Abstract
Geographical subspecies of wild silkmoths, Samia cynthia ssp. (Lepidoptera: Saturniidae), differ considerably in sex chromosome constitution owing to sex chromosome fusions with autosomes, which leads to variation in chromosome numbers. We cloned S. cynthia orthologues of 16 Bombyx mori genes and mapped them to chromosome spreads of S. cynthia subspecies by fluorescence in situ hybridization (FISH) to determine the origin of S. cynthia neo-sex chromosomes. FISH mapping revealed that the Z chromosome and chromosome 12 of B. mori correspond to the Z chromosome and an autosome (A1) of S. c. ricini (Vietnam population, 2n=27, Z0 in female moths), respectively. B. mori chromosome 11 corresponds partly to another autosome (A2) and partly to a chromosome carrying nucleolar organizer region (NOR) of this subspecies. The NOR chromosome of S. c. ricini is also partly homologous to B. mori chromosome 24. Furthermore, our results revealed that two A1 homologues each fused with the W and Z chromosomes in a common ancestor of both Japanese subspecies S. c. walkeri (Sapporo population, 2n=26, neo-Wneo-Z) and S. cynthia subsp. indet. (Nagano population, 2n=25, neo-WZ1Z2). One homologue, corresponding to the A2 autosome in S. c. ricini and S. c. walkeri, fused with the W chromosome in S. cynthia subsp. indet. Consequently, the other homologue became a Z2 chromosome. These results clearly showed a step-by-step evolution of the neo-sex chromosomes by repeated autosome–sex chromosome fusions. We suggest that the rearrangements of sex chromosomes may facilitate divergence of S. cynthia subspecies towards speciation.
Keywords: Lepidoptera, sex chromosomes, fluorescence in situ hybridization, gene mapping, evolution, speciation
Introduction
Moths and butterflies (Lepidoptera) have sex chromosome systems with female heterogamety, which is also characteristic of their closest relatives, caddis flies (Trichoptera). The majority of lepidopteran species show a WZ/ZZ (female/male) pair of sex chromosomes (reviewed by Traut et al. (2007)). Besides the common WZ/ZZ system, variants without the W chromosome (Z/ZZ) and with multiple sex chromosomes, such as W1W2Z/ZZ and WZ1Z2/Z1Z1Z2Z2, have been found (for example, Nilsson et al., 1988; Traut and Marec, 1997; Yoshido et al., 2005b). The multiple sex chromosome systems may have originated from either fission or sex chromosome–autosome fusions (Marec et al., 2010). The latter scenario, giving rise to neo-sex chromosomes, seems to be common not only in vertebrates (Toder et al., 1997; Schmid et al., 2003), but also in some insects, such as flies of the genus Drosophila (Flores et al., 2008) and true bugs of the genus Dysdercus (Bressa et al., 2009). This scenario is also more likely the source of multiple sex chromosomes in Lepidoptera (Yoshido et al., 2005b; reviewed by Marec et al. (2010)).
Wild silkmoths of the genus Samia (Lepidoptera: Saturniidae) represent a complex of closely related species native to Asia (Peigler and Naumann, 2003). In the literature, they are mostly treated as geographical subspecies of Samia cynthia (Drury), the type species of the genus, obviously because of largely allopatric occurrence, morphological similarity and unclear taxonomic relationships. Recently, Peigler and Naumann (2003) reconstructed the genus and identified 19 different Samia species. Nevertheless, here we prefer the former classification of subspecies, which is consistent with that used in our previous cytogenetic study (see Yoshido et al., 2005b). In a previous study, we showed that three different geographic populations of S. cynthia exhibit a unique polymorphism in chromosome number, resulting from variations in the sex chromosome systems. Three different sex chromosome constitutions have been identified: Z/ZZ in a Vietnam population (the Eri silkworm, S. c. ricini) with 2n=27/28, neo-Wneo-Z/neo-Zneo-Z in a Sapporo population (the ailanthus silkworm, S. c. walkeri) with 2n=26/26 and neo-WZ1Z2/Z1Z1Z2Z2 in a Nagano population (the Shinju silkworm, S. cynthia subsp. indet.) with 2n=25/26. On the basis of these findings, it has been proposed that a common ancestor of S. cynthia subspecies had 2n=28 chromosomes and a WZ/ZZ sex chromosome constitution (Yoshido et al., 2005b). The ancestral karyotype could be still preserved in some extant subspecies of S. cynthia, as Kawaguchi (1937) described populations (Kyoto and Fukuoka, Japan) of the Shinju silkworm, S. c. pryeri, with 14 meiotic bivalents in male silkworm, and Song et al. (1996) found 28 chromosomes in mitotic metaphase complements from a specimen collected in China. According to the proposed scheme of the sex chromosome evolution in S. cynthia (Yoshido et al., 2005b), a neo-Wneo-Z sex chromosome pair in S. c. walkeri resulted from a fusion of an autosomal pair with the original W and Z chromosomes. The sex chromosome constitution of S. cynthia subsp. indet. may have derived from the subsequent fusion event of the neo-W chromosome with another autosome; then, the homologue of this autosome became a Z2 chromosome in this subspecies. However, this hypothesis has not yet been proven, mainly because of the lack of suitable cytogenetic and/or molecular markers to elucidate the origin of individual parts of the neo-sex chromosomes.
Gene-based chromosome maps have been constructed only in four lepidopteran species: the silkworm (Bombyx mori), the tobacco hornworm (Manduca sexta) and two butterflies, Heliconius melpomene and Bicyclus anynana. Gene mapping in these species was carried out either by genetic linkage analysis (B. mori, H. melpomene and B. anynana) or by physical localization of genes using bacterial artificial chromosomes-fluorescence in situ hybridization (FISH with BAC of B. mori and M. sexta as probes) (Jiggins et al., 2005; Yoshido et al., 2005a; Yasukochi et al., 2006, 2009; Pringle et al., 2007; Beldade et al., 2009). Linkage analysis is a laborious and time-consuming procedure, especially in genetically uncharacterized species, and BAC-FISH mapping requires a BAC library, which is not available in S. cynthia. Here we present an alternative strategy of gene mapping that is based on chromosomal localization of S. cynthia orthologues of known B. mori genes by conventional FISH with PCR-generated probes. We cloned S. cynthia orthologous DNA fragments of both the sex-linked and autosomal B. mori genes. By FISH mapping, we then successfully identified autosomes involved in the evolution of neo-sex chromosomes in geographical populations of this species. This is the first application of FISH for the identification of neo-sex chromosomes by direct gene mapping in Lepidoptera, and most probably in all invertebrates.
Materials and methods
Insects
We used larvae of three S. cynthia (Drury) subspecies originating from distant geographical regions: S. c. walkeri (Felder and Felder) from Sapporo (Hokkaido, Japan), S. cynthia subsp. indet. from Nagano (Honshu, Japan) and S. c. ricini (Donovan) from a Vietnam population (for details, see Yoshido et al., 2005b). The larvae were reared on Ailanthus altissima trees at the Field Science Center for Northern Biosphere, Hokkaido University, Sapporo, Japan.
Cloning and sequencing
Genomic DNA was extracted separately from female and male larvae of S. c. walkeri by a standard phenol–chloroform procedure and used as a template for PCR amplification of S. cynthia orthologues of B. mori genes with primers listed in Table 1. The sequence-tagged site primers were designed for genes encoding 3-hydroxy-3-methylglutaryl-CoA reductase, DOPA decarboxylase, elongation factor 1α subunit (EF-1α), attacin, chitinase, lysozyme, hemolin and storage protein 1 according to S. c. ricini sequences available in a public database. To design further sequence-tagged site primers, sequence information of S. c. ricini gene orthologues for Y-box protein (BYB), ribosomal protein L4 (RpL4), ribosomal protein L18 (RpL18), cytoplasmic actin (A4) and translation elongation factor 2 (eEF-2) was obtained from SilkBase (http://silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi). For the orthologues of kettin, topoisomerase II (Topo II) and xanthine dehydrogenase I (XDH I), degenerate oligonucleotide primers were designed from the nucleotide sequences in the conserved regions of B. mori and Drosophila melanogaster. PCR amplifications were carried out with an initial denaturation step at 94 °C for 2 min, followed by 40 cycles of 1 min denaturation at 94 °C, 1 min annealing at 52–60 °C and 2 min extension at 72 °C, and a final extension step at 72 °C for 5 min. Amplified gene fragments were cloned into pGEM T-easy vector (Promega KK, Tokyo, Japan), and their nucleotide sequences were determined with an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Tokyo, Japan), following the sequencing protocol supplied by the manufacturer.
Table 1. Primers used in this study and the size of amplified fragments of Samia cyntia walkeri orthologues of Bombyx mori genes, and corresponding orthologous genes and/or ESTs of S. cynthia.
|
PCR amplification of S. c. walkeri orthologues |
S. cynthia orthologues and ESTs |
Bombyx mori genes |
||||||
|---|---|---|---|---|---|---|---|---|
| Forward primer sequence | Reverse primer sequence | Product size (bp) | Accession no. | EST (SilkBase code)a | Accession no. | Putative function | Chr. no. | Location in KAIKObaseb |
| GGNCCNGGNGAYGARGGNGARTAYAC | GGNGGRTANACNCGRTCYTCYTCNCC | 1820 | AB543309c | Not found | AB079865 | Kettin | 1 (Z) | 6 505 696– 6 533 895 |
| TAAAACAGTTAAACAAAAGCAGGTCAT | CTCATCTCCYTGTGCACCCCCTTGGTT | 1663 | AB543308 | S06A01NCLL0010_M07 | AB098537 | BYB | 1 (Z) | 10 855 943– 10 857 680 |
| GTTTGGGGAGCACATGGAGAAT | CATTTCCGTCTGCGTTCGCTTGG | ca. 2500 | DQ465407 | S06A01NCLL0005_L15 | AB274990 | 3-Hydroxy-3- methylglutaryl- CoA reductase | 2 | 916 956– 919 459 |
| ACCGAATTAGAAGTGGTGATG | GCTTTCTTCACTCATACGTGA | ca. 2300 | AF015065 | Not found | AF372836 | DDC | 4 | 12 504 927– 12 521 471 |
| GGTATCGACAAACGTACCATCG | ATGGATTTAGGGTTGTCCTCTG | 1085 | AF015084 | I09A02NGRL0007_G07 | D13338 | EF-1α | 5 | 17 106 041– 17 107 432 |
| GTTCGCTAAGTTGTTCCTAGTG | GTGCTGGGTTCCCACGAGGACTT | 921 | AB059394 | I09A02NGRL0003_D23 | D76418 | Attacin | 6 | 18 601 317– 18 602 129 |
| CATATTCCGCACGATGCGGG | GTTACCTAAGCCCATGTTCAT | ca. 2000 | AB201280 | Not found | AB104488 | Chitinase | 7 | 14 117 862– 14 119 517 |
| GTCCAGGAGATCAACAAGACCAAG | CAGCTTCAGGAGGTTCAGCTTGTT | ca. 1800 | AB543313, AB543314 | S13A01NGRL0010_F15 | AY769271 | RpL4 | 11 | 2 852 029– 2 862 665 |
| GGNGCNAARCTNTGYAAYATHTTYTC | TGRAARTAYTCYTTNGCYTCYTTNGA | 1391 | AB543310c | Not found | AF013277 | Topo II | 11 | 8 589 427– 8 602 332 |
| CATAAACACGACAGGAAAGTTCGGC | CTTGGTGCGCACGTATGGTTTAGT | ca. 2000 | AB543311, AB543312 | S06A01NCLL0020_P12 | AY769287 | RpL18 | 11 | 9 399 580– 9 401 164 |
| GCTTTGCATTGCGATGCGAAACGT | ACAGTCGCTAATATCTGGCAGTCC | ca. 3000 | AB048258 | I10A02NGRL0003_P23 | L37416 | Lysozyme | 12 | 3 312 565– 3 309 259 |
| GCNCAYGGNTCNCARTGYGGNTTYTGYAC | TARAARTGYTCYTGNCCNCCCATNCGRCA | 1985 | AB543315c | Not found | D38159 | XDH I | 12 | 8 502 165– 8 493 509 |
| GTTGCCGCGTTGGTAGTAGACAA | CCAGACTCGTCGTACTCCTGTTT | 1169 | AB543316 | I10A02NGRL0006_L21 | U49644 | Cytoplasmic actin A4 | 17 | 2 936 956– 2 938 173 |
| ATTCAGCGTACAATTCTTATGATGGG | CCAATGATCAAATACGCATTGTGGGAA | 1113 | AB543317 | S06A01NCLL0007_L10 | DQ443396 | eEF-2 | 19 | 12 802 871– 12 811 573 |
| AAAATGGGCATGCTACCGCGTGG | GCTCATCAACATGACGTCTTTGTA | ca. 2300 | AB288051 | S13A02NGRL0010_N13 | X12978 | SP1 | 23 | 16 906 448– 16 911 216 |
| GGGTAAACAATGGCGTCTAAG | GCTCTGCAGCCGTAGTATCCC | ca. 3000 | AB220992 | I09A02NGRL0003_K24 | AY515321 | Hemolin | 24 | 13 879 059– 13 884 127 |
Abbreviations: BYB, Y-box protein; DDC, DOPA decarboxylase; DOP-PCR, degenerate oligonucleotide-primed PCR; EF-1α, elongation factor 1α eEF-2, translation elongation factor 2; EST, expressed sequence tag; RpL4, ribosomal protein L4; RpL18, ribosomal protein L18; SP1, sex-specific storage-protein 1; Topo II, topoisomerase II; XDH I, xanthine dehydrogenase I.
ESTs showing the highest score were taken from http://silkbase.ab.a.u-tokyo.ac.jp/cgi-bin/index.cgi.
Inferred from http://sgp.dna.affrc.go.jp/KAIKObase/.
An orthlogous sequence was obtained using DOP-PCR.
Chromosome preparations
Spread preparations were made from pachytene oocytes, which were obtained from ovaries of last instar larvae as described by Yoshido et al. (2005b). Briefly, the ovaries were dissected in a saline solution and fixed for 10–15 min in Carnoy's fixative (ethanol, chloroform, acetic acid, 6:3:1). Cells were dissociated in 60% acetic acid and spread on the slide using a heating plate at 50 °C. The preparations were then passed through a graded ethanol series (70, 80 and 98%) and stored in a −30 °C freezer until further use.
Fluorescence in situ hybridization
Genomic in situ hybridization (GISH) was carried out according to the methods described by Sahara et al. (2003). Female genomic DNA was labelled by nick translation using a Nick Translation Mix (Roche Diagnostics, Mannheim, Germany) with Green-dUTP (Abbott Molecular Inc., Des Plaines, IL, USA) and used as a probe. For FISH mapping of orthologous genes, the cloned fragments were labelled with Cy3-dCTP (GE Healthcare, Piscataway, NJ, USA) by PCR under conditions described above, except for a 4 min extension. We purified the probes using a Wizard SV Gel and PCR Clean-Up System (Promega KK) as per the manufacturer's protocol.
Chromosome preparations were removed from the freezer, passed through the graded ethanol series and air-dried. They were denatured at 72 °C for 3.5 min in 70% formamide, 2 × standard sodium citrate. FISH mapping and GISH were simultaneously performed for one preparation with a probe cocktail containing 500 ng Green-labelled female DNA (green), 100 ng Cy3-labelled gene fragment (red), 3 μg unlabelled sonicated male genomic DNA and 25 μg sonicated salmon sperm DNA (Sigma-Aldrich, Tokyo, Japan) in 10 μl hybridization solution (50% formamide, 10% dextran sulphate, 2 × standard sodium citrate). After hybridization in a moist chamber at 37 °C for 3 days, the slides were washed at 62 °C in 0.1 × standard sodium citrate containing 1% Triton X-100, and then counterstained and mounted in antifade solution containing 1,4-diazabicyclo[2,2,2]octan (DABCO; for composition, see Traut et al., 1999) and 0.5 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Preparations were observed in a Leica DMRE HC fluorescence microscope. Digital images were acquired and processed as described in Sahara et al. (2003).
Construction of molecular phylogenetic trees
To examine relationships between S. cynthia subspecies, a molecular phylogenetic analysis was carried out using (i) a fragment of mitochondrial DNA (COI–COII) containing complete sequences of genes encoding cytochrome oxidase subunit I, tRNALeu, and cytochrome oxidase subunit II, tRNALys, and (ii) a partial coding sequence of the nuclear gene EF-1α. In each subspecies, the COI–COII fragment was amplified from the respective mitochondrial DNA by degenerate oligonucleotide-primed PCR using two primers, 5′-CGAAAATGACTTTAYTCNACTAATC-3′ (forward) and 5′-TCATTAGAAGTANTTGNTAATTTAC-3′ (reverse). The EF-1α sequence was amplified from the respective genomic DNA using primers given in Table 1. PCR conditions were as described above. PCR products (2355 bp COI–COII and 1085 bp EF-1α fragments) were sequenced with an ABI PRISM 3100 Genetic Analyzer following the sequencing protocol supplied by the manufacturer.
Neighbour-joining trees of S. cynthia subspecies were constructed using the Molecular Evolutionary Genetics Analysis 4 software (Tamura et al., 2007; http://www.megasoftware.net/). Bootstrap values were estimated with 1000 replicates. The Chinese oak silkmoth, Antheraea pernyi (Saturniidae), and B. mori were used as outgroups. The corresponding COI–COII and EF-1α sequences of these two species were obtained from a public database. GenBank accession numbers of all sequences used are given in Supplementary Table S1.
Results
S. cynthia orthologues of B. mori genes
We cloned S. c. walkeri (Sapporo population) orthologues of 16 B. mori and/or S. c. ricini genes (Table 1). BlastN searches of the coding region in S. c. walkeri orthologues of 13 genes (except for kettin, Topo II and XDH I) against expressed sequence tag libraries of S. c. ricini in SilkBase and public databases indicated very high sequence identities (at least 96%). TblastX searches of S. c. walkeri orthologues of three B. mori genes, Topo II, kettin and XDH I, showed 91, 89 and 76% identities, respectively.
FISH mapping of S. cynthia orthologues of B. mori Z-linked genes
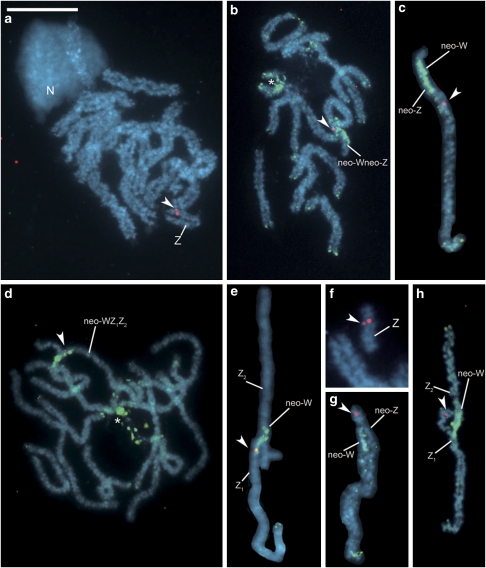
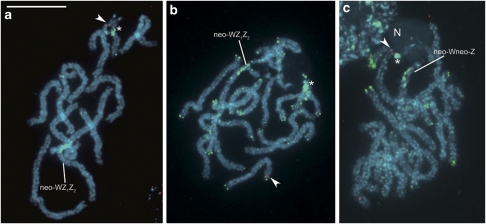
We carried out FISH mapping of the S. cynthia orthologues of B. mori Z-linked genes (BYB and kettin) to female pachytene complements in three S. cynthia subspecies/populations. In these complements, the Z chromosome was easily identified as the only univalent among autosome bivalents of S. c. ricini (Figure 1a), whereas the neo-Wneo-Z bivalent of S. c. walkeri (Figure 1b) and the neo-WZ1Z2 trivalent of S. cynthia subsp. indet. (Figure 1d) were recognized according to green hybridization signals of the female genomic probe, which bound to the ancestral W-heterochromatin segment of the neo-W chromosome. In S. c. ricini, a probe made from the orthologue of B. mori BYB hybridized to the Z-chromosome univalent (Figure 1a). A clear hybridization signal of the probe was found on the neo-Z chromosome in the neo-Wneo-Z bivalent of S. c. walkeri (Figures 1b and c) and on the Z1 chromosome in the neo-WZ1Z2 trivalent of S. cynthia subsp. indet. (Figures 1d and e). The kettin orthologous probe also hybridized to the Z univalent in S. c. ricini (Figure 1f, Supplementary Figure S1a), the original Z part of the neo-Z chromosome in S. c. walkeri (Figure 1g, Supplementary Figure S1b) and the Z1 chromosome in S. cynthia subsp. indet. (Figure 1h, Supplementary Figure S1c). In the latter two subspecies, hybridization signals of the kettin orthologous probe were observed on unpaired regions of the sex chromosomes.
Figure 1.
FISH mapping of S. cynthia orthologues of B. mori Z-linked genes on female pachytene chromosomes of three subspecies of S. cynthia. Red signals (arrowheads) are Cy3-labelled probes of BYB (a–e) and kettin (f–h) orthologous genes. GISH with Green-labelled female genomic probe (green signals) identified the original W compartment composed of heterochromatin (b–e, g, h) and also highlighted a block of heterochromatin in the NOR autosome (asterisk in b, d). Chromosomes were counterstained with DAPI (light blue). S. c. ricini (a, f): the U-shaped, apparently self-paired univalent of the Z chromosome with BYB orthologue signals on both chromatids is seen in the pachytene complement besides 13 autosome bivalents; also note a conspicuous nucleolus (N) associated with one end of the NOR autosome (a); and a Z-chromosome univalent with kettin orthologue signals on both chromatids (f). S. c. walkeri (b, c, g): a pachytene complement composed of a neo-Wneo-Z sex chromosome bivalent and 12 autosome bivalents (b); signals of the BYB (b, c) and kettin (g) orthologous probes are located on the neo-Z chromosome. S. cynthia subsp. indet. (d, e, h): a pachytene complement consists of a neo-sex chromosome trivalent (neo-WZ1Z2) and 11 autosome bivalents (d); signals of the BYB (d, e) and kettin (h) orthologous probes are located on the Z1 chromosome. Bar represents 10 μm (a–e, h), 7.5 μm (g) and 5 μm (f).
Identification of autosomal parts of the neo-sex chromosomes
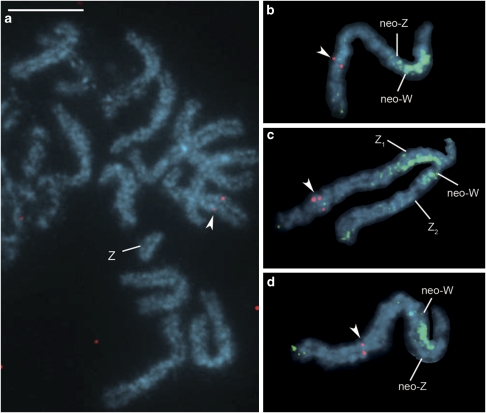
We searched for S. cynthia orthologues that would map to autosomal parts of the neo-sex chromosomes in S. c. walkeri and S. cynthia subsp. indet. These autosomal parts correspond to A1 and A2 autosomes of S. c. ricini, as shown in the previously proposed scheme of the sex chromosome evolution in this species (see Figure 4 in Yoshido et al., 2005b). Among randomly selected orthologues of B. mori autosomal genes, a probe prepared from the S. cynthia orthologue of the XDH I gene, which is located on B. mori chromosome 12, hybridized to an autosome bivalent in S. c. ricini (Figure 2a). The probe mapped to the putative A1 part of the neo-W and neo-Z sex chromosomes in S. c. walkeri (Figure 2b, Supplementary Figure S1d) and to the neo-W and Z1 sex chromosomes in S. cynthia subsp. indet. (Figure 2c, Supplementary Figure S1e). Lysozyme is another representative of the gene located on the B. mori chromosome 12. The S. cynthia orthologous probe of this gene also hybridized to the A1 segments of neo-sex chromosomes in S. c. walkeri (Figure 2d, Supplementary Figure S1f) and S. cynthia subsp. indet. (data not shown). These results indicated that at least a part of the A1 segment of the two subspecies with neo-sex chromosomes corresponds to a part of the B. mori chromosome 12.
Figure 2.
FISH identification of autosomal parts of the S. c. walkeri and S. cynthia subsp. indet. neo-sex chromosomes on female pachytene chromosomes of three subspecies of S. cynthia. The Cy3-labelled orthologous probe of the B. mori XDH 1 gene (red signals, arrowheads) hybridized to an autosome bivalent, but not to the ‘U-shaped' univalent of the Z chromosome in the female pachytene complement of S. c. ricini (a), whereas the XDH 1 orthologous probe mapped to autosomal parts of both the neo-W and neo-Z chromosomes in S. c. walkeri (b) and to autosomal parts of both the neo-W and Z1 chromosomes in S. cynthia subsp. indet. (c). The Cy3-labelled orthologous probe of the B. mori lysozyme gene (red signals, arrowhead) hybridized to the same autosomal segment of both the neo-W and neo-Z chromosomes in S. c. walkeri (d). GISH with Green-labelled female genomic probe (green signals) identified the original W-heterochromatin parts of the neo-W chromosome in S. c. walkeri (b, d) and S. cynthia subsp. indet. (c). Chromosomes were counterstained with DAPI (light blue). Bar represents 10 μm.
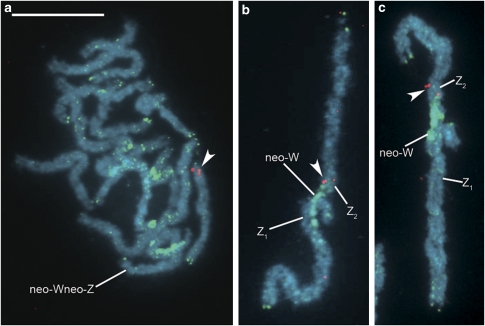
In B. mori, the Topo II and RpL18 genes are located on chromosome 11. A probe prepared from the S. cynthia orthologue of the Topo II gene mapped to an autosome in S. c. walkeri (Figure 3a), whereas in S. cynthia subsp. indet., hybridization signals of the probe were observed on the neo-W and Z2 sex chromosomes (Figure 3b, Supplementary Figure S1g). Similarly, the RpL18 orthologous probe hybridized to autosomal segments of both the neo-W and Z2 sex chromosomes in S. cynthia subsp. indet. (Figure 3c, Supplementary Figure S1h), whereas in S. c. walkeri, the probe hybridized to a pair of autosomes (data not shown). Hence, a part of the B. mori chromosome 11 corresponds to the putative A2 chromosome of S. cynthia (see Figure 4 in Yoshido et al., 2005b). Further FISH experiments showed that the orthologue of the RpL4 gene located on B. mori chromosome 11 does not map to the neo-sex chromosomes, but to the nucleolar organizer region (NOR) bivalent in S. cynthia subsp. indet. (Figure 4a). This NOR bivalent was easily identified by a specific heterochromatin block highlighted by GISH with the S. cynthia female genomic probe (cf. Yoshido et al., 2005b). The RpL4 orthologue also mapped to the NOR bivalents of S. c. walkeri and S. c. ricini (data not shown). This suggests that the NOR chromosome has the same origin in all three S. cynthia subspecies.
Figure 3.
FISH identification of the autosomal segment of the neo-W chromosome homologous to the Z2 chromosome in S. cynthia subsp. indet. on female pachytene chromosomes of S. c. walkeri (a) and S. cynthia subsp. indet. (b, c). The Cy3-labelled orthologous probe of the B. mori Topo II gene (red signals, arrowheads) hybridized to an autosomal bivalent but not to the neo-Wneo-Z bivalent in a female pachytene complement of S. c. walkeri (a), whereas in S. cynthia subsp. indet., the probe mapped to the Z2 chromosome and the neo-W chromosome of the neo-WZ1Z2 trivalent (b). The Cy3-labelled orthologous probe of the B. mori RpL18 gene (red signal, arrowhead) also hybridized to the Z2 chromosome and the neo-W chromosome of the neo-WZ1Z2 trivalent in S. cynthia subsp. indet. (c). GISH with Green-labelled female genomic probe (green signals) identified the original W-heterochromatin parts of the neo-W chromosome in S. c. walkeri (a) and S. cynthia subsp. indet. (b, c). Chromosomes were counterstained with DAPI (light blue). Bar represents 10 μm.
Figure 4.
FISH mapping of S. cynthia orthologues of B. mori autosomal genes on female pachytene chromosome complements of S. cynthia subspecies. Red signals (arrowheads) are Cy3-labelled orthologous probes of the B. mori genes RpL4 (a), eEF-2 (b) and hemolin (c). GISH with Green-labelled female genomic probe (green signals) identified the original W-heterochromatin compartment of the neo-W chromosome and also highlighted a heterochromatin block on the NOR bivalent (asterisk) in S. cynthia subsp. indet. (a, b) and S. c. walkeri (c). Chromosomes were counterstained with DAPI (light blue). In S. cynthia subsp. indet., the RpL4 orthologue mapped to the NOR bivalent (a), whereas the eEF-2 orthologue mapped to an autosome bivalent (b). The hemolin orthologue mapped to the NOR bivalent in S. c. walkeri (c); note a conspicuous nucleolus (N) associated with the NOR bivalent. Bar represents 10 μm.
S. cynthia orthologous probes of genes located on B. mori chromosomes 2, 4, 5, 6, 7, 17, 19 and 23 (Table 1) hybridized to unspecified autosomes in two subspecies with the neo-sex chromosomes, S. c. walkeri and S. cynthia subsp. indet. (an example is shown in Figure 4b). Only the orthologue of the hemolin gene, a representative of B. mori chromosome 24, mapped to the NOR bivalents in all three S. cynthia subspecies (an example is shown in Figure 4c). These results suggest that the NOR chromosome of S. cynthia is composed of two segments, one corresponding to a part of chromosome 11 and the other to a part of chromosome 24 of B. mori.
Evolutionary relationship between geographical subspecies of S. cynthia
The molecular phylogenetic analysis carried out using both the mitochondrial COI–COII and the nuclear EF-1α sequences confirmed the closer relationship of two Japanese subspecies, S. c. walkeri and S. cynthia subsp. indet., and separation of the Vietnam subspecies, S. c. ricini. A higher support for separation of the two Japanese subspecies was obtained with the COI–COII sequences, whereas the separation of S. c. ricini was obvious in both trees (Supplementary Figure S2a, b).
Discussion
Neo-sex chromosomes that originated by autosome–sex chromosome fusions have been reported in various animals with male heterogamety (for example, Schmid et al., 2003; Král, 2007; Zhou et al., 2008; Bressa et al., 2009). However, reports on neo-sex chromosomes in animals with female heterogamety are limited to a few species of fish (Ueno et al., 2001) and moths (reviewed by Marec et al. (2010)). S. cynthia silkmoths represent a special case. Their sex chromosome constitution was altered by a series of chromosomal rearrangements in geographical subspecies after they differentiated from a putative ancestor with the diploid karyotype of 2n=28 and a WZ sex chromosome pair. Thus, a loss of the W chromosome resulted in a Z0 system as found in S. c. ricini (Vietnam population), whereas fusion of sex chromosomes with autosomes resulted in the neo-Wneo-Z and neo-WZ1Z2 systems as found in S. c. walkeri (Sapporo population) and S. cynthia subsp. indet. (Nagano population), respectively (Yoshido et al., 2005b).
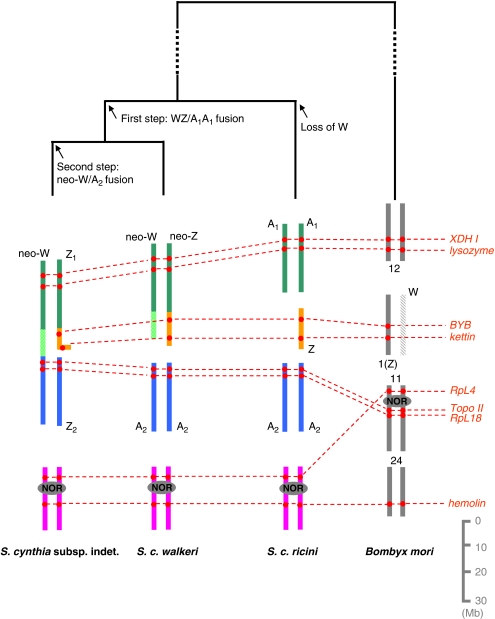
In this study, we identified autosomes involved in the evolution of the complex sex chromosome systems in S. cynthia by comparative mapping of S. cynthia orthologues of B. mori genes. Figure 5 shows a schematic representation of the process of chromosomal rearrangements that occurred between B. mori and three geographical subspecies of S. cynthia. Our findings support a hypothetical scenario of the sex chromosome evolution in S. cynthia mentioned previously (see Figure 4 in Yoshido et al., 2005b). In addition, the findings point towards conserved synteny of genes between Z chromosomes of B. mori and S. c. ricini and also to homology of the S. c. ricini Z chromosome with parts of the neo-Z chromosome of S. c. walkeri and the Z1 chromosome of S. cynthia subsp. indet. Our data suggest that the first step of neo-sex chromosome evolution was a fusion between an autosome pair (A1), which corresponds to B. mori chromosome 12, and the original W and Z chromosomes in a common ancestor of both Japanese subspecies, S. c. walkeri and S. cynthia subsp. indet., whereas no such event occurred in S. c. ricini (Figure 5). In the next step, the neo-W chromosome of S. cynthia subsp. indet. was formed by a fusion of the neo-W of S. c. walkeri with another autosome (A2) corresponding to B. mori chromosome 11, and therefore the other A2 corresponds to the Z2 sex chromosome in S. cynthia subsp. indet. Phylogenetic trees constructed with nucleotide sequences of mitochondrial and nuclear genes (Supplementary Figure S2a, b) also support the scenario of karyotype evolution in the geographic populations of S. cynthia.
Figure 5.
Schematic drawing of step-by-step evolution of the sex chromosome constitution in female silkmoth of Samia cynthia subspecies, based on comparative mapping of orthologues of Bombyx mori genes. In two Japanese subspecies, the neo-sex chromosomes evolved in two steps by fusions of the ancestral sex chromosomes with autosomes. In the first step, the W and Z chromosomes each fused with an autosome of the A1A1 pair, corresponding to chromosome 12 of B. mori; this resulted in the neo-Wneo-Z constitution of S. c. walkeri female silkmoth. In the second step, only the neo-W chromosome fused with an autosome (A2), partly corresponding to chromosome 11 of B. mori; this resulted in a neo-WZ1Z2 constitution of S. cynthia subsp. indet. female silkmoth. On the contrary, loss of the W chromosome in the putative S. cynthia ancestor generated a Z0 constitution of S. c. ricini. A segment corresponding to another part of the B. mori chromosome 11 containing the NOR plus a segment corresponding to a part of B. mori chromosome 24 formed the only NOR chromosome in all three S. cynthia subspecies. B. mori chromosomes are represented by grey vertical bars drawn to relative scale in Mb taken from KAIKObase.
The genetic content of the lepidopteran Z chromosomes is little known except for B. mori. Molecular linkage analyses revealed only a few orthologues of B. mori Z-linked genes that were also located on the Z chromosomes in other lepidopteran species, such as the European corn borer Ostrinia nubilalis (Dopman et al., 2005), Heliconius butterflies (Jiggins et al., 2005; Pringle et al., 2007) and the butterfly B. anynana (Van't Hof et al., 2008; Beldade et al., 2009). Recently, BAC clones containing M. sexta orthologues of four B. mori Z-linked genes were mapped to the Z chromosomes of two species of the family Sphingidae, M. sexta and Agrius convolvuli, indicating the conserved synteny of the Z-linked genes between the three species (Yasukochi et al., 2009). These results suggest that Z chromosomes are conserved in three large clades of Lepidoptera: the Pyraloidea (O. nubilalis), Papilionoidea (Heliconius sp. and B. anynana) and Bombycoidea (B. mori and two sphingids). In this study, FISH mapping of two orthologues of the B. mori Z-linked genes (kettin and BYB; Table 1) in S. cynthia clearly showed that both genes are located on the Z chromosome of S. c. ricini and also on the original Z compartment in S. c. walkeri and S. cynthia subsp. indet (Figures 1a–h and 5). Our approach, which is based on the identification of conserved gene fragments and their comparative FISH mapping, is applicable to a wide range of species and thus it could facilitate studies on the evolution of sex chromosomes in Lepidoptera and their closest relatives, the Trichoptera. This method could also accelerate the construction of comparative maps of Lepidoptera, which has so far been performed for a limited number of species with BAC-FISH mapping (Sahara et al., 2007; Yasukochi et al., 2009).
Our study revealed that S. cynthia orthologues of two genes (Topo II and RpL18) of the B. mori chromosome 11 are located in the autosomal compartment of the neo-W chromosome in S. cynthia subsp. indet. (Nagano population) (Figure 3). However, another S. cynthia orthologue of the B. mori chromosome 11-linked gene, RpL4, did not map to the neo-W chromosome but to the NOR chromosome in all three S. cynthia subspecies (Figure 4). B. mori chromosome 11 can be easily recognized by an interstitially located nucleolus, which divides the chromosome into a shorter arm and a longer arm (Yoshido et al., 2005a). The RpL4 gene is located on the shorter arm (approximately 2.86 Mb in KAIKObase), and the Topo II and RpL18 genes are located on the longer arm (approximately 8.60 and 9.40 Mb, respectively). Besides the orthologue of the RpL4 gene, an orthologue of the hemolin gene of B. mori chromosome 24 mapped to the NOR chromosome of S. cynthia, indicating homology of this chromosome with the shorter arm of chromosomes 11 and 24 of B. mori (Figure 5). These chromosomal rearrangements can be explained by two fission/fusion events involving two chromosomes of S. cynthia (A2 and the NOR chromosome) and two chromosomes of B. mori (11 and 24).
In sex chromosome systems with male heterogamety, the Y chromosome is thought to originate from a homologous autosomal pair through the acquisition of a dominant, male-sex-determining role. The absence of meiotic recombination between X and Y chromosomes accelerates the accumulation of mutations on the Y chromosome, which leads to the functional inactivation of Y-linked genes followed by genetic erosion and accumulation of repetitive sequences (Charlesworth, 1996). However, the conception of the Y chromosome as a ‘graveyard' (Steinemann et al., 1993; Kjellman et al., 1995) has been revised recently. In the highly evolved Y chromosomes of primates, the palindrome-driven sister chromatid and/or intrachromatid recombination prevents degradation and loss of Y-chromosome genes, and thus contributes to continuing evolution of the Y chromosome (Lange et al., 2009; Hughes et al., 2010). Moreover, recent findings in Drosophila suggest that the Y chromosome has an important role in male fitness and also contributes to adaptive phenotypic variation through a regulatory role of Y-linked polymorphic elements in gene expression (Lemos et al., 2008). In contrast, a gradual degeneration appears to be the primary driving force in evolutionarily young Y chromosomes as shown in the plant models Carica papaya and Silene latifolia (Jamilena et al., 2008). In addition, once the degenerated Y chromosome fuses or translocates to an autosome, a new cycle of its evolution starts. Hence, organisms carrying a primitive Y chromosome or an evolutionarily young neo-Y chromosome represent valuable models for the study of sex chromosome differentiation. Recent studies using genomic approaches provided supporting evidence for the hypothesis on early steps of sex chromosome evolution in several plants and animals with the XY system (Liu et al., 2004; Yamato et al., 2007; Bachtrog et al., 2008; Zhou et al., 2008). However, similar approaches have not been applied yet in the W chromosome of Lepidoptera. Geographic populations of S. cynthia examined in this study are the only representatives of lepidopteran species in which the autosomal origin of the neo-sex chromosomes has been molecularly determined. As S. c. ricini (Vietnam population) has a Z0 sex chromosome constitution with the original set of autosomes, comparative analyses between autosomal parts of the neo-W chromosomes in Japanese subspecies of S. cynthia and the original set of autosomes in S. c. ricini may significantly contribute to understanding the early steps of sex chromosome evolution in organisms with female heterogamety.
A question remains whether the evolutionary changes in sex chromosome constitution of S. cynthia populations have a role in speciation. A disproportionate association between traits that distinguish closely related species and the Z chromosome, found in several moths and butterflies, suggests that the lepidopteran Z chromosome has a large effect on species divergence (Sperling, 1994; Prowell 1998). Similarly, the Z-linkage of key components of reproductive isolation in flycatchers, such as species recognition, species-specific male traits and hybrid incompatibilities, suggests an important role of the avian Z chromosome in adaptive speciation (Saether et al., 2007). However, it seems that the so-called ‘Large Z-effect' (or ‘Large X-effect' in systems with male heterogamety) on speciation can often be attributed to the fact that the Z/X-linked traits evolve faster than autosomal traits leading to incompatibilities between the Z/X-linked and the interacting autosomal genes (Mank et al., 2007; Tang and Presgraves, 2009). Recent results suggest that the so-called ‘Faster-Z effect' in a female heterogametic system is greater than the ‘Faster-X effect' in organisms with male heterogamety (Mank et al., 2010). In S. cynthia, owing to the tendency to autosome–sex chromosome fusions, different sets of originally autosomal genes become sex-linked and subjected to a faster rate of evolution. These fusions also multiply the number of genes exposed to faster divergence, which may accelerate the accumulation of genetically based incompatibilities between populations. The above considerations are consistent with recent evidence on a significant role of neo-sex chromosomes in reproductive isolation of a vertebrate system, the threespine stickleback fish (Gasterosteus aculeatus) (Kitano et al., 2009). Thus, we suggest that the step-by-step evolution of neo-sex chromosomes in geographical populations of S. cynthia by repeated autosome–sex chromosome fusions may facilitate population/species divergence and contribute to the formation of reproductive barriers.
Acknowledgments
We thank Y Yamada, Field Science Center for Northern Biosphere, Hokkaido University, Sapporo, Japan, for enabling us to rear Samia cynthia subspecies on Ailanthus altissima trees. Our thanks also go to H Saito (Kyoto, Japan) and J Kobayashi (Yamaguchi, Japan) for valuable information about S. cynthia; M Žurovcová (České Budějovice, Czech Republic) for valuable advice on molecular phylogenetic analysis; and three anonymous reviewers for useful comments. AY received a grant, 19-1114, from the Japan Society for the Promotion of Science (JSPS). FM was supported by grant IAA600960925 of the Grant Agency of the Academy of Sciences of the Czech Republic, and from the Entomology Institute Project Z50070508. We acknowledge the research fellowship 21-7147 given to KS from JSPS.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade P, Saenko SV, Pul N, Long AD. A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 2009;5:e1000366. doi: 10.1371/journal.pgen.1000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressa MJ, Papeschi AG, Vítková M, Kubíčková S, Fuková I, Pigozzi MI, et al. Sex chromosome evolution in cotton stainers of the genus Dysdercus (Heteroptera: Pyrrhocoridae) Cytogenet Genome Res. 2009;125:292–305. doi: 10.1159/000235936. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Dopman EB, Pérez L, Bogdanowicz SM, Harrison RG. Consequences of reproductive barriers for genealogical discordance in the European corn borer. Proc Natl Acad Sci USA. 2005;102:14706–14711. doi: 10.1073/pnas.0502054102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores SV, Evans AL, McAllister BF. Independent origins of new sex-linked chromosomes in the melanica and robusta species groups of Drosophila. BMC Evol Biol. 2008;8:33. doi: 10.1186/1471-2148-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamilena M, Mariotti B, Manzano S. Plant sex chromosomes: molecular structure and function. Cytogenet Genome Res. 2008;120:255–264. doi: 10.1159/000121075. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, Mavarez J, Beltran M, MacMillan WO, Johnston JS, Bermingham E. A genetic linkage map of the mimetic butterfly Heliconius melpomene. Genetics. 2005;171:557–570. doi: 10.1534/genetics.104.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi E. Chromosomenketten beim Rassenbastarde des Seidenspinners Philosamia cynthia. (Zytologische Untersuchungen am Seidenspinner und seinen Verwandten. III) Cytologia Fujii Jub. 1937;II:1023–1033. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellman C, Sjogren H-O, Widegren B. The Y chromosome: a graveyard for endogenous retroviruses. Gene. 1995;73:86–91. doi: 10.1016/0378-1119(95)00248-5. [DOI] [PubMed] [Google Scholar]
- Král J. Evolution of multiple sex chromosomes in the spider genus Malthonica (Araneae: Agelenidae) indicates unique structure of the spider sex chromosome systems. Chromosome Res. 2007;15:863–879. doi: 10.1007/s10577-007-1169-3. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the Faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Marec F, Sahara K, Traut W.2010Rise and fall of the W chromosome in LepidopteraIn Goldsmith MR, Marec F (eds).Molecular Biology and Genetics of the Lepidoptera CRC Press: Boca Raton, FL; 49–63. [Google Scholar]
- Nilsson N-O, Löfstedt C, Dävring L. Unusual sex chromosome inheritance in six species of small ermine moths (Yponomeuta, Yponomeutidae, Lepidoptera) Hereditas. 1988;108:259–265. [Google Scholar]
- Peigler RS, Naumann S. A Revision of the Silkmoth Genus Samia. University of the Incarnate Word: San Antonio, TX; 2003. [Google Scholar]
- Pringle EG, Baxter SW, Webster CL, Papanicolaou A, Lee SF, Jiggins CD. Synteny and chromosome evolution in the Lepidoptera: evidence from mapping in Heliconiusmelpomene. Genetics. 2007;177:417–426. doi: 10.1534/genetics.107.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowell DP.1998Sex linkage and speciation in LepidopteraIn: Howard DJ, Berlocher SH (eds).Endless Forms: Species and Speciation Oxford University Press: New York; 309–319. [Google Scholar]
- Saether SA, Saetre GP, Borge T, Wiley C, Svedin N, Andersson G, et al. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318:95–97. doi: 10.1126/science.1141506. [DOI] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Kawamura N, Ohnuma A, Abe H, Mita K, et al. W-derived BAC probes as a new tool for identification of the W chromosome and its aberrations in Bombyx mori. Chromosoma. 2003;112:48–55. doi: 10.1007/s00412-003-0245-5. [DOI] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Marec F, Fuková I, Zhang HB, Wu CC, et al. Conserved synteny of genes between chromosome 15 of Bombyx mori and a chromosome of Manduca sexta shown by five-color BAC-FISH. Genome. 2007;50:1061–1065. doi: 10.1139/g07-082. [DOI] [PubMed] [Google Scholar]
- Schmid M, Feichtinger W, Steinlein C, Garcia RV, Badillo AF. Chromosome banding in Amphibia. XXVIII. Homomorphic XY sex chromosomes and a derived Y-autosome translocation in Eleutherodactylus riveroi (Anura, Leptodactylidae) Cytogenet Genome Res. 2003;101:62–73. doi: 10.1159/000073420. [DOI] [PubMed] [Google Scholar]
- Song F, Banno Y, Kawaguchi Y, Koga K, Xiang Z. Dimorphism in chromosome number of Samia cynthia walkeri in China. Int J Wild Silkmoth Silk. 1996;2:11–13. [Google Scholar]
- Sperling FAH. Sex-linked genes and species differences in Lepidoptera. Can Entomol. 1994;126:807–818. [Google Scholar]
- Steinemann M, Steinmann S, Lottspeich F. How Y chromosomes become genetically inert. Proc Natl Acad Sci USA. 1993;90:5737–5741. doi: 10.1073/pnas.90.12.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toder R, O'Neill RJ, Wienberg J, O'Brien PC, Voullaire L, Marshall-Graves JA. Comparative chromosome painting between two marsupials: origins of an XX/XY1Y2 sex chromosome system. Mamm Genome. 1997;8:418–422. doi: 10.1007/s003359900459. [DOI] [PubMed] [Google Scholar]
- Traut W, Marec F. Sex chromosome differentiation in some species of Lepidoptera (Insecta) Chromosome Res. 1997;5:283–291. doi: 10.1023/B:CHRO.0000038758.08263.c3. [DOI] [PubMed] [Google Scholar]
- Traut W, Sahara K, Marec F. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 2007;1:332–346. doi: 10.1159/000111765. [DOI] [PubMed] [Google Scholar]
- Traut W, Sahara K, Otto TD, Marec F. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma. 1999;108:173–180. doi: 10.1007/s004120050366. [DOI] [PubMed] [Google Scholar]
- Ueno K, Ota K, Kobayashi T. Heteromorphic sex chromosomes of lizardfish (Synodontidae): focus on the ZZ–ZW1W2 system in Trachinocephalus myops. Genetica. 2001;111:133–142. doi: 10.1023/a:1013749027620. [DOI] [PubMed] [Google Scholar]
- Van't Hof AE, Marec F, Saccheri IJ, Brakefield PM, Zwaan BJ. Cytogenetic characterization and AFLP-based genetic linkage mapping for the butterfly Bicyclus anynana, covering all 28 karyotyped chromosomes. PLoS ONE. 2008;3:e3882. doi: 10.1371/journal.pone.0003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, et al. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci USA. 2007;104:6472–6477. doi: 10.1073/pnas.0609054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Ashakumary LA, Baba K, Yoshido A, Sahara K. A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects. Genetics. 2006;173:1319–1328. doi: 10.1534/genetics.106.055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukochi Y, Tanaka-Okuyama M, Shibata F, Yoshido A, Marec F, Wu C, et al. Extensive conserved synteny of genes between the karyotypes of Manduca sexta and Bombyx mori revealed by BAC-FISH mapping. PLoS ONE. 2009;4:e7465. doi: 10.1371/journal.pone.0007465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshido A, Bando H, Yasukochi Y, Sahara K. The Bombyx mori karyotype and the assignment of linkage groups. Genetics. 2005a;170:675–685. doi: 10.1534/genetics.104.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshido A, Marec F, Sahara K. Resolution of sex chromosome constitution by genomic in situ hybridization and fluorescence in situ hybridization with (TTAGG)n telomeric probe in some species of Lepidoptera. Chromosoma. 2005b;114:193–202. doi: 10.1007/s00412-005-0013-9. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang J, Huang L, Nie W, Wang J, Liu Y, et al. Neo-sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol. 2008;9:R98. doi: 10.1186/gb-2008-9-6-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.