Abstract
The speciation model of divergence by monobrachially homologous fusions (that is, with one arm in common) benefits from a wide conceptual acceptance, because heterozygotes between populations carrying such fusions suffer from high levels of meiotic dysfunction. The same meiotic configurations can also be generated by WART (whole-arm reciprocal translocation), rearrangements that are known to occur in mammals. Estimating the disadvantage of heterozygotes carrying monobrachially homologous fusions is required to evaluate the relevance of this mode of chromosomal evolution in diversification and speciation. House mice are an excellent study models because chromosomal races exist carrying monobrachially homologous fusions, and WARTs have been documented in this species. The fertility of heterozygote mice carrying the smallest number of monobrachially homologous fusions (that is, a chain of four chromosomes, C4) was investigated in laboratory-bred hybrids between two parapatric chromosomal races from the island of Madeira. Meiotic nondisjunction analyses and histological sections of testes showed that aneuploidy (16.7%) and germ cell death (50.9%) rates reached significantly higher mean values in hybrids than in homozygotes. In females, however, the histological analysis of ovarian follicle parameters revealed no significant differences between hybrid and homozygous individuals. Overall, the reproductive assays indicated that these C4-carrying hybrids were not sterile but showed an approximately 50% decrease in fertility compared to homozygous parental mice. Implications for modes of chromosomal evolution involving monobrachially homologous fusions are discussed.
Keywords: Robertsonian fusions, WART, fertility, Madeira, hybrid, house mouse
Introduction
Chromosomal speciation models have recently become the focus of renewed interest (Rieseberg, 2001; Navarro and Barton, 2003; Ayala and Coluzzi, 2005). The traditional model—called ‘hybrid dysfunction model'—is based on the underdominance of chromosomal hybrids between differentiated taxa (White, 1978; King, 1993; Delneri et al., 2003). This underdominance generates a partial barrier to gene flow between populations and, ultimately, favors the evolution of complete reproductive isolation through selective processes. Heterozygous disadvantage for chromosomal rearrangements results from the incorrect segregation of chromosomes during meiosis, leading to the formation of fertile aneuploid gametes and embryonic mortality (King, 1993; Marchetti et al., 1999). Premeiotic perturbations have been also observed in some cases causing a reduction in germ cell number (Garagna et al., 1990; Hauffe and Searle, 1998; Banaszek et al., 2000; Wallace et al., 2002). Data from different mammalian species (including human) have shown that values of underdominance vary according to the nature of the rearrangement, the chromosomes concerned, the breakpoint site, the gender and also the genetic context and age (King, 1993; Searle, 1993; Djelati et al., 1997; Hauffe and Searle, 1998; Castiglia and Capanna, 2000; Wallace et al., 2002; Pellestor et al., 2005; Anton et al., 2006). Most experimental analyses of the effects of chromosomal heterozygosity on reproductive fitness stem from studies on humans, the house mouse and the common shrew. The two latter taxa show extensive chromosomal diversity through the fixation of centric fusions, which are the most frequent macromutation detected in chromosomal evolution (Searle and Wójcik, 1998; Piálek et al., 2005). This rearrangement, also called Robertsonian (Rb) translocation, involves the fusion by the centromere of two acrocentric chromosomes forming one metacentric chromosome and thereby reducing the diploid number.
House mice are noteworthy for their high rate of karyotypic change with more than 90 Rb-carrying populations originating in <3000 years (Piálek et al., 2005). Whereas the standard karyotype of the house mouse consists of 2n=40 acrocentric chromosomes, such races differ by the number (2n=38–22) and arm combination of Rb metacentrics they exhibit. It is generally held that these races have diverged by the sequential accumulation of Rb fusions (White, 1978; Capanna, 1982). Heterozygotes for fusions produce trivalents at meiosis, formed by the pairing of each metacentric and its two acrocentric homologs. These meiotic configurations usually result in very low nondisjunction (NDJ) rates when few in number, but the degree of gametogenetic impairment increases when many are present (Garagna et al., 1990; Hauffe and Searle, 1998; Castiglia and Capanna, 2000; Wallace et al., 2002). In some instances, however, parapatric Rb races carry monobrachially homologous fusions; that is, Rb metacentrics that have one chromosome arm in common. In hybrids between these races, meiotic pairing will lead to the formation of complex multivalent configurations (chains or rings of chromosomes), the segregation of which can be highly perturbed, considerably reducing their reproductive fitness (Gropp et al., 1982; Garagna et al., 1990; Hauffe and Searle, 1998; Piálek et al., 2001). Such races may have two origins. The first is the traditional one through the independent fixation of Rb fusions with one arm in common in different populations within the ancestral taxon. This represents the central tenet of the monobrachial chromosomal speciation model proposed by Baker and Bickham (1986). This ‘hybrid dysfunction' model benefits from wide conceptual acceptance because the fixation of only a few Rb fusions in each race can lead to an efficient and rapid mechanism of reproductive isolation (King, 1993; Rieseberg, 2001). The second mode of divergence is through arm exchanges occurring between Rb metacentrics or between Rb metacentric and acrocentric chromosomes, resulting in new arm combinations and thus, a new race. These events are named WARTs (whole-arm reciprocal translocations) and have been documented in mice and other species (Catalan et al., 2000; Hirai et al., 2005; Veyrunes et al., 2007; Fedyk and Chętnicki, 2009). In the house mouse, chromosomal phylogenetic reconstructions of related groups of Rb populations have supported the involvement of such events and suggested that they may have contributed to the extensive diversification of Rb metacentrics and races (Piálek et al., 2005; Britton-Davidian et al., 2005a).
Whatever the mode of evolution, however, interracial hybrids carrying monobrachially homologous fusions or individuals in which a WART has occurred may be heterozygous for the same complex chain or ring configurations at meiosis, and are expected to suffer from impaired gametogenesis. The strong selective disadvantage of such complex heterozygotes while supporting the monobrachial model of speciation will conversely tend to discredit the contribution of WARTs to chromosomal diversification, as their fixation rate would then be expected to be very low. To determine the relevance of monobrachially homologous fusions to speciation and diversification processes in the house mouse, it is thus critical to measure the reproductive fitness of Rb heterozygotes carrying complex meiotic configurations. Data from the mouse literature indicate that gametogenetic dysfunction is usually higher for chains than for rings, and that this increases with the number of Rb metacentrics involved, reaching sterility when many chromosomes are present (Gropp et al., 1982; Garagna et al., 1990). Less is known for smaller complex chains, particularly for the smallest one, that is, a chain of four chromosomes (C4). The few studies that exist show that reproduction of C4 heterozygotes varies from normal to complete sterility depending on the sex of the individuals and the arm combinations (Gropp et al., 1982; Mahadevaiah et al., 1990). These data were measured in laboratory-bred heterozygotes carrying wild Rb metacentrics introgressed into house mouse strains. As the interaction between the strain and wild genomes is known to affect fertility scores, the reproductive fitness of such heterozygotes needs to be reassessed in wild genomes (Wallace et al., 2002). However, natural situations in which hybrids between related races carry a single chain of four chromosomes are scarce (Piálek et al., 2005).
The aim of this study was to evaluate the reproductive performance of C4-carrying hybrids in Rb mice from the island of Madeira in which an impressive chromosomal radiation has taken place in <1500 years (Britton-Davidian et al., 2000; Förster et al., 2009). The rationale for focusing on this type of meiotic configuration is threefold. First, within the chromosomal races uncovered in this island, the two westernmost ones have seven fusions in common (Rb(2.4)(3.14)(5.18)(8.11)(9.12)(10.16)(13.17)), but differ by two additional monobrachially homologous ones involving chromosome 7, that is, Rb(6.7) and Rb(7.15), respectively, in each race. Second, these races are parapatric and share a contact zone in which hybrids are present with an expected C4 meiotic chain (Nunes et al., 2005). Third, in a recent study on the chromosomal phylogeny of Madeiran Rb mice, allowance for WART events yielded the best supported and most parsimonious trees (Britton-Davidian et al., 2005a). In particular, this analysis postulated that the race carrying Rb(7.15) would have evolved from the one carrying Rb(6.7) by a WART between Rb(6.7) and chromosome 15, through a C4 heterozygous state. In this study, the extent of chromosomal underdominance (germ cell death (GCD) and NDJ rates) in male and female hybrids between these two races was evaluated to estimate the contribution of this type of event to speciation and diversification processes in Rb-evolving species.
Materials and methods
Specimens
A total of 131 house mice were trapped in the island of Madeira (32°37′-32°52′ N, 16°39′-17°15′ W) in 2001–2002 using Sherman and Longworth livetraps. Thirteen localities were sampled in the areas occupied by the westernmost races Estreito da Calheta (E. Calheta) and Achadas da Cruz (A. Cruz; see Figure 1 in Nunes et al., 2005). Both wild males and females of these two races were considered in this study, as well as hybrid individuals between them. The animals were brought to the facilities of the Centre for Environmental Biology of the Faculty of Science of Lisbon, where they were maintained in captivity under a 12 h light/dark regime at ambient temperature and provided with water and food ad libitum. Due to the scarcity of interracial hybrids in the wild (only two individuals captured, one male and one female), hybrid mice were bred in the laboratory from crosses between wild animals, homozygous for the diagnostic fusions, Rb(6.7) in race E. Calheta and Rb(7.15) in race A. Cruz (hereafter designated 6.72 and 7.152, respectively). As chromosomal polymorphism was present particularly in race A. Cruz (see Nunes et al., 2005), selection of homozygous individuals for the breeding program required assessing the karyotype before the set up of the crosses. Two procedures were used. The first one involved short-term blood cultures that provided results for roughly 50% of the individuals. This procedure was complemented by crosses in which the diploid number (2n) of the tested parent was inferred from those of its progeny (3–6 individuals). Sixty-two crosses were set up between A. Cruz mice and 2n=40 individuals from the Balb laboratory strain, as well as between wild specimens of race E. Calheta (for verification of homozygous state). A total of 438 progeny were produced, 203 of which were karyotyped. Screening and selection of homozygous individuals took 6 months (November 2002–April 2003). Twenty-two homozygous individuals (2n=24) of both races were selected and used to set up 11 interracial reciprocal pairs: 6 (male 6.72 × female 7.152) and 5 (male 7.152 × female 6.72). Interracial crosses were maintained for a period of 7 months (June–December 2003). Two litters of 3–4 hybrid mice were obtained from each pair. Animals were killed by cervical dislocation, after which body weight (BW) and testis weight (TW) was recorded. Gonads (left testis in males and both ovaries in females) were fixed in Bouin for at least 1 month before processing. The right testis was used for meiotic chromosome analyses. Necropsies of hybrids were performed at the age of 7–8 weeks, at which time the mice were considered to be sexually mature.
Fertility estimates
Fertility estimates of wild individuals, as well as of laboratory-bred interracial hybrids, were performed (38 males, 10 females). Comparisons involved mice with different karyotypes (Figure 1): (1) those that were homozygous for 6.72 or 7.152, (2) homozygotes carrying neither of these fusions (hereafter 002), (3) simple heterozygotes for the fusions carrying trivalents (designated 6.7 or 7.15) and finally (4) hybrids (complex heterozygotes; 6/6.7/7.15/15). Four additional specimens were analyzed: a simple heterozygote for Rb(9.12) and the others for two fusions (1.15 and 6.7; 7.15 and 9.12; 7.15 and 5.18). These heterozygotes were not included in the statistical analyses. Two complementary approaches were used. First, estimates of aneuploidy rates (incorrect segregation of the homologous chromosomes during meiosis) and of univalency (XY or autosomal) were assessed in males only. Second, gametogenesis was examined through a histological study of gonads of both sexes.
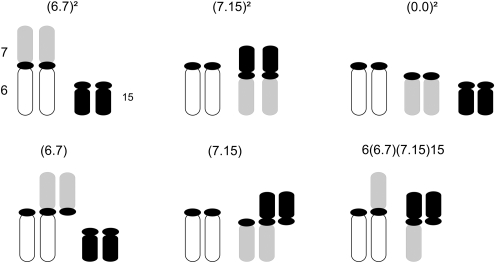
Figure 1.
Schematic representation of partial karyotypes involving chromosomes 6, 7 and 15: homozygotes (6.7)2, (7.15)2 and (0.0)2; simple heterozygotes (6.7) and (7.15); complex heterozygotes carrying the tetravalent 6/6.7/7.15/15.
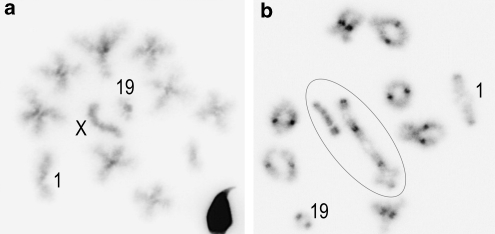
Meiotic cellular suspensions from the right testis were prepared using the air-drying method (Evans et al., 1964). Chromosome preparations were stained with the DAPI fluorescent dye (stock solution: 1 mg ml−1 of 2 × SSC diluted to 1 μg per 200 μl 2 × SSC) that specifically stained the centromeric heterochromatic regions. Twenty males were analyzed; NDJ rates were estimated from the observation of metaphase II plates (11–87 per individual; Figure 2a) and the frequency of univalency of sex or autosomal chromosomes was scored in metaphase I spreads (15–90 per mouse; Figure 2b). NDJ rates were estimated from counts of hyperhaploid cells (that is, with a chromosome arm number n>20), because hypohaploid cells may be due to artifactual chromosome loss. In all cases, aneuploidy was calculated by doubling the frequency of hyperhaploid cells: (2(n+1)). In simple heterozygotes for more than one Rb metacentric, false pseudo-euploid cells may be generated by multiple malsegregation (that is, these aneuploid cells carry the correct n=20). To take this error factor into account, we quadrupled the number of cells with (n+2) in the 7.15/9.12 heterozygote. The segregation of the C4 meiotic configuration is more complex consisting in three main patterns according to the number of chromosomes distributed into each gamete (2:2, 3:1, 4:0). Among the unbalanced cells for each segregation pattern, hyperhaploid cells were easily identified by counting the chromosome arm number (n+1), and their frequency was doubled as for simple heterozygotes. The 2:2 segregation pattern, however, includes balanced cells (n), hyperhaploid cells (n+1) and false pseudo-euploid cells (n) in which an extra chromosome is compensated by the loss of another one. Although these pseudo-euploid cells cannot be distinguished from balanced ones, no correction was applied because they were found to be extremely rare (<1%) in a previous study using chromosome probes (see Marchetti et al., 1999 for details of segregation patterns).
Figure 2.
Metaphase plates of hybrid male mice (inverted DAPI stain). (a) Metaphase II cell showing a euploid cell with n=20 chromosome arms (8 metacentrics, 4 acrocentrics corresponding to chromosomes 1, 19, X, and acrocentric 6 or 15). Chromosomes 1, 19 and X are indicated. (b) Metaphase I cell showing 11 paired elements (7 metacentrics, acrocentric pairs 1 and 19, sex bivalent) and the chain-of-four tetravalent corresponding to the pairing of chromosomes 6, Rb(6.7), Rb(7.15) and 15. Circled chromosomes correspond to the tetravalent and the XY pair. Magnification × 125.
The histological analyses of testes followed the procedure detailed by Britton-Davidian et al. (2005b). The numbers of primary spermatocytes and round spermatids were scored at stages I–VIII of the seminiferous cycle in 15 transverse cross sections of the seminiferous tubules (Leblond and Clermont, 1952; Oakberg, 1956). The Abercrombie correction (Abercrombie, 1946) was applied to the cell counts to calculate the mean spermatid-to-spermatocyte ratio (SSR). This ratio provides an indication of the overall GCD occurring between the primary spermatocyte and round spermatid stages in the testis, the expected ratio being 4:1 (spermatids/spermatocyte) if spermatogenesis proceeds unimpaired. GCD percentages were calculated as follows: 100(1−(SSR/4)). In addition, in each cross section, the number of Sertoli cells was counted and expressed per 100 μm of perimeter of the seminiferous tubule (S/100 μm). Diameters of the seminiferous tubules (D) are also provided.
The histological analyses in females involved counting the number of corpora lutea and of follicles at different stages—primordial I, II and III following the procedure by Peters and McNatty (1980) in 25 consecutive sections of the right ovary. The mean number of follicles at different stages was calculated per section after applying the Abercrombie correction (Abercrombie, 1946) where appropriate.
Mitotic chromosome preparations
The blood cell culture procedure followed the improved method by Davisson and Akeson (1987). Peripheral blood was collected from the retro-orbital sinus using heparinized sterile Pasteur pipettes. Three culture tubes were collected for each individual, adding approximately 150 μl of blood into a culture medium composed of RPMI, purified phytohemagglutinin, glutamine, gentamicin, fetal bovine serum and lipopolysaccharide.
Direct chromosome preparations were obtained from a suspension of yeast-stimulated bone marrow cells from femurs and tibias (for details see Nunes et al., 2005). Chromosome arm identification was achieved by the G-banding method. A minimum of three metaphase plates was analyzed for each individual. Observations were made under a Zeiss Axiophot fluorescent photomicroscope equipped with an image analyzer (Cytovision 3.93.2, Genetix Europe Ltd, Hampshire, UK).
Data treatment
NDJ were compared among chromosomal groups (homozygotes, simple and complex heterozygotes) using χ2-tests. Differences between chromosomal groups, regarding the histological parameters considered (males: S/100 μm, Sertoli, SSR, GCD, BW, TW, RTW and D; females: primordial, I, II, III follicles, corpora lutea and BW), were assessed with analyses of variance (males) and Mann–Whitney (U) nonparametric tests (females). Mean values are provided as mean±standard error (s.e.).
Results
The incidence of Rb heterozygosity on the segregation of chromosomes at meiosis was evaluated by the analysis of metaphase II spreads in males. No aneuploidy was detected in homozygous individuals, whereas NDJ rates in heterozygotes for single trivalents ranged between 0 and 16% with an overall low mean value (3.00%±1.1; Table 1). The analysis of a specimen carrying two trivalents yielded an NDJ score within the range of the single trivalent heterozygotes (8%). In contrast, aneuploidy rates reached significantly higher values in C4 hybrids than in homozygotes or simple heterozygotes with a maximum of 42.9% (mean: 16.7%±6.6; P=0.001; Table 2). Univalency was observed in all chromosomal groups and mostly involved the sex bivalents. However, the frequency of unpaired XY chromosomes was low (below 5%) and did not differ between karyotypes. Autosomal univalency was exceptional (one cell) and present only in C4 hybrids, which also displayed two cells in which associations between the C4 chain and the X or an autosome were observed.
Table 1. Meiotic analysis of male homozygotes and trivalent-carrying heterozygotes for different Rb fusions.
| Rb fusion |
MII |
MI |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | n+1 | n+2 | NDJ% | XY | X/Y | A/A | Total | X/Y% | ||
| Homozygotes | 6.72 | 52 | 0 | 0 | 0 | 67 | 0 | 0 | 67 | 0 |
| 6.72 | 22 | 0 | 0 | 0 | 90 | 3 | 0 | 93 | 3.2 | |
| 6.72 | 61 | 0 | 0 | 0 | 49 | 1 | 0 | 50 | 2.0 | |
| 7.152 | 53 | 0 | 0 | 0 | 52 | 11 | 0 | 63 | 17.5 | |
| 7.152 | 47 | 0 | 0 | 0 | 37 | 0 | 0 | 37 | 0.0 | |
| 0.02 | 52 | 0 | 0 | 0 | 42 | 0 | 0 | 42 | 0.0 | |
| 0.02 | 52 | 0 | 0 | 0 | 42 | 0 | 0 | 42 | 0.0 | |
| Mean | 0 | 3.2±2.4 | ||||||||
| Heterozygotes | 6.7 | 50 | 0 | 0 | 0 | 36 | 0 | 0 | 36 | 0.0 |
| 6.7 | 49 | 0 | 0 | 0 | 37 | 6 | 0 | 42 | 14.3 | |
| 6.7 | 65 | 6 | 0 | 15.6 | 53 | 1 | 0 | 54 | 1.9 | |
| 7.15 | 86 | 1 | 0 | 2.3 | 38 | 1 | 0 | 39 | 2.6 | |
| 7.15 | 56 | 0 | 0 | 0.0 | 46 | 1 | 0 | 47 | 2.1 | |
| 7.15 | 61 | 0 | 0 | 0.0 | 56 | 2 | 0 | 58 | 3.4 | |
| Mean | 3.0±1.1 | 4.0±2.1 | ||||||||
| 9.12 | 49 | 1 | 0 | 3.9 | ||||||
| 7.15/9.12 | 46 | 0 | 1 | 8.0 | ||||||
Haploid chromosome arm complement is indicated by the number of cells with n, n+1, n+2. Aneuploidy rates (NDJ%) are estimated from metaphase II plates (MII), and sex chromosome (X/Y%) as well as autosome pair (A/A) dissociation from metaphase I plates (MI). XY records the number of cells with paired sex chromosomes. Mean values are provided with standard error.
Table 2. Meiotic analysis in C4-carrying heterozygote males.
| Individual | Segregation |
N | MII |
MI |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | n | NDJ% | XY | X/Y | A/A | C/4 | Total | X/Y% | Other% | ||
| H12 | 2:2 | n | 33 | 46 | 0 | 0 | 0 | 46 | 0 | 0 | |
| 2:2 | n+1 | 1 | |||||||||
| 3:1 | n+1 | 1 | |||||||||
| Total | 35 | 10.8 | |||||||||
| H44 | 2:2 | n | 51 | 32 | 1 | 1 | 1 | 33 | 3.0 | 6.1 | |
| 2:2 | n+1 | 3 | |||||||||
| 3:1 | n+1 | 1 | |||||||||
| Total | 55 | 13.5 | |||||||||
| MK607* | 2:2 | n | 54 | 63 | 6 | 0 | 1 | 69 | 8.7 | 1.4 | |
| 2:2 | n+1 | 2 | |||||||||
| 3:1 | n+1 | 1 | |||||||||
| Total | 57 | 8.4 | |||||||||
| H1 | 2:2 | n | 24 | 22 | 0 | 0 | 0 | 22 | 0 | 0 | |
| 2:2 | n+1 | 1 | |||||||||
| 3:1 | n+1 | 0 | |||||||||
| Total | 25 | 7.7 | |||||||||
| H40 | 2:2 | n | 8 | 15 | 0 | 0 | 0 | 15 | 0 | 0 | |
| 2:2 | n+1 | 3 | |||||||||
| 3:1 | n+1 | 0 | |||||||||
| Total | 11 | 42.9 | |||||||||
| Mean | 16.7±6.6 | ||||||||||
Abbreviations: N, number of cells analyzed; n, haploid chromosome arm complement.
Aneuploidy rates (NDJ%) are estimated from metaphase II plates (MII), and sex-chromosome (X/Y%) as well as autosome pair dissociation (Other%) from metaphase I plates (MI). Dissociations are indicated by a slash between the chromosomal elements, whereas XY records cells with paired sex chromosomes. For details on segregation type, see text. The asterisk indicates the wild hybrid. Mean values are provided with standard error. No cells showing a 4:0 segregation pattern were observed.
The histological analyses were performed on a total of 34 individuals, 24 males and 10 females. In males, comparisons between homozygotes, simple and complex heterozygotes revealed significant differences concerning the number of Sertoli cells (F=6.162, P=0.008), S/100 μm (F=10.492, P=0.001), SSR (F=25.127, P=0.000) and GCD (F=25.374, P=0.000; Table 3 and Figure 3), but only marginal ones in tubule diameter (F=3.245, P=0.06). Whereas homozygous and simple heterozygous males of each race showed a similar mean value of GCD (homozygotes, GCD=28.3%±2.6; simple heterozygotes, GCD=24.8%±1.6; Table 3; Figure 3a), the percentage was twice as high in interracial C4 hybrids (GCD=50.9%±3.0; Table 3; Figure 3b). No significant differences in BW or TW were found between chromosomal groups (BW: F=1.763, P=0.19; TW: F=1.580, P=0.23; RTW: F=0.935, P=0.41).
Table 3. Testis histological parameters of male mice carrying different karyotypes.
| Type | 2n | Karyotype | BW | TW | RW | S/100 | Sertoli | D | SSR | GCD% |
|---|---|---|---|---|---|---|---|---|---|---|
| Homozygotes | 24 | 6.72 | 23.68 | 0.152 | 0.0064 | 1.63 | 10.31 | 201.50 | 2.93 | 26.8 |
| 24 | 6.72 | 23.12 | 0.173 | 0.0075 | 1.59 | 10.14 | 203.70 | 3.03 | 24.3 | |
| 24 | 6.72 | 20.90 | 0.150 | 0.0073 | 1.6 | 9.70 | 193.87 | 2.62 | 34.5 | |
| 24 | 7.152 | 23.18 | 0.191 | 0.0082 | 1.66 | 9.53 | 183.47 | 3.01 | 24.9 | |
| 24 | 7.152 | 13.44 | 0.092 | 0.0068 | 1.93 | 10.33 | 172.00 | 2.74 | 31.4 | |
| 24 | 7.152 | 18.26 | 0.143 | 0.0078 | 1.61 | 9.27 | 183.47 | 3.38 | 15.4 | |
| 24 | 7.152 | 20.72 | 0.136 | 0.0066 | 1.54 | 9.38 | 194.00 | 2.76 | 31.1 | |
| 24 | 7.152 | 16.56 | 0.096 | 0.0058 | 1.92 | 10.20 | 166.00 | 2.15 | 46.3 | |
| 26 | 002 | 14.67 | 0.137 | 0.0093 | 1.90 | 10.80 | 181.73 | 2.60 | 34.9 | |
| 26 | 002 | 20.12 | 0.185 | 0.0092 | 1.75 | 9.63 | 175.00 | 3.25 | 18.8 | |
| 26 | 002 | 19.97 | 0.163 | 0.0082 | 1.60 | 9.27 | 184.00 | 3.07 | 23.3 | |
| Mean | 28.3±2.6 | |||||||||
| Heterozygotes | 25 | 6.7 | 20.62 | 0.172 | 0.0083 | 1.46 | 7.93 | 173.33 | 2.81 | 29.6 |
| 25 | 6.7 | 20.54 | 0.153 | 0.0074 | 1.57 | 9.44 | 191.88 | 3.31 | 20.0 | |
| 25 | 6.7 | 23.00 | 0.126 | 0.0055 | 1.52 | 8.93 | 188.40 | 2.86 | 28.5 | |
| 25 | 7.15 | 19.52 | 0.149 | 0.0076 | 1.63 | 10.03 | 195.86 | 3.04 | 24.0 | |
| 25 | 7.15 | 17.25 | 0.080 | 0.0046 | 1.61 | 9.20 | 182.00 | 2.95 | 26.3 | |
| 25 | 7.15 | 14.25 | 0.091 | 0.0064 | 1.71 | 9.53 | 178.40 | 3.18 | 20.6 | |
| Mean | 24.8±1.6 | |||||||||
| Hybrids | 26 | 7.15/9.12 | 20.70 | 0.201 | 0.0097 | 1.43 | 9.07 | 203.20 | 3.08 | 23.0 |
| 26 | 7.15/5.18 | 19.34 | 0.127 | 0.0066 | 1.60 | 9.33 | 186.60 | 2.75 | 31.3 | |
| 27 | 9.12 | 26.82 | 0.154 | 0.0057 | 1.59 | 9.19 | 184.88 | 3.07 | 23.3 | |
| 24 | 6.7/7.15* | 18.99 | 0.114 | 0.0060 | 1.95 | 9.41 | 156.24 | 1.87 | 53.3 | |
| 24 | 6.7/7.15 | 17.01 | 0.111 | 0.0076 | 1.80 | 9.93 | 175.73 | 1.79 | 55.2 | |
| 24 | 6.7/7.15 | 17.52 | 0.154 | 0.0088 | 1.94 | 10.5 | 173.00 | 2.47 | 38.3 | |
| 24 | 6.7/7.15 | 18.67 | 0.09 | 0.0048 | 2.17 | 10.87 | 158.40 | 1.49 | 62.7 | |
| 24 | 6.7/7.15 | 12.33 | 0.092 | 0.0075 | 2.00 | 11.00 | 176.53 | 1.92 | 51.9 | |
| 24 | 6.7/7.15 | 18.44 | 0.140 | 0.0076 | 1.71 | 9.80 | 183.07 | 1.95 | 51.2 | |
| 24 | 6.7/7.15 | 14.49 | 0.148 | 0.0102 | 1.87 | 10.87 | 185.20 | 2.24 | 44.0 | |
| Mean | 50.9±3.0 |
Abbreviations: BW, body weight (g); D, tubule diameter (μm); GCD%, percentage germ cell death; RW, relative testis weight; S/100, number of Sertoli cells per 100 μm; Sertoli, number of Sertoli cells; SSR, spermatid-to-spermatocyte ratio; TW, testis weight (g).
The asterisk indicates the wild hybrid. Mean values are provided with standard error.
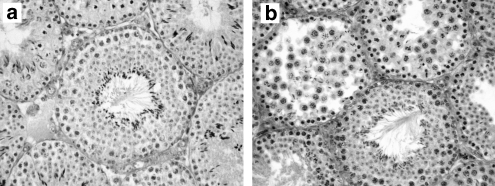
Figure 3.
Testis histological cross sections. (a) Laboratory-bred simple heterozygous male showing a functional seminiferous tubule (SSR=3.08). (b) Wild hybrid male showing the presence of both defective and functional seminiferous tubules (SSR=1.87). Magnification × 25.
With respect to the fertility parameters estimated in females, comparisons between homozygotes and complex heterozygotes did not highlight any significant difference between chromosomal groups in mean follicle number whatever the maturation phase (0.14<P<1; Table 4; Figures 4a and b).
Table 4. Ovary histological parameters in female mice carrying different karyotypes.
| Type | Karyotype | PF | FI | FII | FIII | CL | BW |
|---|---|---|---|---|---|---|---|
| Homozygotes | 6.72 | 10.12±1.98 | 3.25±0.66 | 4.8±1.06 | 3.95±1.32 | 0 | 15.64 |
| 6.72 | 2.67±0.94 | 2.06±0.67 | 3.17±0.87 | 3.33±0.70 | 3.89±0.83 | 23.88 | |
| 7.152 | 4.60±0.92 | 3.26±1.03 | 5.75±2.06 | 6.31±1.03 | 2.13±1.45 | 15.85 | |
| 0.02 | 4.86±1.36 | 5.15±1.67 | 3.26±1.54 | 5.04±1.24 | 4.08±1.03 | 16.78 | |
| 0.02 | 10.43±3.08 | 4.65±094 | 2.65±1.13 | 2.46±1.48 | 2.08±0.84 | 17.56 | |
| Heterozygote | 1.15/6.7 | 5.18±1.90 | 2.63±0.58 | 4.35±2.08 | 5.40±1.79 | 0.4±0.50 | 17.56 |
| Hybrids | 6.7-7.15* | 5.40±1.58 | 3.57±1.00 | 3.09±1.38 | 1.94±1.13 | 2.07±0.61 | 17.83 |
| 6.7-7.15 | 3.65±1.52 | 3.15±0.88 | 5.2±1.47 | 5.37±0.81 | 0.13±0.35 | 14.25 | |
| 6.7-7.15 | 5.65±2.16 | 2.87±1.60 | 4.67±2.13 | 5.33±0.49 | 0 | 12.54 | |
| 6.7-7.15 | 7.78±2.54 | 5.53±1.97 | 3.24±1.62 | 5.19±1.15 | 1.52±1.67 | 11.72 |
Abbreviations: BW, body weight (g); FI, FII, FII, primary, secondary, and tertiary follicles, respectively; PF, primordial follicles.
The asterisk indicates the wild hybrid. Mean values are provided with standard error.
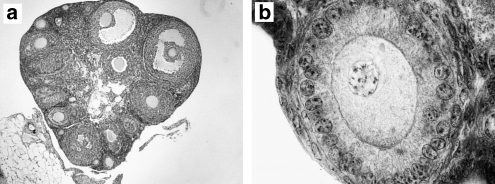
Figure 4.
Ovarian cross sections of a laboratory-bred hybrid female. (a) View of ovary showing follicles at different stages of development ( × 6.25 magnification). (b) Oocyte with nucleus surrounded by the zona pellucida ( × 125 magnification).
Discussion
Fertility estimates
The assessment of fertility in Rb mice from the island of Madeira provided a comparative estimate of chromosomal underdominance related to different meiotic configurations. Male homozygotes and single trivalent-carrying heterozygotes showed very low levels of aneuploidy and GCD in accordance with data reported for wild house mice elsewhere (Hauffe and Searle, 1998; Castiglia and Capanna, 2000). In contrast, the presence of a chain-of-four meiotic pairing configuration (that is, complex heterozygosity) reduced the reproductive performance of male hybrids, due to both a higher aneuploidy rate and spermatogenetic dysfunction. Although such a result was expected, the extent of the impairment was surprisingly moderate (mean NDJ=16.7%±6.6; mean GCD=50.9%±3.0) compared to that previously recorded in studies of Rb progeny with laboratory/wild mixed genomes, most of which highlighted severe perturbations generally leading to complete gametogenetic arrest and sterility (Gropp et al., 1982; Mahadevaiah et al., 1990). Our results are more in agreement with data obtained for wild C5 male heterozygotes from northern Italy. Although massive GCD was present in several C5 individuals (GCD=70–98% Piálek et al., 2001), another specimen showed a value similar to those presented here (GCD=55.5% Hauffe and Searle, 1998). Thus, that a shorter meiotic chain (for example, C4) leads to a more moderate degree of spermatogenetic impairment in the original genomic background is not surprising. The fertility of the C4 hybrids can be estimated at 50% of that of the Rb homozygotes, considering a relative 67% gametogenetic production, 20% of which (at most) is expected to comprise aneuploid sperm. The contrast between the data for the Madeiran hybrids, none of which were sterile, and previous analyses of non-wild C4 heterozygotes supports the incidence of genetic incompatibilities between genomic backgrounds in decreasing fertility scores of chromosomal heterozygotes (Wallace et al., 2002). Differences found in body mass between the parental and hybrid mice had no impact on the histological parameters analyzed as TW did not vary significantly between chromosomal groups or types. The lack of a significant variation in body mass between the wild mice and laboratory-reared interracial hybrids may be related to the young age of hybrids (7–8 weeks old) and/or the maintenance of the wild animals in captive conditions, as the reduced mobility and ad libitum feeding generally leads to an increase in weight.
The histological analysis of the ovaries of female hybrids did not reveal an effect of complex meiotic heterozygosity on reproductive function. These results are similarly in accordance with data from the literature that indicate that chromosomally related infertility in females is rarely due to GCD (Gropp et al., 1982; Hauffe and Searle, 1998). In contrast, aneuploidy rates tend to be higher in females (not measured in this study) than in males (Gropp et al., 1982; Hauffe and Searle, 1998). The reasons for the differences between sexes are not clear, but may be related to more efficient meiotic checkpoints in males than in females, that trigger an apoptotic pathway more readily during spermatogenesis than oogenesis (Kouznetsova et al., 2007). In chain-carrying male heterozygotes, reproductive impairment is thought to result from altered gene expression due to inappropriate interaction with the XY pair, or possibly to epigenetic causes (Manterola et al., 2009). Only one C4/XY association was observed in this study, although more cells may initially have been present and eliminated during earlier stages of meiosis (that is, prophase). Notwithstanding, data for wild C4-carrying females are nonexistent; it seems reasonable to assume that aneuploidy rates would be less pronounced in C4 females than in C5 females for which estimates are available (NDJ=38% in Hauffe and Searle, 1998). Given these observations, and in the absence of aneuploidy rates for C4 females, a decrease in fertility of <50% appears as a conservative estimate for hybrid females.
Consequences for Rb models of chromosomal evolution
Phylogenetic reconstructions of geographically related Rb systems have suggested that chromosomal evolution in house mice occurs by three processes: de novo Rb fusion, WARTs and introgression by hybridization (coined zonal raciation; see Searle, 1993; Piálek et al., 2001). In particular, the most parsimonious chromosomal phylogeny of the Maderian races included 11–15 Rb fusions as well as 5–9 WARTs, all of which would have involved an intermediate C4 heterozygosity step (Britton-Davidian et al., 2005a). The results of this study confirm the marginal incidence of single Rb heterozygosity on the reproductive performance of house mice, and thus the low negative impact on fixation probabilities. In this analysis, the fertility of wild C4 hybrids, which is the smallest complex chain configuration, is estimated on a wild genomic background for the first time. These hybrids exhibit moderate subfertility (a 50% decrease at the most), suggesting that these mice are able to reproduce. Thus, the fixation of a new metacentric following a C4-type WART (that is, an exchange between an Rb metacentric and an acrocentric) can occur with a reasonable probability under specific conditions such as random drift in small demes. In essence, our results provide support for the role of WARTs as a mechanism of chromosomal diversification in house mice (Britton-Davidian et al., 2005a; Piálek et al., 2005; Mitsainas and Giagia-Athanasopoulou, 2009). These conclusions can be extended to the other well-studied European Rb model organism, the common shrew (Sorex araneus), in which small meiotic chains or rings are known to have a remarkably low impact on the fertility of heterozygotes (Searle, 1993; Banaszek et al., 2000). In addition, WARTs have also been documented in this species (Fedyk and Chętnicki, 2009), and several authors have argued and recently tested their implication in shrew race formation (Fredga, 1996; Andersson et al., 2005). The relevance of WART events in chromosomal evolution has also been investigated in other mammalian taxa characterized by multiple Rb-type rearrangements. Chromosomal and molecular phylogenetic reconstructions suggested the involvement of WARTs in the karyotypic diversification of bats (Mao et al., 2008); this was not the case for Tenrecs in which a chromosomal parsimony analysis did not favor WARTs over recurrent Rb fusions and fissions (Gilbert et al., 2007).
If the reproductive cost of complex chromosomal heterozygosity is moderate, is it nevertheless sufficient to act as a barrier to gene flow between populations? The subfertility of C4 hybrids observed in this analysis suggests that their reproductive performance is approximately half that of homozygotes. Such a level of underdominance would undoubtedly contribute to limit gene flow between populations, and may provide the theoretical requirements for reinforcement and finally speciation to occur (Baker and Bickham, 1986). However, the analysis of several hybrid zones between monobrachially differentiated populations of the house mouse and the common shrew shows that the situation is more complex than previously thought. In most cases, these contact zones are characterized by the presence of an acrocentric peak in which most of the individuals are heterozygotes for trivalents and not chain-carrying hybrids (Searle and Wójcik, 1998; Britton-Davidian et al., 2002; Nunes et al., 2005). This situation is thought to result from the differential reproductive fitness of simple vs complex heterozygotes. In effect, when Rb polymorphism is present in one of the races (house mouse, common shrew) and/or fissions are possible (common shrew), the two types of heterozygotes (trivalent, chain or ring-carrying) co-occur in the hybrid zone. The lower fitness of complex hybrids compared to trivalent-bearing heterozygotes will favor the latter, thereby increasing the frequency of acrocentrics in the center of the contact zone. The pattern observed here, that is, higher level of underdominance in hybrids compared to Rb heterozygotes, is in agreement with the occurrence of such a process, and is in fact the case in the contact zone between the E. Calheta (6.7) and A. Cruz (7.15) races in Madeira, in which only 2 out of 131 individuals captured were hybrids and 39% were trivalent-carrying heterozygotes (Nunes et al., 2005). The evolutionary outcome in terms of the build up of reproductive isolation in such a situation is not intuitive, as this acrocentric peak may act as a buffer zone limiting the contact between races and the production of unfit hybrids (Searle, 1993; Searle and Wójcik, 1998; Britton-Davidian et al., 2002). Nonetheless, reproductively isolated parapatric Rb races carrying monobrachially homologous fusions have been described in the house mouse (Franchini et al., 2008). The extent of divergence between these races is such that potential hybrids would carry long meiotic chains; this suggests that at least in house mice, the speciation by monobrachial homology model is relevant, but would require the accumulation of a sufficient number of incompatible Rb fusions in the two races. In contrast, no case of chromosomally mediated complete genetic isolation has been documented so far between an Rb race and a standard population (Britton-Davidian et al., 1989; Brahim et al., 2005). Thus, these studies support the monobrachial speciation model as a process that will accelerate the acquisition of reproductive isolation through Rb change, whether by WARTs or independent Rb fusion fixation.
Acknowledgments
We thank the collaboration of Guila Ganem, Ruben Capela and Marco Perriat-Sanguinet for field support; Luísa Ganço for field support and assessment of mouse karyotypes; Bruno Oliveira for help in the maintenance of mice in captivity and Terry J Robinson as well as Fred Veyrunes for critical comments on the article. This study was supported by FCT–Fundação para a Ciência e a Tecnologia (Project POCTI/BSE/47019/02—Evolução de barreiras cromossómicas e especiação no ratinho-caseiro (Mus musculus domesticus) da ilha da Madeira, involving European FEDER funds), a PhD Grant (FCT: SFRH/BD/3114/2000) and by the French Government (Ministère de l' Education et Ministère de la Recherche: Convention de Cotutelle de Thèse; Ministère des Affaires Etrangères: coopération France-Portugal). ISEM contribution No 2010-036.
The authors declare no conflict of interest.
References
- Abercrombie M. Estimation of nuclear population from microtome sections. Anat Record. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- Andersson A-C, Alström-Rapaport C, Fredga K. Lack of mitochondrial DNA divergence between chromosome races of the common shrew, Sorex araneus, in Sweden. Implications for interpreting chromosomal evolution and colonization history. Mol Ecol. 2005;14:2703–2716. doi: 10.1111/j.1365-294X.2005.02584.x. [DOI] [PubMed] [Google Scholar]
- Anton E, Vidal F, Egozcue J, Blanco J. Genetic reproductive risk in inversion carriers. Fertil Steril. 2006;85:661–666. doi: 10.1016/j.fertnstert.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Ayala FJ, Coluzzi M. Chromosome speciation: humans, Drosophila and mosquitoes. Proc Natl Acad Sci USA. 2005;102:6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RJ, Bickham JW. Speciation by monobrachial centric fusions. Proc Natl Acad Sci USA. 1986;83:8245–8248. doi: 10.1073/pnas.83.21.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszek A, Fedyk S, Szalaj KA, Chętnicki W. A comparison of spermatogenesis in homozygotes, simple Robertsonian heterozygotes and complex heterozygotes of the common shrew (Sorex araneus L.) Heredity. 2000;84:570–577. doi: 10.1046/j.1365-2540.2000.00701.x. [DOI] [PubMed] [Google Scholar]
- Brahim IO, Chatti N, Britton-Davidian J, Said K. Origin and evolution of the Robertsonian populations of the house mouse (Rodentia, Muridae) in Tunisia based on allozyme studies. Biol J Linn Soc. 2005;84:515–521. [Google Scholar]
- Britton-Davidian J, Catalan J, Belkhir K. Chromosomal and allozyme analysis of a hybrid zone between parapatric Robertsonian races of the house mouse: a case of monobrachial homology. Cytogenet Genome Res. 2002;96:75–84. doi: 10.1159/000063040. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Catalan J, Ramalhinho MdG, Ganem G, Auffray J-C, Capela R, et al. Rapid chromosomal evolution in island mice. Nature. 2000;403:158. doi: 10.1038/35003116. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Catalan J, Ramalhinho MG, Auffray J-C, Nunes AC, Gazave E, et al. Chromosomal phylogeny of Robertsonian races of the house mouse on the island of Madeira: testing between alternative mutational processes. Genet Res. 2005a;86:171–183. doi: 10.1017/S0016672305007809. [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J, Fel-Clair F, Lopez J, Alibert P, Boursot P. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol J Linn Soc. 2005b;84:379–393. [Google Scholar]
- Britton-Davidian J, Nadeau JH, Croset H, Thaler L. Genic differentiation and origin of Robertsonian populations of the house mouse (Mus musculus domesticus Rutty) Genet Res. 1989;53:29–44. doi: 10.1017/s0016672300027841. [DOI] [PubMed] [Google Scholar]
- Capanna E.1982Robertsonian numerical variation in animal speciation: Mus musculus, an emblematic modelIn: Barigozzi C (ed).Mechanisms of Speciation Alan R Liss: New York; 155–177. [PubMed] [Google Scholar]
- Castiglia R, Capanna E. Contact zone between chromosomal races of Mus musculus domesticus. 2. Fertility and segregation in laboratory-reared and wild mice heterozygous for multiple Robertsonian rearrangements. Heredity. 2000;85:147–156. doi: 10.1046/j.1365-2540.2000.00743.x. [DOI] [PubMed] [Google Scholar]
- Catalan J, Auffray J-C, Pellestor F, Britton-Davidian J. Spontaneous occurrence of a Robertsonian fusion involving chromosome 19 by single whole-arm reciprocal translocation (WART) in wild-derived house mice. Chrom Res. 2000;8:593–601. doi: 10.1023/a:1009281823488. [DOI] [PubMed] [Google Scholar]
- Davisson MT, Akeson EC. An improved method for preparing G-banded chromosomes from mouse peripheral blood. Cytogenet Cell Genet. 1987;45:70–74. doi: 10.1159/000132432. [DOI] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, Oliver SG. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Djelati R, Brun B, Rumpler Y. Meiotic studies of hybrids in the genus Eulemur and taxonomic considerations. Am J Primat. 1997;42:235–245. doi: 10.1002/(SICI)1098-2345(1997)42:3<235::AID-AJP6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Evans EP, Breckon G, Ford CE. An air-drying method for meiotic preparations from mammalian testes. Cytogenet. 1964;3:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
- Fedyk S, Chętnicki W. Whole-arm reciprocal translocation in a hybrid population of Sorex araneus. Chrom Res. 2009;17:451–454. doi: 10.1007/s10577-009-9036-z. [DOI] [PubMed] [Google Scholar]
- Förster DW, Gündüz I, Nunes AC, Gabriel S, Ramalhinho MG, Mathias ML, et al. Molecular insights into the colonization and chromosomal diversification of Madeiran house mice. Mol Ecol. 2009;18:4477–4494. doi: 10.1111/j.1365-294X.2009.04344.x. [DOI] [PubMed] [Google Scholar]
- Franchini P, Castiglia R, Capanna E. Reproductive isolation between chromosomal races of the house mouse Mus musculus domesticus in a parapatric contact area revealed by an analysis of multiple unlinked loci. J Evol Biol. 2008;21:502–513. doi: 10.1111/j.1420-9101.2007.01492.x. [DOI] [PubMed] [Google Scholar]
- Fredga K. The chromosome races of Sorex araneus in Scandinavia. Hereditas. 1996;125:123–135. [Google Scholar]
- Garagna S, Redi CA, Zuccotti M, Britton-Davidian J, Winking H. Kinetics of oogenesis in mice heterozygous for Robertsonian translocations. Differentiation. 1990;42:167–171. doi: 10.1111/j.1432-0436.1990.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Goodman SM, Soarimalala V, Olson LE, O'Brien PCM, Elder FFB, et al. Chromosomal evolution in tenrecs (Microgale and Oryzorictes, Tenrecidae) from the Central Highlands of Madagascar. Chrom Res. 2007;15:1075–1091. doi: 10.1007/s10577-007-1182-6. [DOI] [PubMed] [Google Scholar]
- Gropp A, Winking H, Redi C.1982Consequences of Robertsonian heterozygosity: segregational impairment of fertility versus male-limited sterilityIn: Crosignani PG (ed).Serono Clinical Colloquia on ReproductionVol. 3.Grune and Stratton: London; 115–134. [Google Scholar]
- Hauffe HC, Searle JB. Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Genetics. 1998;150:1143–1154. doi: 10.1093/genetics/150.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Wijayanto H, Tanaka H, Mootnick AR, Hayano A, Perwitasari-Farajallah D, et al. A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features. Chrom Res. 2005;13:123–133. doi: 10.1007/s10577-005-7475-8. [DOI] [PubMed] [Google Scholar]
- King M. Species Evolution. The Role of Chromosome Change. Cambridge University Press: Cambridge; 1993. [Google Scholar]
- Kouznetsova A, Lister L, Nordenskjöld M, Herbert M, Höög C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Clermont Y. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the ‘periodic acid-fuchsin sulfurous acid' technique. Am J Anat. 1952;90:167–215. doi: 10.1002/aja.1000900202. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Setterfield LA, Mittwoch U. Pachytene pairing and sperm counts in mice with single Robertsonian translocations and monobrachial compounds. Cytogenet Cell Genet. 1990;53:26–31. doi: 10.1159/000132889. [DOI] [PubMed] [Google Scholar]
- Manterola M, Page J, Vasco C, Berrios S, Parra MT, Viera A, et al. A high incidence of meiotic silencing of unsynapsed chromatin is not associated with substantial pachytene loss in heterozygous male mice carrying multiple Robertsonian translocations. PloS Genet. 2009;5:e1000625. doi: 10.1371/journal.pgen.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Nie W, Wang J, Su W, Feng Q, Wang Y, et al. Comparative cytogenetics of bats (Chiroptera): the prevalence of Robertsonian translocations limits the power of chromosomal characters in resolving interfamily phylogenetic relationships. Chrom Res. 2008;16:155–170. doi: 10.1007/s10577-007-1206-2. [DOI] [PubMed] [Google Scholar]
- Marchetti F, Lowe X, Bishop J, Wyrobek AJ. Absence of selection against aneuploid mouse sperm at fertilization. Biol Reprod. 1999;61:948–954. doi: 10.1095/biolreprod61.4.948. [DOI] [PubMed] [Google Scholar]
- Mitsainas GP, Giagia-Athanasopoulou EB. Possible involvement of whole-arm reciprocal translocations (WARTs) in the evolution of a Mus musculus domesticus Robertsonian system from Greece. Rend Fis Acc Lincei. 2009;20:153–162. [Google Scholar]
- Navarro A, Barton NH. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Nunes AC, Britton-Davidian J, Catalan J, Ramalhinho MG, Capela R, Mathias ML, et al. Influence of physical environmental characteristics and anthropogenic factors on the position and structure of a contact zone between chromosomal races of the house mouse on the island of Madeira (North Atlantic, Portugal) J Biogeo. 2005;32:2123–2134. [Google Scholar]
- Oakberg EF. A description of spermatogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat. 1956;99:391–413. doi: 10.1002/aja.1000990303. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Anahory T, Hamamah S. Effect of maternal age on the frequency of cytogenetic abnormalities in human oocytes. Cytogenet Genome Res. 2005;111:206–212. doi: 10.1159/000086891. [DOI] [PubMed] [Google Scholar]
- Peters H, McNatty K. The Ovary. Granada Publishing: London; 1980. [Google Scholar]
- Piálek J, Hauffe HC, Rodriguez-Clark KM, Searle JB. Raciation and speciation in house mice from the Alps: the role of chromosomes. Mol Ecol. 2001;10:613–625. doi: 10.1046/j.1365-294x.2001.01209.x. [DOI] [PubMed] [Google Scholar]
- Piálek J, Hauffe HC, Searle JB. Chromosomal variation in the house mouse. Biol J Linn Soc. 2005;84:535–563. [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Searle JB.1993Chromosomal hybrid zones in eutherian mammalsIn: Harrison RG (ed).Hybrid Zones and the Evolutionary Process Oxford University Press: Oxford; 309–353. [Google Scholar]
- Searle JB, Wojcik JM.1998Chromosomal evolution: the case of Sorex araneusIn: Wojcik JM, Wolsan M (eds).Evolution of Shrews Mammal Research Institute, Polish Academy of Sciences: Bialowieza, Poland; 219–268. [Google Scholar]
- Veyrunes F, Watson J, Robinson TJ, Britton-Davidian J. Accumulation of rare sex chromosome rearrangements in the African pygmy mouse, Mus (Nannomys) minutoides: a whole-arm reciprocal translocation (WART) involving a X-autosome fusion. Chrom Res. 2007;15:223–230. doi: 10.1007/s10577-006-1116-8. [DOI] [PubMed] [Google Scholar]
- Wallace BMN, Searle JB, Everett CA. The effect of multiple simple Robertsonian heterozygosity on chromosome pairing and fertility of wild-stock house mice (Mus musculus domesticus) Cytogenet Genome Res. 2002;96:276–286. doi: 10.1159/000063054. [DOI] [PubMed] [Google Scholar]
- White MJD. Chain processes in chromosomal speciation. Syst Zool. 1978;27:285–298. [Google Scholar]