois R, Coulon A, Frantz AC. Statistical methods in spatial genetics. Mol Ecol. 2009;18:4734–4756. doi: 10.1111/j.1365-294X.2009.04410.x. [DOI] [PubMed] [Google Scholar]
- Hall SJG, Clutton-Brock J. Two Hundred Years of British Farm Livestock. British Museum (Natural History): London; 1988. [Google Scholar]
- Hartl D, Clark A. Principles of Population Genetics. Sinauer Associates; 1989. [Google Scholar]
- Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process prior. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen MT. Individual assignment using microsatellite DNA reveals unambiguous breed identification in the domestic dog. Animal Genet. 2003;4:297–301. doi: 10.1046/j.1365-2052.2003.01005.x. [DOI] [PubMed] [Google Scholar]
- Latch E, Dharmarajan G, Glaubitz J, Rhodes O. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet. 2006;7:295–302. [Google Scholar]
- Li M, Tapio I, Vilkki J, Ivanova Z, Kiselyova T, Marzanov N, et al. The genetic structure of cattle populations (Bos taurus) in northern Eurasia and the neighbouring Near Eastern regions: implications for breeding strategies and conservation. Mol Ecol. 2007;16:3839–3853. doi: 10.1111/j.1365-294X.2007.03437.x. [DOI] [PubMed] [Google Scholar]
- MacHugh D, Loftus R, Cunningham P, Bradley D. Genetic structure of seven European cattle breeds assessed using 20 microsatellite markers. Animal Genet. 1998;29:333–340. doi: 10.1046/j.1365-2052.1998.295330.x. [DOI] [PubMed] [Google Scholar]
- MacHugh DE, Bradley DG. Livestock genetic origins: goats buck the trend. Proc Natl Acad Sci USA. 2001;98:5382–5384. doi: 10.1073/pnas.111163198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megens H-J, Crooijmans RPMA, Cristobal MS, Hui X, Li N, Groenen MAM. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: differences in microsatellite variation between two areas of domestication. Genet Sel Evol. 2008;40:103–128. doi: 10.1186/1297-9686-40-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Pflueger SM, Lindblad-Toh K, Wade CM, O'Brien SJ, et al. Patterns of molecular genetic variation among cat breeds. Genomics. 2008;91:1–11. doi: 10.1016/j.ygeno.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Menozzi P, Piazza A, Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201:786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- Minch E, Ruiz-Linares A, Goldstein D, Feldman M, Cavalli-Sforza L.1997pp Microsat v.1.5d : a computer program for calculating various statistics on microsatellite allele data ( http://hpgl.stanford.edu/projects/microsat ).
- O'Connor B. SPSS and SAS programs for determining the number of components using parallel analysis and velicer's MAP test. Behav Res Meth Instr Comput. 2000;32:396–402. doi: 10.3758/bf03200807. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse DE, Crandall KA. Beyond FST: analysis of population genetic data for conservation. Conserv Genet. 2004;5:585–602. [Google Scholar]
- Porter V. Pigs. A handbook to the Breeds of the World. Helm Information, Ltd; 1993. [Google Scholar]
- Pritchard J, Wen W.2004. Department of Human Genetics, Univerity of Chicago, 920 E 58th st, CLCS 507, Chicago IL 60637, USA.
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2009R: A language and environment for statistical computingR Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org .
- Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MAM, et al. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics. 2001;159:699–713. doi: 10.1093/genetics/159.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genepop ′007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rowe G, Beebee JC. Defining population boundaries: use of three Bayesian approaches with microsatellite data from British natterjack toads (Bufo calamita) Mol Ecol. 2007;16:785–796. doi: 10.1111/j.1365-294X.2006.03188.x. [DOI] [PubMed] [Google Scholar]
- SanCristobal M, Chevalet C, Haley CS, Joosten R, Rattink AP, Harlizius B, et al. Genetic diversity within and between European pig breeds using microsatellite markers. Animal Genet. 2006;37:189–198. doi: 10.1111/j.1365-2052.2005.01385.x. [DOI] [PubMed] [Google Scholar]
- Toro MA, Caballero A. Characterization and conservation of genetic diversity in subdivided populations. Phil Trans Roy Soc Lond, B. 2005;360:1367–1378. doi: 10.1098/rstb.2005.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples R, Gaggiotti O. What is a population? an empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.