Abstract
Coexistence allows growers and consumers the choice of producing or purchasing conventional or organic crops with known standards for adventitious presence of genetically engineered (GE) seed. Flax (Linum usitatissimum L.) is multipurpose oilseed crop in which product diversity and utility could be enhanced for industrial, nutraceutical and pharmaceutical markets through genetic engineering. If GE flax were released commercially, pollen-mediated gene flow will determine in part whether GE flax could coexist without compromising other markets. As a part of pre-commercialization risk assessment, we quantified pollen-mediated gene flow between two cultivars of flax. Field experiments were conducted at four locations during 2006 and 2007 in western Canada using a concentric donor (20 × 20 m) receptor (120 × 120 m) design. Gene flow was detected through the xenia effect of dominant alleles of high α-linolenic acid (ALA; 18:3cisΔ9,12,15) to the low ALA trait. Seeds were harvested from the pollen recipient plots up to a distance of 50 m in eight directions from the pollen donor. High ALA seeds were identified using a thiobarbituric acid test and served as a marker for gene flow. Binomial distribution and power analysis were used to predict the minimum number of seeds statistically required to detect the frequency of gene flow at specific α (confidence interval) and power (1−β) values. As a result of the low frequency of gene flow, approximately 4 million seeds were screened to derive accurate quantification. Frequency of gene flow was highest near the source: averaging 0.0185 at 0.1 m but declined rapidly with distance, 0.0013 and 0.00003 at 3 and 35 m, respectively. Gene flow was reduced to 50% (O50) and 90% (O90) between 0.85 to 2.64 m, and 5.68 to 17.56 m, respectively. No gene flow was detected at any site or year >35 m distance from the pollen source, suggesting that frequency of gene flow was ⩽0.00003 (P=0.95). Although it is not possible to eliminate all adventitious presence caused by pollen-mediated gene flow, through harvest blending and the use of buffer zones between GE and conventional flax fields, it could be minimized. Managing other sources of adventitious presence including seed mixing and volunteer populations may be more problematic.
Keywords: coexistence, adventitious presence, conventional, organic, pollen-mediated gene flow, regulation
Introduction
With the introduction of genetically engineered (GE) crops, pollen-mediated gene flow has become easier to detect and important to mitigate. Detection of adventitious presence of transgenes at low frequency using real time PCR has become a routine testing procedure to detect GE seeds of major crops including canola (Brassica napus L.), maize (Zea mays L.) and soybean (Glycine max L.) and their products (Hubner et al., 2001; Pla et al., 2006). In addition, conventional products with minimal GE content are in demand in some regions, notably the European Union (EU), but worldwide, adventitious presence thresholds either vary or have yet to be established (Demeke et al., 2006). Concurrent with the commercialization of GE crops, there has been increased demand for organic products in which there is a zero threshold for the presence of transgenes. Pollen-mediated gene flow is only one source of adventitious presence. Contamination of seed for sowing, gene persistence in volunteer populations (Jhala et al., 2010; Dexter et al., 2010a, 2010b) and seed mixing during transport must also be considered. Traditional isolation distances and other management practices designed to segregate crop varieties and production systems to ensure seed and product purity may be insufficient given these constraints. Quantification of pollen-mediated gene flow is required to determine if GE, conventional and organic crops can coexist without constraints to international trade and to develop practices for mitigation of transgene movement (Devos et al., 2004).
Flax, Linum usitatissimum L., an oilseed crop, is predominantly a self-pollinated species (Gill, 1987). Flax has a perfect flower with stamens and pistils present in the same flower. As the flax flower opens, the stamens bend inward, discharging pollen on stigma, usually resulting in self-pollination (Dillman, 1938). Gene flow in flax not only depends on the position of anthers in relation to stigma, receptivity of stigma, viability of pollen, availability of pollinators (Henry and Tu, 1928; Yermanos and Kostopoulos, 1970) but may also vary with genotype and environment (Dillman, 1938). Previous reports have indicated outcrossing rates in flax in the range of 1–5%, when flax plants were grown in close proximity (Howard et al., 1919; Graham and Roy, 1924; Bolley, 1927; Dillman and Stoa, 1935; Dillman and Goar, 1937; Joshi, 1994). However, pollen-mediated gene flow beyond 7.0 m was not documented. There is no recent information on pollen vectors, although insect pollinators have been reported to favor gene flow in flax (Dillman, 1938; Gubin, 1945).
Flax has been transformed with several novel genes (McSheffrey et al., 1992; Dong and McHughen, 1993; McHughen and Holm, 1995; Wijayanto and McHughen, 1999; McHughen, 2002) but currently no GE flax cultivars are commercially grown. Sulfonylurea-resistant flax (CDC Triffid) (Event PF967) was approved as safe for release in Canada and the United States but was withdrawn before full commercial release at the request of the Flax Council of Canada, primarily to avoid trade concerns with the EU, the primary export market. Approval of Event PF967 was not completed in the EU and there is a zero tolerance for contamination with an unapproved trait. Recently Event PF967 was identified in Germany in a shipment of Canadian flaxseeds (Bedard, 2009). This incident led to a disruption in the trade of Canadian flax to many countries including the EU.
The development of GE flax is under consideration in Canada and several parts of the world (Wrobel et al., 2004; Wrobel-Kwiatkowska et al., 2007). Development of these cultivars is predicated on the approval of food, feed and environmental biosafety. In the EU, crop products that contain approved GE events must be labeled if the content exceeds 0.9%. If thresholds can be met, GE flax could coexist along with conventional and organic flax.
Quantifying pollen-mediated gene flow before commercialization of GE flax allows trait developers and the flax industry to assess the risks (in part) of releasing a GE flax cultivar at a commercial scale. However, because pollen movement is dependent on the size of the donor and recipient populations, small trials may underestimate the distance and frequency of pollen-mediated gene flow. For example, Hanson et al. (2005) detected pollen-mediated gene flow in wheat (Triticum aestivum L.) up to a maximum of 42 m in a 45.7 m diameter pollen source, while Matus-Cadiz et al. (2007) reported occurrence of gene flow up to 2.75 km from a 33-ha pollen source.
As no GE flax cultivars were available to be used as the donor population, various methods have been used to identify gene flow. Dillman (1938) used flax flower petal color as a marker, but it was not reliable or accurate. Several markers have been used in other crops to detect pollen-mediated gene flow in donor–receptor experiments including molecular markers based on quantitative PCR (Weekes et al., 2005, 2007; Pla et al., 2006; Weber et al., 2007) or micro-satellite markers (Chaix et al., 2003; Dje et al., 2004; Isagi et al., 2004). Other techniques like green fluorescent protein (Halfhill et al., 2003) and blue aleurone seed color in wheat (Hanson et al., 2005) and triticale (Hills et al., 2007) have also been reported. Flax cultivars with high ALA (α-linolenic acid: 18:3cisΔ9,12,15) (Kenaschuk and Rashid, 1994) and low ALA ‘Solin' (<3% ALA) cultivars have been developed (Dribnenki et al., 2003). Two independently inherited genes, LuFAD3A and LuFAD3B control the ALA trait in flax (Vrinten et al., 2005). Seeds formed from the hybridization between conventional and Solin cultivars express high levels of ALA.
A thiobarbituric acid (TBA) test has been described for rapid screening of individual seed of flax for the content of ALA (Bhatty and Rowland, 1990). The TBA test is sensitive in determining peroxides of ALA and arachidonic acid (C20:4cisΔ5,8,11,14) because autoxidation of monoenes and dienes do not yield products that react with TBA, only trienes and more highly unsaturated fatty acids yield such products (Dahle et al., 1962). Polyunsaturated fatty acids form five-membered peroxide ring on oxidation (McGregor, 1974). In this study, the dominant high ALA trait in combination with the sensitive TBA test was exploited to determine pollen-mediated gene flow between two flax cultivars.
An additional constraint to detect pollen-mediated gene flow is sample size. Without testing every seed in a population, it is not possible to confirm the absence of transgenes. A strategy is required for testing the minimum number of seeds to provide a statistically meaningful conclusion. Power, broadly defined as the probability of getting a positive result when one is present, provides a decision-making framework. A sampling strategy was developed using power analysis and binomial distribution to define the minimum number of seeds required to measure the pollen-mediated gene flow for a specified null hypothesis between two crop cultivars. Three basic parameters affect power: proposed sample size, significance level used to determine whether or not to accept the study's hypothesis and theoretical effect size (Bausell and Li, 2002). Generally, a power value of 0.8 at 95% confidence interval is statistically accepted. However, for longer distances (>25 m in this study) it is advisable to screen more seeds to provide a higher power (>0.8). A larger sample size generally leads to parameter estimates with smaller variances, giving a greater ability to detect small differences.
The objective of this study was to measure pollen-mediated gene flow in flax under field conditions using high and low ALA flax cultivars in western Canada. Frequency of gene flow and distance from the pollen source was quantified up to a distance of 50 m. We also determined the minimum number of seeds required to provide a statistically meaningful conclusion on detecting gene flow at various distances by using the theoretical values of gene flow frequencies and various confidence intervals (α). This ensured that sufficient power (1−β) existed to measure the pollen-mediated gene flow.
Materials and methods
Plant material
Breeder seed of two flax cultivars, ‘AC McDuff' and ‘SP 2047' were provided by Vittera, Calgary, Alberta, Canada. AC McDuff, released by the Agriculture and Agri-Food Canada, is a late maturing cultivar with high ALA content (∼70% ALA) (Kenaschuk and Rashid, 1994). SP 2047, is a solin cultivar with low ALA (<3%), developed by Vittera and registered in 2002 (Dribnenki et al., 2003). The reproductive biology of these two cultivars was representative of flax cultivars grown in western Canada.
Field experiments
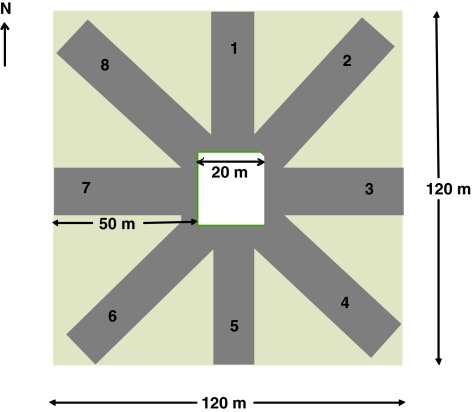
Field experiments were conducted at two locations, Edmonton Research Station (EdRS) and Ellerslie Research Station (ElRS), University of Alberta, Canada in 2006 and 2007. The soil at EdRS was a clay loam, which consisted of 34.2% sand, 37.5% silt, 28.3% clay, 12.9% organic matter with a pH of 5.6. Soil at ElRS was a clay loam with 28.2% sand, 41.1% silt, 30.7% clay, 11.2% organic matter with a pH 6.5. The high ALA flax cultivar AC McDuff was seeded in a 20 × 20 m area in the center, surround by the low ALA cultivar SP 2047, for a total area of 120 × 120 m in a concentric ring experimental design (Figure 1) at both locations and years. Flax was seeded on 16 and 18 May in 2006; and 26 and 28 May in 2007, at EdRS and ElRS sites, respectively. Fertilizers applied at each site were at the recommended rates based on soil tests (data not shown). Both flax cultivars flowered synchronously at each location-year. Just before harvesting, the field was divided into eight directional blocks and the crop between blocks was removed to facilitate accurate sampling (Figure 1). Harvesting took place on 17 and 21 September at EdRS and ElRS, respectively in 2006; and on 23 and 27 September at EdRS and ElRS, respectively in 2007. Harvesting began distal to the source and continued inward in a clockwise direction to reduce cross contamination. Samples were harvested with a Wintersteiger 2001 Elite research combine in 1.2 m wide swaths from each block from 50 to 3 m from the source in all blocks. A binder was used to harvest plants from 3.0 m to the source to reduce sample contamination. Samples were combined at longer distances to increase sample size. Seeds were threshed (if required), cleaned and stored at room temperature before screening.
Figure 1.
Design of pollen-mediated gene flow experiment in flax at all the locations and years. The pollen source flax cultivar (AC MCDuff) was seeded in the area of 20 × 20 m in the center of the field. The pollen receptor solin flax cultivar (SP 2090) was grown in the surrounding 120 × 120 m area. After flowering but before harvesting, the pollen receptor area was divided in eight blocks, crop between blocks removed and flax seed samples were collected at specific distances.
Seed screening
The TBA test described in Bhatty and Rowland (1990) was modified to accommodate approximately 500 flax seeds on a 13 × 30 cm rectangle paper, facilitating the screening of nearly 20 000 seeds per day. Before planting in 2006 and 2007, 1.3 million SP 2047 seeds were tested for purity and five and three seeds, respectively were identified with high ALA content, a frequency of <0.00005, α=0.05, power=0.95. This level of contamination was considered as a minor source of variance. The known standards of high and low ALA seeds were tested with every 500 seed lot and no false positives or negatives were identified. Seeds harvested from pollen receptor plots were screened in laboratory using TBA test and those containing high ALA were considered the product of gene flow from high ALA to low ALA flax cultivars.
Statistical analysis
A power analysis, using binomial probabilities was used to determine the minimum sample size required to accept the outcome of a statistical test with a particular level of confidence. If sample size is too low, the experiment will lack the precision and if sample size is too large, time and resources will be wasted, often for minimal gain. We estimated the minimum sample size for different theoretical frequencies at three different confidence intervals (α) and power values (1−β) (Zar, 1999; McPherson et al., 2008). The theoretical frequencies at the different α values served as the null hypotheses levels used to declare significance of gene flow. The observed frequencies from the study were then compared with the theoretical frequencies and if the observed frequencies were above the theoretical frequency, there is significant gene flow. Sample size was calculated with the following equation using software R (Chambers, 2008):
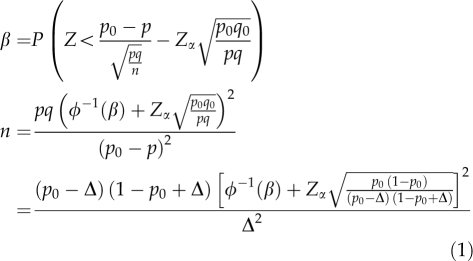 |
where, n, minimum number of seeds required; β, power; Z, random variable following N(0, 1); q, 1−p; q0=1−p0; p0, true value in null hypothesis-hypothesized parameter; p, theoretical frequency of gene flow; Δ, effect size (p0−p); Zα, critical value for significant level α; φ−1, anti-function of the normal curve.
Compared with other equations that also estimate the minimum sample size, such as the binomial estimator (ln(α)/ln(1−p)) (Mcpherson, 2008), the above equation has two notable advantages: (i) the researcher/worker is capable of determining the effect size (Δ), thus the sensitivity of the test and (ii) it affords the researcher the flexibility of determining (fixing) the ideal power (β) which, in the context of this study, is the probability of detecting gene flow when present. The sensitivity of the test is manipulated by varying p; and if for example high sensitivity, which is small Δ (the ability to detect low level gene flow), is required, the p is moved closer to p0. The p0 (the true value of the null hypothesis) could be a set frequency such as a threshold established by commodity importers and p the theoretical frequency of gene flow would be the frequency the researcher requires in a sample. Thus, moving p closer to p0, which shrinks Δ (making the test more sensitive) enables the researcher/worker to detect the required gene flow frequencies, which could be those set by commodity importers. Hence the two advantages, ability to manipulate the sensitivity (Δ) and fix the power (β) give certainty to the tests and therefore confidence to markets, especially in which strict restrictions to certain levels of contamination in some commodities are applied. With the binomial estimator, such certainty is lacking. The assumptions with the binomial estimator being that once the type I errors are controlled, the statistical requirements have been met and the sample size is adequate. This is not the case with the equation above, in which the sample size is derived by controlling three important parameters, α, Δ and β, which enhances degree of accuracy by reducing assumptions.
Experiments were analyzed separately and as combined, with gene flow frequency calculated as the ratio of high ALA seeds to the total seeds analyzed divided by 100. A 95% confidence interval for the mean frequency of gene flow at each distance was calculated assuming a binomial distribution (McPherson et al., 2008). The mean frequency of gene flow at each mean distance was subjected to nonlinear regression using a mixed model (PROC NLMIXED) (SAS Institute Inc., 2007). The dependent variable was estimated using a binomial distribution by fitting the data to an exponential decay function (Hanson et al., 2005),
where, p is the predicted frequency of gene flow; a the intercept; b the curve parameter; and d the mean distance from the source (m). Standard errors and 95% confidence intervals were calculated for each parameter estimate. The distances wherein frequency of gene flow was reduced by 50 and 90% were estimated from the exponential decay function following the equations (McPherson et al., 2008):
where, a is the intercept and b the slope.
To test for heterogeneity of gene flow between directional blocks for each experiment, a log-likelihood ratio test using the χ2 distribution was calculated using the –2 log-likelihood ratio provided by the regression analysis in Statistical Analysis System (SAS). In measuring low frequencies of gene flow, care was taken to reduce seed contamination. However, potential sources of sample contamination must be considered, including low frequencies (<0.00005) of high ALA seed in the low ALA seed source, and potential for cross contamination of samples during combine harvesting.
Results
Uncertainty resulting from testing random samples can be reduced by combining binomial probability distribution and power analysis to define a sampling strategy (Table 1). Seeds were screened at each distance until the minimum power of 0.8 (α=0.05) was obtained. The objective of the sampling strategy was to allow researchers to confirm the sample size necessary to enable statistical judgments that are accurate and reliable. This strategy is useful wherein large samples are available but sampling costs are not insignificant. Alternatively, a sample size can be determined a priori if the theoretical frequency of gene flow can be estimated. To further interpret the data, samples containing zero outcrossing frequencies would be described as having a frequency equal to or less than the null hypothesis (Ho) frequency level (Table 1).
Table 1. Power analysis assuming a binomial distribution to determine minimum number of seeds required to detect at least one GE seed for different frequency of gene flow; four α values (α=5, 2.5, 1.0, 0.05%); and three power (1−β) values (0.80, 0.85, 0.95).
| Hoa frequency of gene flow p(x) |
α Value (α) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
α=5% |
α=2.5% |
α=1% |
α=0.05% |
α=5% |
α=2.5% |
α=1% |
α=0.05% |
α=5% |
α=2.5% |
α=1% |
α=0.05% |
|
| Minimum sample size (n)b for power=0.80c | Minimum sample size (n)b for power=0.85c | Minimum sample size (n)b for power=0.95c | ||||||||||
| 0.01 | 1989 | 2588 | 3383 | 3985 | 2242 | 2875 | 3710 | 4340 | 3128 | 3869 | 4829 | 5544 |
| 0.005 | 3996 | 5199 | 6797 | 8007 | 4503 | 5776 | 7454 | 8719 | 6282 | 7771 | 9700 | 11136 |
| 0.0025 | 8010 | 10 422 | 13 625 | 16 051 | 9026 | 11 577 | 14 941 | 17 477 | 12 590 | 15 573 | 19 441 | 22 319 |
| 0.001 | 20 052 | 26 090 | 34 109 | 40 183 | 22 595 | 28 980 | 37 402 | 43 750 | 31 513 | 38 982 | 48 664 | 55 870 |
| 0.0005 | 40 122 | 52 204 | 68 248 | 80 402 | 45 209 | 57 985 | 74 837 | 87 540 | 63 051 | 77 995 | 97 368 | 111 788 |
| 0.00025 | 80 262 | 104 431 | 136 528 | 160 841 | 90 437 | 115 996 | 149 707 | 175 120 | 126 127 | 156 023 | 194 777 | 223 624 |
| 0.0001 | 200 681 | 261 114 | 341 367 | 402 159 | 226 123 | 290 028 | 374 319 | 437 860 | 315 356 | 390 106 | 487 005 | 559 132 |
| 0.00005 | 401 379 | 522 251 | 682 765 | 804 355 | 452 266 | 580 082 | 748 671 | 875 759 | 630 737 | 780 244 | 974 051 | 1 118 312 |
| 0.000025 | 802 776 | 1 044 526 | 1 365 561 | 1 608 746 | 904 551 | 1 160 190 | 1 497 376 | 1 751 558 | 1 261 500 | 1 560 519 | 1 948 144 | 2 236 672 |
| 0.00001 | 2 006 968 | 2 611 350 | 3 413 949 | 4 021 921 | 2 261 406 | 2 900 514 | 3 743 490 | 4 378 955 | 3 153 787 | 3 901 347 | 4 870 422 | 5 591 751 |
| 0.000005 | 4 013 954 | 5 222 724 | 6 827 930 | 8 043 879 | 4 522 833 | 5 801 054 | 7 487 013 | 8 757 949 | 6 307 600 | 7 802 726 | 9 740 884 | 11 183 549 |
| 0.0000025 | 8 027 925 | 10 445 471 | 13 655 891 | 16 087 794 | 9 045 685 | 11 602 134 | 14 974 060 | 17 515 937 | 12 615 225 | 15 605 485 | 19 481 810 | 22 367 146 |
Abbreviation: GE, genetically engineered.
Null hypothesis: theoretical value of frequency of gene flow.
Minimum sample size required to detect one or more GE seeds at a given theoretical frequency of gene flow and different values of α and power (1−β). Null hypothesis that the frequency X⩾P is rejected at a given α value, when no GE seeds were found in a sample size of n or greater.
Value of power to ascertain how much power (1−β) would be available for a given sample size and confidence interval.
To evaluate differences in the gene flow frequency among directional blocks at each experimental site, the maximum likelihood ratio was calculated (data not shown, see Supplementary material). Results suggest that frequency of gene flow between various blocks was relatively similar; indicating that wind or wind direction did not have a significant role in pollen dissemination from source to receptor plant population. Regular observations recorded during the growing season suggest that flowering of recipient and donor flax cultivars were synchronous and uniform at all the sites and years (data not shown). Therefore, it is unlikely that flowering time or flower density influenced gene flow frequency at different experimental sites.
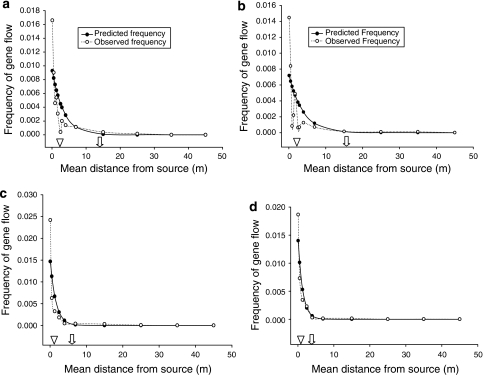
Gene flow rates in flax followed an exponential decline with distance in these small plot experiments. However, maximum gene flow and gene flow distance was affected by differences between sites and years (Figure 2). Gene flow was lower in 2006 than 2007. At EdRS in 2006, a total 647 hybrids were identified by TBA testing 769 600 seeds (Table 2, Figure 2). A maximum gene flow of 0.0166% was observed at the minimum mean distance of 0.1 m from the pollen source. At 35 and 45 m, no positive seeds were detected in the sample, and therefore gene flow was <0.00005. At ElRS in 2006, the frequency of gene flow immediately adjacent to the source was 0.0145 (Table 3, Figure 2). Gene flow of a 0.00011 was detected at 35 m but was <0.00005 at 45 m. In some instances, the sample size exceeded the minimum requirements.
Figure 2.
Pollen-mediated gene flow in flax at various locations and years (a) EdRS, 2006; (b) EIRS, 2006; (c) EdRS, 2007; and (d) EIRS, 2007. The triangle indicates the distance, in which 50% (O50) reduction in the frequency of gene flow and the arrow indicates the distance in which 90% (O90) reduction in gene flow from the pollen source (m).
Table 2. Frequency of flax gene flow from 0.1–45 m distance from the pollen source at EdRS in 2006.
| Mean distancea (m) | Seeds screenedb | Seeds with high ALA | Actual frequency of gene flow in sample | Ho frequency of gene flow p(x) | Power, (1−β); xα=0.05%c |
|---|---|---|---|---|---|
| 0.1 | 14 400 | 239 | 0.016597 | 0.005 | 0.95 |
| 0.5 | 14 400 | 130 | 0.009028 | 0.005 | 0.95 |
| 0.9 | 7200 | 33 | 0.004583 | 0.01 | 0.95 |
| 1.3 | 2400 | 13 | 0.005417 | 0.01d | <0.8 |
| 1.7 | 12 000 | 37 | 0.003083 | 0.005 | 0.95 |
| 2.5 | 4800 | 2 | 0.000417 | 0.005d | <0.8 |
| 2.9 | 14 400 | 29 | 0.002014 | 0.005 | 0.95 |
| 4 | 26 400 | 37 | 0.001402 | 0.0025 | 0.95 |
| 7 | 40 800 | 46 | 0.001127 | 0.0025 | 0.95 |
| 15 | 105 600 | 41 | 0.000388 | 0.001 | 0.95 |
| 25 | 244 800 | 40 | 0.000163 | 0.00005 | 0.95 |
| 35 | 138 400 | 0 | 0.000000 | 0.00005 | 0.95 |
| 45 | 144 000 | 0 | 0.000000 | 0.00005 | 0.95 |
Abbreviations: ALA, α-linolenic acid; EdRS, Edmonton Research Station.
Mean distance from the pollen source was used in analysis for all observations.
Total number of seeds screened from all (eight) blocks for a specific distance from the pollen source.
Value of power (1−β) was calculated for 95% confidence interval (α=5%) using equation (1) (see text).
α=5%.
Frequency of gene flow was calculated from the number of seeds screened and the number of high ALA seeds. To determine gene flow significance, the null hypothesis frequency, based on the sample size and power, was compared with the sample frequency.
Table 3. Frequency of flax gene flow from 0.1–45 m distance from the pollen source at ElRS in 2006.
| Mean distancea (m) | Seeds screenedb | Seeds with high ALA | Actual frequency of gene flow in sample | Ho frequency of gene flow p(x) | Power, (1−β); α=0.05%c |
|---|---|---|---|---|---|
| 0.1 | 24 000 | 347 | 0.014458 | 0.005 | 0.95 |
| 0.5 | 24 000 | 201 | 0.008375 | 0.005 | 0.95 |
| 0.9 | 4800 | 4 | 0.000833 | 0.005 | <0.8 |
| 1.3 | 9600 | 21 | 0.002188 | 0.01 | 0.95 |
| 1.7 | 9600 | 47 | 0.004896 | 0.01 | 0.95 |
| 2.5 | 9600 | 6 | 0.000625 | 0.005d | <0.8 |
| 2.9 | 14 400 | 10 | 0.000694 | 0.005d | <0.8 |
| 4 | 48 000 | 60 | 0.00125 | 0.025 | 0.95 |
| 7 | 52 800 | 37 | 0.000701 | 0.001 | 0.85 |
| 15 | 148 800 | 22 | 0.000148 | 0.0005 | 0.95 |
| 25 | 168 000 | 17 | 0.000101 | 0.0005 | 0.95 |
| 35 | 100 800 | 11 | 0.000109 | 0.001 | 0.95 |
| 45 | 134 400 | 0 | 0.000000 | 0.0005 | 0.95 |
Abbreviations: ALA, α-linolenic acid; ElRS, Ellerslie Research Station.
Frequency of gene flow was calculated from the number of seeds screened and the number of high ALA seeds. To determine gene flow significance, the null hypothesis frequency, based on the sample size and power, was compared with the sample frequency.
Mean distance from the pollen source was used in analysis for all observations.
Total number of seeds screened from all (eight) blocks for a specific distance from the pollen source.
Value of power (1−β) was calculated for 95% confidence interval (α=5%) using equation (1) (see text) and Table 1.
α=5%.
In 2007 at EdRS, a maximum frequency of gene flow 0.0242 was recorded at the mean distance of 0.1 m from the pollen source (Table 4; Figure 2). Gene flow of 0.000063 was detected as far as 25 m distance at this site in 2007 with 13 seeds with high ALA out of 460 600 seeds screened to achieve the power value of 0.85 (Table 4). Gene flow at 35 and 45 m was <0.00005. Immediately adjacent to the pollen source, at ElRS gene flow was 0.018646 (Table 5; Figure 2). Some rare gene flow was observed (0.000028) at a power value of 0.85 after screening 476 800 seeds at 35 m distance from the pollen source at ElRS site in 2007.
Table 4. Frequency of flax gene flow from 0.1–45 m distance from the pollen source at EdRS in 2007.
| Mean distancea (m) | Seeds screenedb | Seeds with high ALA | Actual frequency of gene flow in sample | Ho Frequency of gene flow p(x) | Powerc, (1−β); α=0.05% |
|---|---|---|---|---|---|
| 0.1 | 9600 | 232 | 0.024167 | 0.01 | 0.95 |
| 0.5 | 9600 | 60 | 0.00625 | 0.01 | 0.95 |
| 1.3 | 9600 | 31 | 0.003229 | 0.01 | 0.95 |
| 2.5 | 14 400 | 26 | 0.001806 | 0.005 | 0.95 |
| 4 | 91 200 | 40 | 0.000439 | 0.001 | 0.95 |
| 7 | 115 200 | 44 | 0.000382 | 0.0005 | 0.95 |
| 15 | 96 000 | 29 | 0.000302 | 0.0005 | 0.85 |
| 25 | 460 600 | 13 | 0.000063 | 0.0001 | 0.85 |
| 35 | 176 800 | 0 | 0 | 0.00005 | 0.95 |
| 45 | 176 800 | 0 | 0 | 0.00005 | 0.95 |
Abbreviations: ALA, α-linolenic acid; EdRS, Edmonton Research Station.
Frequency of gene flow was calculated from the number of seeds screened and the number of high ALA seeds. To determine gene flow significance, the null hypothesis frequency, based on the sample size and power, was compared with the sample frequency.
Mean distance from the pollen source was used in analysis for all observations.
Total number of seeds screened from all (eight) blocks for a specific distance from the pollen source.
Value of power (1−β) was calculated for 95% confidence interval (α=5%) using equation (1) (see text) and Table 1.
Table 5. Frequency of flax gene flow from 0.1–45 m distance from the pollen source at ElRS in 2007.
| Mean distancea (m) | Seeds screenedb | Seeds with high ALA | Actual frequency of gene flow in sample | Ho frequency of gene flow p(x) | Power, (1−β); α = 0.05%c |
|---|---|---|---|---|---|
| 0.1 | 9600 | 179 | 0.018646 | 0.01 | 0.95 |
| 0.5 | 9600 | 70 | 0.007292 | 0.01 | 0.95 |
| 1.3 | 9600 | 33 | 0.003438 | 0.01 | 0.95 |
| 2.5 | 9600 | 22 | 0.002292 | 0.001 | 0.85 |
| 4 | 100 800 | 27 | 0.000268 | 0.001 | 0.95 |
| 7 | 76 800 | 11 | 0.000143 | 0.001 | 0.95 |
| 15 | 91 200 | 13 | 0.000143 | 0.001 | 0.95 |
| 25 | 176 800 | 0 | 0 | 0.0005 | 0.95 |
| 35 | 476 800 | 17 | 0.000028 | 0.00001 | 0.85 |
| 45 | 170 000 | 0 | 0 | 0.00005 | 0.95 |
Abbreviations: ALA, α-linolenic acid; ElRS, Ellerslie Research Station.
Frequency of gene flow was calculated from the number of seeds screened and the number of high ALA seeds. To determine gene flow significance, the null hypothesis frequency, based on the sample size and power, was compared with the sample frequency.
Mean distance from the pollen source was used in analysis for all observations.
Total number of seeds screened from all (eight) blocks for a specific distance from the pollen source.
Value of power (1−β) was calculated for 95% confidence interval (α=5%) using equation (1) (see text) and Table 1.
The exponential decay curve models for each site and year suggest that differences between locations are less important than differences between years, presumably because of the influence of weather on gene flow (Figure 2). The distance wherein gene flow was decreased by 50% ranged from <1 m (0.85 m at ElRS in 2007 to a maximum of over 2 m (2.6 m at ElRS in 2006) (Table 6). The variability of gene flow increased with distance from the source and this was reflected in increase in confidence intervals at the distance that 90% of the gene flow occurred (O90). The O90 ranged from 5.7 m at ElRS in 2007 to 17.5 m at ElRS in 2006. Average across all locations and years, the O50 was 1.61 m and the O90 was 5.37 m (Figure 2). Although average values from small plot research are valuable, a more conservative approach of using worse case scenarios should be taken when regulations for isolation distances are established.
Table 6. Parameter estimates and the distances where 50% (O50) and 90% (O90) reduction in frequency of gene flow occurred with their respective standard errors and confidence intervals from the regression analysis at various sites in 2006 and 2007.
| Experiment | Parametera | Estimateb | Standard error | Dfc |
95% Confidence interval |
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Edmonton, 2006 | a | 0.009570 | 0.03162 | 60 | −0.05368 | 0.07282 |
| b | 0.3024 | 1.1767 | 60 | −2.0514 | 2.6563 | |
| O50 | 2.29191 | 8.91769 | 60 | −15.5461 | 20.1299 | |
| O90 | 15.2271 | 59.2478 | 60 | −103.286 | 133.74 | |
| Ellerslie, 2006 | a | 0.007363 | 0.02665 | 63 | −0.04589 | 0.06062 |
| b | 0.2622 | 1.1557 | 63 | −2.0473 | 2.5717 | |
| O50 | 2.64364 | 11.6528 | 63 | −20.6427 | 25.9300 | |
| O90 | 17.5640 | 77.4198 | 63 | −137.147 | 172.275 | |
| Edmonton, 2007 | a | 0.01566 | 0.05022 | 64 | −0.08466 | 0.1160 |
| b | 0.6588 | 2.1599 | 64 | −3.6561 | 4.9737 | |
| O50 | 1.05209 | 3.44916 | 64 | −5.83840 | 7.94258 | |
| O90 | 6.98994 | 22.9157 | 64 | −38.7895 | 52.7694 | |
| Ellerslie, 2007 | a | 0.01518 | 0.05221 | 64 | −0.08911 | 0.1195 |
| b | 0.8106 | 2.8400 | 64 | −4.8630 | 6.4842 | |
| O50 | 0.85513 | 2.99613 | 64 | −5.13032 | 6.84058 | |
| O90 | 5.68136 | 19.9058 | 64 | −34.0851 | 45.4478 | |
Parameters a and b were estimated using equation (2). The distances (O50 and O90) where gene flow was reduced by 50 and 90% were estimated using equations (3) and (4), respectively.
Estimates of the parameters for intercept (a), slope (b) and the estimates of the distances where gene flow was reduced by 50 and 90%.
Degree of freedom.
Discussion
Results indicate that maximum gene flow, ranging from 0.0144 to 0.024 was observed at the closest distance (a mean distance of 0.1 m). At 0.5 m from the source, average gene flow was reduced to 0.0077; and further reduced at 1.0 m to 0.0027. Similar results were obtained in earlier studies in short distance gene flow between flax cultivars. Henry and Tu (1928) studied the extent of gene flow in flax by growing blue and white flowered cultivars in adjacent rows. Frequency of gene flow was reduced from 0.0126 to 0.0033, when flax cultivars were grown 0.25 to 1.25 m apart, respectively. In a similar experiment, (Robinson, 1937) reported that frequency of gene flow in flax varied from 0 to 0.003, depending on the spacing between plants and climatic conditions. Kadam et al. (1938) reported the frequency of gene flow was in the range of 0–0.06, however, the average frequency was <0.03.
Flax gene flow did not differ significantly by direction (block), suggesting that wind may not have had an important role in flax gene flow. Flax is an indeterminate species and may keep flowering up to 33–63 days (Chopde and Thakre, 1969) during which time wind speed and direction may vary considerably. Early reports stated that flax flowers generally produce pollen in small quantities and it is relatively heavy and therefore, pollen dissemination by wind seems unlikely (Eyre and Smith, 1916). In several studies insect-mediated pollination was considered to determine the effect of insects on seed production or yield. Dillman (1938) reported that sticky flax pollen was primarily disseminated by honey bees and thrips. Subsequent experiments suggest that the honey bee was the most frequent visitor of flax flowers followed by bumble bees and thrips (Smirnov, 1956). Although insects visit flax flowers, the importance to gene flow has been disputed (Dillman, 1938; Gubin, 1945; Gill, 1987). Most recently, Williams et al. (1991) reported no increase in flax seed production in the presence of honey bees suggesting limited gene flow mediated by insects. In this study, pollinators were observed at each location and year but there role in promoting gene flow between flax cultivars was not quantified.
Gene flow declined exponentially with distance, with the value of O90 being <18 m at all site-year. No gene flow was detected beyond 35 m and from the power analysis we would accept the null hypothesis that the frequency of gene flow from these samples was equal to or <0.00005 at 95% confidence interval. Cultivar, environment and their interaction may have effect on the gene flow and inferences cannot be made to field scale gene flow because the size of the pollen source may affect distance of pollen movement (Matus-Cadiz et al., 2004, 2007). A simple empirical modeling approach for these initial small-scale trials was chosen rather than the alternative of dispersal kernel modeling because of its ease of interpretation (Beckie and Hall, 2008). As part of a tiered approach to quantifying gene flow, it should facilitate decisions on the feasibility of continued development of GE flax, assuming a strategy of coexistence. Given the small size of the pollen source and receptor; inferences drawn from complex mechanical or determinist models would be suspect.
Although pollen-mediated gene flow has been studied in many crops including canola (Beckie et al., 2003; Knispel et al., 2008), maize (Messeguer et al., 2006; Goggi et al., 2007; Weber et al., 2007; Mercer and Wainwright, 2008), wheat (Gaines et al., 2007; Gatford et al., 2007; Matus-Cadiz et al., 2007), soybean (Yoshimura et al., 2006), safflower (Carthamus tinctorius L.) (McPherson et al., 2008) and other crops reviewed by Mallory-Smith and Zapiola (2008), it is difficult to directly compare the gene flow between crops because of study differences. Soybean is an annual, self-pollinating, oilseed species. Gene flow in soybean was measured in previous studies in the range of 0.03–0.44% with some rare gene flow events (0.004%) observed at 14 m distance from the pollen source (Caviness, 1966). In this study, gene flow in flax was higher than reported for soybean, but lower than reported in similar scale experiments for wheat (Hanson et al., 2005), and safflower (McPherson et al., 2008). In contrast, gene flow in the obligate outcrossing, insect-pollinated alfalfa (Medicago sativa L.) was detected as high as 0.2% at 1.5 km from the pollen source using a glyphosate-resistant trait (Van Deynze et al., 2004).
When developing a GE crop, one factor to consider may be the proclivity for long-distance gene flow. Although pollen-mediated gene flow contributes to gene flow model, for highly self-pollinated species such as flax, the seed-mediated gene flow (occurring primarily during harvest loss, transportation or trade) will have an important role and therefore require careful management operations to reduce admixture.
Strategies to reduce pollen-mediated gene flow in flax
As recent as 2009, flax international trade was interrupted and market prices reduced following the detection by the EU of an unapproved GE flax in a shipment of Canadian flax. The variety CDC Triffid had not been registered to grow in Canada since 2001 and the source of contamination has not been established (Bedard, 2009). Unapproved GE crops have a zero threshold for adventitious presence in the EU (Devos et al., 2004). In Canada, approved GE crops are considered substantially equivalent to non-GE crops and segregation is not required between two cropping systems or their products (Smyth and McHughen, 2008). However, Europe has defined a 0.9% labeling threshold for the EU approved GE seeds in organic or conventional crop seeds (European Commission, 2003). Adventitious presence above this threshold triggers product labeling as originating from GE material (Devos et al., 2008). Before the commercial release of GE flax, a strategy is required to mitigate transgene movement.
The current isolation distance for certified flax seed growers is 3 m (Canadian Seed Grower's Association, 2010). Although this research does not specifically address isolation distances, we suggest isolations be increased and routine testing of certified flax seed should be implemented (Jhala et al., 2008). For coexistence of GE, conventional and organic flax, the use of non-GE buffer zone around GE flax fields and discarding buffer zones around organic flax fields after flowering should be considered (Jhala et al., 2009). The maximum gene flow recorded in this research between cultivars was <3% immediately adjacent to the source and 90% of gene flow occurred within 6 to 18 m. In combination with harvest blending of fields, the adventitious presence through pollen-mediated gene flow could be reduced but not eliminated. Recent GENESYS modeling on gene flow containment in canola (Brassica napus L.) suggest that buffer zones are more effective than isolation zones in reducing harvest admixture because they increased the distance between GE and non-GE fields and diminished the proportion of GE pollen in the total pollen cloud (Colbach et al., 2009). These measures, in addition to practices designed to reduce seed-mediated gene flow (Jhala et al., 2010; Dexter et al., 2010a) may be useful to develop the best management practices to grow GE flax in western Canada.
Conclusions
Pollen-mediated gene flow alone should not prevent the coexistence of conventional, organic and GE flax if the novel trait is approved in the EU and consumers will accept a non-zero threshold. However, pollen is only one possible source of adventitious presence. Other sources that must be mitigated include seed-mediated gene flow through certified flax seed; volunteer flax and inadvertent mixing of products within the transportation system. Risk mitigation through agronomic practices, product testing and channeled production, all impose costs that must be balanced by the benefits of GE varieties to flax growers.
Acknowledgments
We would like to acknowledge the financial contribution of the Alberta Ingenuity Graduate Student Scholarship Fund, Alberta Advanced Education and Technology, University of Alberta and Alberta Agriculture and Rural Development. We thank Dr Paul Dribnenki and Susan McEachern of Vittera, Canada for providing breeder seeds of flax cultivars used in this study and also for providing training to first author for the TBA test. Additional technical support provided by Marc McPherson, Cara Kozak, Judy Irving, Jaime Crowe, Lisa Raatz, Vanessa Kavanagh, Alex Fedko, Cam Stevenson and Susan Jess is appreciated. We thank Debby Topinka and Alexander Pswarayi for their technical assistance to prepare this paper.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Bausell RB, Li YF. Power Analysis for Experimental Research: A Practical Guide for the Biological, Medical and Social Sciences. Cambridge University Press: West Nyack, NY; 2002. [Google Scholar]
- Beckie HJ, Hall LM. Simple to complex: modelling crop pollen-mediated gene flow. Plant Sci. 2008;175:615–628. [Google Scholar]
- Beckie HJ, Warwick SI, Nair H, Seguin-Swartz G. Gene flow in commercial fields of herbicide-resistant canola (Brassica napus L.) Ecol Appl. 2003;13:1276–1294. [Google Scholar]
- Bedard D.2009Deregistered GM flax pops up in Europe [on-line]Available at http://www.albertafarmexpress.ca/issues/ISArticle.asp?aid=1000340642&PC =FBC&issue=0102009 (accessed 18 September 2009)Alberta Business Communication: Winnipeg, MB, Canada [Google Scholar]
- Bhatty RS, Rowland GG. Measurement of alpha-linolenic acid in the development of edible oil flax. J Am Oil Chem Soc. 1990;67:364–367. [Google Scholar]
- Bolley HL. Indications of the transmission of an acquired character in flax. Science. 1927;66:301–302. doi: 10.1126/science.66.1709.301. [DOI] [PubMed] [Google Scholar]
- Canadian Seed Grower's Association 2010Foundation, registered and certified production of barley, buckwheat, canaryseed, durum, flax, oat, rye, triticale and wheat [on-line]Available at http://www.seedgrowers.ca/pdfs/Circular%206_2010/Circ6-SECTION %2002-ENGLISH_Rev01-5_20100126.pdf Canadian Seed Grower's Association: Ottawa, ON, Canada [Google Scholar]
- Caviness CE. Estimates of natural cross-pollination in Jackson soybeans in Arkansas. Crop Sci. 1966;6:211–212. [Google Scholar]
- Chambers JM.2008Software for data analysis: programming with R Springer: Secaucus, NJ; 498 p. [Google Scholar]
- Chaix G, Gerber S, Razafimaharo V, Vigneron P, Verhaegen D, Hamon S. Gene flow estimation with microsatellites in a Malagasy seed orchard of Eucalyptus grandis. Theor Appl Genet. 2003;107:705–712. doi: 10.1007/s00122-003-1294-0. [DOI] [PubMed] [Google Scholar]
- Chopde PR, Thakre JT. Flowering pattern of linseed (Linum usitatissimum L.) Nagpur Agric Coll Mag. 1969;41:36–39. [Google Scholar]
- Colbach N, Devaux C, Angevin F. Comparative study of the efficiency of buffer zones and harvest discarding on gene flow containment in oilseed rape. A modelling approach. Eur J Agron. 2009;30:187–198. [Google Scholar]
- Dahle LK, Hill EG, Holman RT. The thiobarbituric acid reaction and the autoxidations of polyunsaturated fatty acid methyl esters. Arch Biochem Biophys. 1962;98:253–261. doi: 10.1016/0003-9861(62)90181-9. [DOI] [PubMed] [Google Scholar]
- Demeke T, Perry DJ, Scowcroft WR. Adventitious presence of GMOs: scientific overview for Canadian grains. Can J Plant Sci. 2006;86:1–23. [Google Scholar]
- Devos Y, Reheul D, de Schrijver A, Cors F, Moens W. Management of herbicide-tolerant oilseed rape in Europe: a case study on minimizing vertical gene flow. Environ Biosafety Res. 2004;3:135–148. doi: 10.1051/ebr:2005001. [DOI] [PubMed] [Google Scholar]
- Devos Y, Sanvido O, Demont M. Coexistence in the EU-Return of the moratorium on GM crops. Nat Biotechnol. 2008;26:1223–1225. doi: 10.1038/nbt1108-1223. [DOI] [PubMed] [Google Scholar]
- Dexter JE, Jhala AJ, Hills MJ, Yang RC, Topinka KC, Weselake RJ, et al. Quantification and mitigation of adventitious presence of volunteer flax (Linum usitatissimum L.) in wheat (Triticum aestivum L.) Weed Sci. 2010a;58:80–88. [Google Scholar]
- Dexter JE, Jhala AJ, Hills MJ, Yang RC, Topinka KC, Weselake RJ, et al. 2010bEmergence and persistence of volunteer flax in western Canadian cropping systems Agron J 102(in press). [Google Scholar]
- Dillman AC. Natural crossing in flax. Am Soc Agron J. 1938;30:279–286. [Google Scholar]
- Dillman AC, Goar LG.1937Flaxseed production in the far western statesRep. Farmer's Bulletin no. 1792.United States Department of Agriculture: Beltsville, MD [Google Scholar]
- Dillman AC, Stoa TE.1935Flaxseed production in the north central statesFarmer's bul. 1747, pp.18.United States Department of Agriculture: Beltsville, MD [Google Scholar]
- Dje Y, Heuertz M, Ater M, Lefebvre C, Vekemans X. In situ estimation of outcrossing rate in sorghum landraces using microsatellite markers. Euphytica. 2004;138:205–212. [Google Scholar]
- Dong JZ, McHughen A. Transgenic flax plants from Agrobacterium mediated transformation: incidence of chimeric regenerants and inheritance of transgenic plants. Plant Sci. 1993;88:61–71. [Google Scholar]
- Dribnenki JCP, McEachern SF, Chen Y, Green AG, Rashid KY. Linola 2047 low linolenic flax. Can J Plant Sci. 2003;83:81–83. [Google Scholar]
- European Commission 2003Commission recommendation on guidelines for the development of national strategies and best management practices to ensure the co-existence of genetically modified crops with conventional and organic farming [on-line]Available at http://ec.europa.eu/agriculture/coexistence/index_en.htm (accessed 6 November 2007).
- Eyre JV, Smith G. Some notes on the Linaceae — the cross pollination of flax. J Genet. 1916;5:189–197. [Google Scholar]
- Gaines TA, Byrne PF, Westra P, Nissen SJ, Henry WB, Shaner DL, et al. An empirically derived model of field-scale gene flow in winter wheat. Crop Sci. 2007;47:2308–2316. [Google Scholar]
- Gatford KT, Basri Z, Edlington J, Lloyd J, Qureshi JA, Brettell R, et al. Gene flow from transgenic wheat and barley under field conditions. Euphytica. 2007;151:383–391. [Google Scholar]
- Gill KS.(ed.) (1987Linseed Indian Council of Agricultural Research: New Delhi, India; pp 386. [Google Scholar]
- Goggi AS, Lopez-Sanchez H, Caragea P, Westgate M, Arritt R, Clark CA. Gene flow in maize fields with different local pollen densities. Int J Biometeorol. 2007;51:493–503. doi: 10.1007/s00484-007-0088-5. [DOI] [PubMed] [Google Scholar]
- Graham RJD, Roy SC. Linseed (L. usitatissimum) hybrids. Agri J India. 1924;1:28–31. [Google Scholar]
- Gubin AF. Cross-pollination of fiber flax. Bee World. 1945;26:30–31. [Google Scholar]
- Halfhill MD, Millwood RJ, Weissinger AK, Warwick SI, Stewart CN. Additive transgene expression and genetic introgression in multiple green-fluorescent protein transgenic crop x weed hybrid generations. Theor Appl Genet. 2003;107:1533–1540. doi: 10.1007/s00122-003-1397-7. [DOI] [PubMed] [Google Scholar]
- Hanson BD, Mallory-Smith CA, Shafii B, Thill DC, Zemetra RS. Pollen-mediated gene flow from blue aleurone wheat to other wheat cultivars. Crop Sci. 2005;45:1610–1617. [Google Scholar]
- Henry AW, Tu C. Natural crossing in flax. J Am Soc Agron. 1928;20:1183–1192. [Google Scholar]
- Hills MJ, Hall LM, Messenger DF, Graf RJ, Beres BL. Evaluation of crossability between triticale (X Triticosecale Wittmack) and common wheat, durum wheat and rye. Environ Biosafety Res. 2007;6:249–257. doi: 10.1051/ebr:2007046. [DOI] [PubMed] [Google Scholar]
- Howard A, Howard GLC, Khan AR. The economic significance of natural cross fertilization in India. Mem Dep Agric India (Bot Ser. 1919;3:281–230. [Google Scholar]
- Hubner P, Waiblinger HU, Pietsch K, Brodmann P. Validation of PCR methods for quantitation of genetically modified plants in food. J AOAC Int. 2001;84:1855–1864. [PubMed] [Google Scholar]
- Isagi Y, Kanazashi T, Suzuki W, Tanaka H, Abe T. Highly variable pollination patterns in Magnolia obovata revealed by microsatellite paternity analysis. Int J Plant Sci. 2004;165:1047–1053. [Google Scholar]
- Jhala AJ, Hall LM, Hall JC. Potential hybridization of flax with wild and weedy relatives: An avenue for movement of engineered genes. Crop Sci. 2008;48:825–840. [Google Scholar]
- Jhala AJ, Raatz L, Dexter JE, Hall LM.2010Adventitious presence: Volunteer flax (Linum usitatissimum L.) in herbicide resistant canola (Brassica napus L.) Weed Technol 24(in press). [Google Scholar]
- Jhala AJ, Weselake RJ, Hall LM. Genetically engineered flax (Linum usitatissimum L.): potential benefits, risks, regulations and mitigation of transgene movement. Crop Sci. 2009;49:1943–1954. [Google Scholar]
- Joshi DP. Extent of cross pollination in linseed under Punjab conditions. J Res Punjab Agric Univ. 1994;31:167–168. [Google Scholar]
- Kadam BS, Kulkarni RK, Patel SM. Natural cross fertilization in linseed. Madras Agril J. 1938;26:3–9. [Google Scholar]
- Kenaschuk EO, Rashid KY. AC McDuff flax. Can J Plant Sci. 1994;74:815–816. [Google Scholar]
- Knispel AL, McLachlan SM, van Acker RC, Friesen LF. Gene flow and multiple herbicide resistance in escaped canola populations. Weed Sci. 2008;56:72–80. [Google Scholar]
- Mallory-Smith C, Zapiola M. Gene flow from glyphosate-resistant crops. Pest Manag Sci. 2008;64:428–440. doi: 10.1002/ps.1517. [DOI] [PubMed] [Google Scholar]
- Matus-Cadiz MA, Hucl P, Dupuis B. Pollen-mediated gene flow in wheat at the commercial scale. Crop Sci. 2007;47:573–581. [Google Scholar]
- Matus-Cadiz MA, Hucl P, Horak MJ, Blomquist LK. Gene flow in wheat at the field scale. Crop Sci. 2004;44:718–727. [Google Scholar]
- McGregor DI. A rapid and sensitive spot test for linolenic acid levels in rapeseed. Can J Plant Sci. 1974;54:211–213. [Google Scholar]
- McHughen A.2002Transgenic plants and crops Transgenic Linseed FlaxIn Khachatorians GG, McHughen A., Scorza R., Nip WK., Hui YH (eds).Marcel Dekker, Inc.: New York; pp 899. [Google Scholar]
- McHughen A, Holm FA. Development and preliminary field testing of a glufosinate-ammonium tolerant transgenic flax. Can J Plant Sci. 1995;75:117–120. [Google Scholar]
- Mcpherson MA.2008Environmental biosafety of field scale plant molecular farming with safflower (Cartamus tinctorius L.)A Ph.D. dissertation submitted to theUniversity of Alberta: Canada [Google Scholar]
- McPherson MA, Good AG, Topinka AKC, Yang RC, McKenzie RH, Cathcart JR, et al. Pollen-mediated gene flow from transgenic safflower (Carthamus tinctorius L.) intended for plant molecular farming to conventional safflower. Environ Biosafety Res. 2008;8:19–32. doi: 10.1051/ebr/2008023. [DOI] [PubMed] [Google Scholar]
- McSheffrey SA, McHughen A, Devine MD. Characterization of transgenic sulfonylurea-resistant flax (Linumusitatissimum L.) Theor Appl Genet. 1992;84:480–486. doi: 10.1007/BF00229510. [DOI] [PubMed] [Google Scholar]
- Mercer KL, Wainwright JD. Gene flow from transgenic maize to landraces in Mexico: an analysis. Agric, Ecosyst Environ. 2008;123:109–115. [Google Scholar]
- Messeguer J, Salvia J, Palaudelmàs M, Melé E, Serra J, Peñas G, et al. Pollen-mediated gene flow in maize in real situations of coexistence. Plant Biotech J. 2006;4:633–645. doi: 10.1111/j.1467-7652.2006.00207.x. [DOI] [PubMed] [Google Scholar]
- Pla M, La Paz JL, Penas G, Garcia N, Palaudelmas M, Esteve T, et al. Assessment of real-time PCR based methods for quantification of pollen-mediated gene flow from GM to conventional maize in a field study. Transgenic Res. 2006;15:219–228. doi: 10.1007/s11248-005-4945-x. [DOI] [PubMed] [Google Scholar]
- Robinson BB. Natural cross pollination in fiber flax. J American Soc Agron. 1937;29:644–649. [Google Scholar]
- SAS Institute Inc. 2007SAS/STAT User's Guide: Statistics SAS Institute: Cary, NC; pp 956. [Google Scholar]
- Smirnov VM. Cross pollination in flax by bees. Pchlevodstvo. 1956;9:53–55. [Google Scholar]
- Smyth S, McHughen A. Regulating innovative crop technologies in Canada: the case of regulating genetically modified crops. Plant Biotechnol. 2008;6:213–225. doi: 10.1111/j.1467-7652.2007.00309.x. [DOI] [PubMed] [Google Scholar]
- Van Deynze A, Putnam DH, Orloff S, Lanini T, Canevari M, Vergas R, et al. 2004University of California, Davis, Division of Agricultural and Natural Resources: Oakland, CA; (Agricultural Biotechnology in California Series), publication no. 8153. [Google Scholar]
- Vrinten P, Hu Z, Munchinsky MA, Rowland G, Qiu X. Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiol. 2005;139:79–87. doi: 10.1104/pp.105.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber WE, Bringezu T, Broer I, Eder J, Holz F. Coexistence between GM and non-GM maize crops - tested in 2004 at the field scale level (Erprobungsanbau 2004) J Agron Crop Sci. 2007;193:79–92. [Google Scholar]
- Weekes R, Allnutt T, Boffey C, Morgan S, Bilton M, Daniels R, et al. A study of crop-to-crop gene flow using farm scale sites of fodder maize (Zea mays L.) in the UK. Transgenic Res. 2007;16:203–211. doi: 10.1007/s11248-006-9036-0. [DOI] [PubMed] [Google Scholar]
- Weekes R, Deppe C, Allnutt T, Boffey C, Morgan D, Morgan S, et al. Crop-to-crop gene flow using farm scale sites of oilseed rape (Brassica napus L.) in the UK. Transgenic Res. 2005;14:749–759. doi: 10.1007/s11248-005-0943-2. [DOI] [PubMed] [Google Scholar]
- Wijayanto T, McHughen A. Genetic transformation of Linum by particle bombardment. In Vitro Cell Development Biol-Plant. 1999;35:456–465. [Google Scholar]
- Williams IH, Simpkins JR, Martin AP. Effect of insect pollination on seed production in linseed (Linumusitatissimum L.) J Agric Sci. 1991;117:75–79. [Google Scholar]
- Wrobel M, Zebrowski J, Szopa J. Polyhydroxybutyrate synthesis in transgenic flax. J Biotech. 2004;107:41–54. doi: 10.1016/j.jbiotec.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Wrobel-Kwiatkowska M, Zebrowski J, Starzycki M, Oszmianski J, Szopa J. Engineering of PHB synthesis causes improved elastic properties of flax fibers. Biotechnol Prog. 2007;23:269–277. doi: 10.1021/bp0601948. [DOI] [PubMed] [Google Scholar]
- Yermanos DM, Kostopoulos SS. Heterostyly and incompatibility in Linum grandiflorum Desf. in the indispensable pollinators. Arkansas Agri Ext Ser (Misc Pub) 1970;127:50–54. [Google Scholar]
- Yoshimura Y, Matsuo K, Yasuda K. Gene flow from GM glyphosate-tolerant to conventional soybeans under field conditions in Japan. Environ Biosafety Res. 2006;5:169–173. doi: 10.1051/ebr:2007003. [DOI] [PubMed] [Google Scholar]
- Zar JH.1999Biostatistical Analysis Prentice-Hall, Inc.: Upper Saddle River: NJ; pp 663. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.