Abstract
Invasive hybrids and their spread dynamics pose unique opportunities to study evolutionary processes. Invasive hybrids of native Spartina foliosa and introduced S. alterniflora have expanded throughout San Francisco Bay intertidal habitats within the past 35 years by deliberate plantation and seeds floating on the tide. Our goals were to assess spatial and temporal scales of genetic structure in Spartina hybrid populations within the context of colonization history. We genotyped adult and seedling Spartina using 17 microsatellite loci and mapped their locations in three populations. All sampled seedlings were hybrids. Bayesian ordination analysis distinguished hybrid populations from parent species, clearly separated the population that originated by plantation from populations that originated naturally by seed and aligned most seedlings within each population. Population genetic structure estimated by analysis of molecular variance was substantial (FST=0.21). Temporal genetic structure among age classes varied highly between populations. At one population, the divergence between adults and 2004 seedlings was low (FST=0.02) whereas at another population this divergence was high (FST=0.26). This latter result was consistent with local recruitment of self-fertilized seed produced by only a few parental plants. We found fine-scale spatial genetic structure at distances less than ∼200 m, further supporting local seed and/or pollen dispersal. We posit a few self-fertile plants dominating local recruitment created substantial spatial genetic structure despite initial long-distance, human dispersal of hybrid Spartina through San Francisco Bay. Fine-scale genetic structure may more strongly develop when local recruits are dominated by the offspring of a few self-fertile plants.
Keywords: invasive species, hybridization, population structure, seed dispersal, microsatellites, Spartina
Introduction
The contemporary origin and spread of invasive hybrids pose unique opportunities to study evolutionary processes. Recently arisen invasive hybrid populations behave as ‘natural laboratories' for studying the processes underlying the expanding margins of a species' range (Parisod and Bonvin, 2008). Natural hybrid zones can move in space and time, and have profound consequences for both evolutionary and conservation biology (Buggs, 2007). Hybrid swarms that have recently arisen from the union of closely related native and nonnative congeners, after the latter has been introduced in the formers range, can result in extremely fit (exceeding parental species) and invasive hybrids by transgressive segregation (Rieseberg et al., 1999). To better understand the processes of invasive spread of an expanding, highly genetically diverse hybrid swarm, the underlying population genetic context and the combined structuring dynamics of hybridization history, varying trajectories of hybrid development, predominant breeding system, dispersal mechanisms and selection intensities have to be evaluated.
Hybridization admixture of two parent species can initially increase allele frequencies at all loci more than any other known type of invasion (Long, 1991). Hybridization can erode spatial genetic structure if hybrids are viable and backcross to parent species or cross with other hybrids. However, over time structure can form in hybrid populations, depending on the genetic endowment of founding populations, the rate of spatial spread, the frequency of cross-hybrid matings and backcrosses, and spatial differences in selection (Abbott, 1992; Ellstrand and Schierenbeck, 2000). Differences in female success in hybrid classes or even single plants can lead to stronger local structure (Schnabel et al., 1998; Valbuena-Carabaña et al., 2007). The amount and spatial pattern of hybrid seedling recruitment can also affect fine-scale spatial genetic structure (Schnabel et al., 1998).
Generally, spatial genetic structure created during colonization can persist on several geographical scales (Austerlitz and Garnier-Géré, 2003; Epperson et al., 2003; Williams et al., 2007). Greater spatial structure will result from an ancestry of colonists that includes few adults, resulting in genetic distinction among populations even if within-population variation is low (Levin, 1981; Slatkin, 1987). Genetic drift, where changes in allele frequencies result from chance mating events in very small populations (Primack and Kang, 1989; Husband and Schemske, 1996), and the effect of distinct environmental optima affecting fitness can also cause structure as populations diverge. Founding events are accompanied by a change from the original population allele frequency (founder effect), as the members of the new colonizing population contain a random allelic subsample that may deviate from the overall allele frequencies in its native range. By and large, the ancestry of local introductions is a biased sample of genotypes from the native range; however, multiple separate introductions and long-distance dispersal can erode the spatial structure arising early in an introduction (Lambrinos, 2004).
Density, dispersion and breeding system of reproducing adults as well as dispersal mechanisms have central roles in generating the spatial genetic structure of populations (Vekemans and Hardy, 2004). For plants, the spatial scale and patterns of seed and pollen dispersal combine in this central role. In many plant species, seed dispersal yields a long-tailed, leptokurtic, distribution, in which most seeds land near the parental plant, with only a few going far away (LeCorre et al., 1997; Donoghue, 1998; Higgins and Richardson, 1999). Species with long-distance seed dispersal, through either biotic or abiotic means, are expected to show significantly less within-population structure than species with limited dispersal (Hamrick and Nason, 1996). So, populations with common long-distance dispersal of either pollen or seeds should tend to be genetically more similar and display less spatial structure, whereas limited dispersal should result in genetically structured populations and eventually lead to pronounced population divergence.
Intertidal Spartina species and their hybrids have invaded worldwide, and their spread after human introduction to new regions shows high potential for long-distance dispersal (Strong and Ayres, 2009). Spartina seed floats for several weeks and can disperse widely on intertidal tides and currents. Seeds remain viable for 6–8 months, and germinate in the spring following late summer or autumn seed set (Huiskes et al., 1995). The spread of S. alterniflora to ca. 6000 hectares and ca. 27% of intertidal lands in Willapa Bay, WA during the ca. 110 years since its introduction was the result of floating seed carried on currents, rather than dispersion of rhizome fragments (Civille et al., 2005). Anemophilous Spartina pollination can be surprisingly local. Field experiments showed dense pollen clouds to be so diluted as to be ineffectual within 100 m, and plants isolated by even a few tens of meters produced no seed in obligately outcrossing S. alterniflora (Davis et al., 2004).
In San Francisco Bay, a rapid invasion of hybrid Spartina occurred after S. alterniflora from the Atlantic and Gulf coast of North America hybridized with the California native species, S. foliosa. S. alterniflora was deliberately planted at a former salt pond at Alameda Creek, Fremont in the 1970s (Figure 1; , 2000); it is inferred that the first hybridizations occurred there (Daehler and Strong, 1997; Ayres et al., 1999). Introgressive hybridization, potentially occurring in parallel at more than one geographic location over time, threatens the native S. foliosa with local extinction, and has left only a few nonhybrid S. alterniflora in San Francisco Bay. Whereas backcrossing produced a diverse array of advanced generation hybrid genotypes within a period of 30 years (Ayres et al., 1999). Hybrids were planted in a few sites and spread by floating seed to the shores of much of South San Francisco Bay and a few spots north of the Golden Gate (Ayres et al., 2004). The subset of genotypes that grew faster, attained larger size, greater culm density, higher pollen production and/or much higher self-fertility than either parental species have driven the invasion of San Francisco Bay (Sloop et al., 2009). Spartina hybrids have further expanded into San Francisco Bay tidal mudflats (lower in elevation than shoreline marshes), where neither parent species can persist.
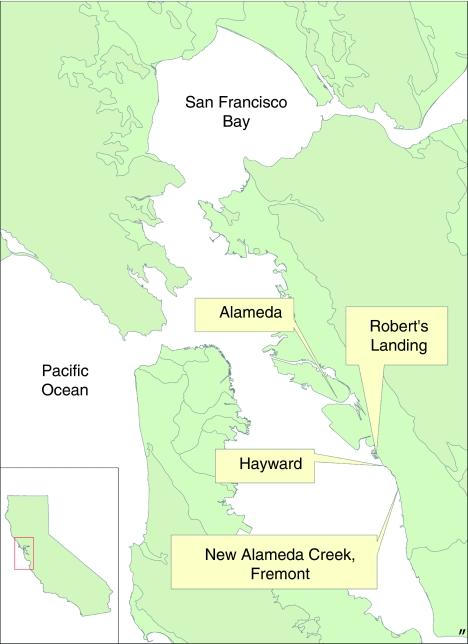
Figure 1.
Map of San Francisco Bay. Study sites: Elsie Roemer Marsh, Alameda; Robert's Landing and Hayward. (The site of the first Spartina alterniflora introduction to San Francisco Bay at Alameda Creek, Fremont is also shown.)
This study explores spatial and temporal genetic structure of expanding hybrid populations early in the invasion. We assessed allelic diversity, and spatial and temporal genetic structure of descendents of founding hybrids, and their progeny in this area. We posited that the high potential for long-distance dispersal of floating seed might preclude local structure among these hybrid populations. Conversely, locally restricted seed and pollen dispersal and plant establishment could generate spatial genetic structure. Microevolutionary development of distinct hybrid populations, in which natural selection acts upon a subset of fit hybrids, may proceed more rapidly when those populations are genetically and spatially isolated. Here we sought to determine the spatial and temporal genetic structure of hybrid populations by examining the genetic relationships between putative hybrid founders along shore and their alleged descendants in adjacent mudflats, and mudflat seedling populations across 2 successive years.
Materials and methods
Study sites
Hybrid Spartina grows from high in the salt marsh, among other terrestrial vegetation, down to open mud in the low intertidal, whereas the parental species grow only in the upper part of the gradient in San Francisco Bay (Ayres et al., 2004, 2008). We studied invading hybrid Spartina and parent species populations at three distinct geographic locations along 18 km of San Francisco Bay shoreline south of Oakland, CA during 2003 and 2004. From north to south the populations were: Elsie Roemer Marsh at Alameda (Figure 1); Robert's Landing (Figure 1) 16 km south of Alameda and Hayward (Figure 1) another 2 km further south (Sloop et al., 2009). The remainder of this 18 km of shoreline was virtually un-invaded by hybrid Spartina and without parent species at that time. We used aerial photographs and visual assessments during field surveys to locate all Spartina plants along 20 km of this shoreline, between Elsie Roemer Marsh to the north and the Cogswell Marsh in Hayward to the south. The site of the original hybridization was New Alameda Creek (Figure 1), 8 km further south of Cogswell Marsh.
The Alameda Spartina population at Elsie Roemer Marsh began in the 1970s when S. alterniflora and Spartina hybrids (S. alterniflora × S. foliosa), which were unknown at that time, were planted by the US Army Corps of Engineers; native S. foliosa also grew at Alameda at this time (US Army Corps of Engineers, 1978; Faber, 2000). These hybrids have since formed a continuous meadow along the shore at Elsie Roemer Marsh, Alameda (Figure 1; Ayres et al., 1999; Sloop et al., 2009). Seven hybrid plants colonized the tidal flats to form isolated, island-like patches in the center of the Alameda estuary.
The Robert's Landing population consisted of scattered plants on a tidal flat that was connected to an inland salt marsh meadow by a narrow tidal channel. Aerial photographs taken in 1986 and 2003 (Supplementary Figure 1) indicated that the tidal flat was colonized before 1986, whereas the inland meadow was opened to tidal action in 1994 (M Taylor, Hayward Regional Shoreline, personal communication). Hybrid Spartina was present in both of these habitats in our earliest surveys (Ayres et al., 1999).
Hayward, the third population, consisted of a few S. foliosa and hybrids along shore and some hybrid plants scattered on the mudflat. Photographic evidence indicates that this shoreline was colonized before 1986.
Genetic analysis
We studied cordgrass spatial genetic structure by assaying all hybrid Spartina adults growing on tidal flats at Alameda, Robert's Landing and Hayward (Table 1) at 17 microsatellite loci: SPAR.01, SPAR.08, SPAR.09, SPAR.10, SPAR.11, SPAR.15, SPAR.16, SPAR.17, SPAR.18, SPAR.20, SPAR.21, SPAR.23, SPAR.26, SPAR.27, SPAR.28, SPAR.29 and SPAR.33 (Blum et al., 2004; Sloop et al., 2005). We extracted DNA from leaf samples using Qiagen DNeasy Plant Mini Kits (Qiagen, Valencia, CA, USA). PCR volumes and genotyping procedures are outlined by Sloop et al. (2005).
Table 1. Population mean observed and expected heterozygosities over all 17 loci.
| Population | n | He (s.d.) | He* (s.d.) | Ho (s.d.) | FIS | N alleles |
|---|---|---|---|---|---|---|
| Parent species: | ||||||
| S. foliosa (range wide) | 50 | 0.11 (0.18) | 0.12 (0.19) | 0.09 (0.25) | NA | 1.6 |
| S. alterniflora (range wide) | 35 | 0.75 (0.12) | 0.78 (0.13) | 0.46 (0.20) | NA | 8.67 |
| Alameda S. alterniflora | 5 | 0.64 (0.12) | 0.71 (0.13) | 0.61 (0.32) | −0.15 | 4.21 |
| Alameda S. foliosa | extinct | — | — | — | — | — |
| Hybrids | ||||||
| Alameda | ||||||
| Meadow | 25 | 0.65 (0.13) | 0.66 (0.12) | 0.59 (0.22) | 0 | 4.76 |
| Tidal flat | 9 | 0.64 (0.12) | 0.69 (0.13) | 0.49 (0.22) | 0.21* | 4.29 |
| Seedlings 2003 | 33 | 0.69 (0.10) | 0.71 (0.10) | 0.54 (0.22) | 0.07* | 5.24 |
| Seedlings 2004 | 67 | 0.69 (0.09) | 0.70 (0.09) | 0.52 (0.20) | 0.09* | 5.65 |
| Robert's Landing | ||||||
| Meadow | 30 | 0.60 (0.17) | 0.61 (0.18) | 0.52 (0.23) | 0.03 | 5.06 |
| Tidal flat | 17 | 0.66 (0.12) | 0.68 (0.13) | 0.40 (0.22) | 0.39* | 4.41 |
| Seedlings 2003 | 42 | 0.60 (0.13) | 0.61 (0.14) | 0.41 (0.18) | 0.11* | 4.59 |
| Seedlings 2004 | 54 | 0.50 (0.14) | 0.51 (0.15) | 0.30 (0.15) | 0.31* | 5.47 |
| Hayward | ||||||
| Tidal flat | 24 | 0.61 (0.11) | 0.62 (0.11) | 0.46 (0.20) | −0.05 | 5.18 |
| Seedlings 2003 | 32 | 0.62 (0.09) | 0.65 (0.10) | 0.36 (0.16) | 0.17* | 3.65 |
| Seedlings 2004 | 69 | 0.54 (0.13) | 0.54 (0.13) | 0.43 (0.19) | 0.12* | 5.12 |
He* calculated without bias (Nei, 1978); n= number of plants sampled, s.d. = standard deviation.
*Significant population specific FIS indices (1023 permutations) at P<0.05; N alleles = mean number of alleles per locus.
We also collected samples from 25 hybrid adults from the adjacent marsh population at Elsie Roemer marsh at Alameda, and 30 adults from the inland Spartina hybrid marsh at Robert's Landing. In addition, we assayed seedlings (distinguished from adults by their size of <1–10 cm, by having only one or a few stems, and occasionally by the presence of a seed coat) growing on tidal flats at these sites. In 2003, we sampled by haphazardly collecting leaf samples from about 11% of all seedlings (n=109) over the full spatial extent of the seedling cloud per tidal flat site (Table 1), whereas in 2004, we collected seedling DNA samples by walking two parallel transects along the length of the shoreline tidal flats, collecting a sample every 20 paces, resulting in 4% of all seedlings (n=190) (Table 1). We used Global Positioning System loggers (Trimble GeoExplorer 3.0, Sunnyvale, CA, USA) to map the geographic locations of all genetically surveyed adults and seedlings.
We further examined the parental species throughout their range: S. foliosa, northern California: 10 plants from the Hayward shoreline (Figure 1) and 29 plants from six sites in San Francisco Bay (other than Alameda, Robert's Landing and Hayward), Bolinas and Tomales Bay; S. foliosa, southern California: Chula Vista, and northern Mexico (Tijuana); S. alterniflora: 27 plants from six Atlantic Coast/Gulf Coast locations (New York, Georgia, Florida, Alabama and Mississippi), and three sites in Willapa Bay, WA (Palix River, Tower Slough and Peninsula), and 5 plants growing on the shoreline at Elsie Roemer Marsh at Alameda (Table 1). S. alterniflora is rare in San Francisco Bay and occurs only at a few sites. A previous analysis using randomly amplified polymorphic DNA (D Ayres, unpublished data) helped to identify S. alterniflora plants in San Francisco Bay. Species-specific loci were developed from genetic surveys of 50 individuals of S. alterniflora and 50 individuals of S. foliosa obtained from throughout their ranges. Species-specific alleles had to be found in at least 10% or greater of sampled individuals per species, and had to be entirely absent in all sampled individuals of the other species. Of 17 microsatellite loci, 9 contained 15 S. foliosa-specific alleles (not found in S. alterniflora, yet introgressed into San Francisco Bay hybrid Spartina), and for 15 loci, 1–7 alleles per locus were found only in hybrid individuals. We found 39 S. alterniflora species-specific alleles at 15 loci. We calculated allelic frequencies, and observed (Ho) and expected (He) heterozygosity using GENETIX (Belkhir et al., 1996–2004). We carried out Hardy–Weinberg (H-W) exact tests (using Markov chain default settings) for all eight adult populations using GENEPOP (Raymond and Rousset, 1995, http://genepop.curtin.edu.au/).
Bayesian cluster analysis
To infer population structure and assign individuals to populations, we applied a model-based Bayesian clustering method to all multilocus genotypes using STRUCTURE (Pritchard et al., 2000; Falush et al., 2003; Pritchard and Wen, 2004). In this analysis, if there is genetic admixture due to hybridization, individuals are probabilistically assigned to either a single cluster (the population of origin), or more than one cluster (the parental populations, Falush et al., 2003). The program assumes the neutral unlinked markers to be in Hardy–Weinberg equilibrium (HWE) and linkage equilibrium and that recent hybridization or migration would likely produce departures from these equilibria. STRUCTURE identifies the K unknown populations (genetic clusters) of individuals' origin and concurrently allocates all individuals to populations, giving their 90% confidence intervals. STRUCTURE was run using the ‘admixture model' and correlated allele frequencies, with a burn-in period of 10 000, followed by 100 000 iterations. To detect the true number of clusters (K), we followed the graphical methods and algorithms outlined by Evanno et al. (2005). Under the assumption that the sampled plants belong to an unknown number of K genetically distinct clusters, we used priors from 2 to 14 to estimate the average posterior probability values for K (log-likelihood; ln L) for 20 runs each. This method established K=4 as the best value of K for our data, distinguishing two parent species clusters and two Spartina hybrid groups.
Analysis of molecular variance
To quantify hybrid population structure, we used analysis of molecular variance using Arlequin 3.11 (Excoffier et al., 2005). To determine the spatial scale of genetic structure, we compared hybrid populations by geographic location, and to ascertain the temporal variation in genetic structure, we evaluated differences between adult descendents and two consecutive generations of seedlings in the tidal flats at each site. Using Arlequin, we further calculated population pairwise FST values and population specific FIS indices across all loci.
Temporal genetic structure
Hybrid ‘descendents' from previously established hybrid ‘founders' were determined using aerial photographs (Supplementary Figure 1; digital orthophoto quadrangles dating from 1993 obtained from the USGS; and historical (1986) and contemporary aerial photos (2003), obtained from the Invasive Spartina Project), historical documents (USACE, 1978) and the personal observation of managers of the East Bay Regional Parks District (M Taylor, Park Ranger). According to these aerial photographs and historical accounts the primary hybrid founders were meadow plants at Alameda and specific tidal flat plants at both Hayward and Robert's Landing. The descendents are those plants that established subsequent to these initial founders; at Alameda these are the plants growing on the tidal flats; and at Robert's Landing these are the plants growing in the adjacent meadow, opened to tidal action in 1994. Seedlings are those we surveyed on tidal flats during 2003 and 2004. Because in wind pollinated Spartina, pollination distances are relatively short (Davis et al., 2004), and our previous work in this system has shown that seed dispersal is local (Sloop et al., 2009), we feel confident that many of our hypothesized descendents really are descended from the early founders.
Spatial genetic analysis
To determine fine-scale spatial genetic structure within populations, we used spatial genetic analysis. This analysis shows the relationship between geographical distance and genetic relatedness between individuals. This type of analysis is based on the mean genetic distance between pairs of individuals of the same distance class (Degen et al., 2001). Within-population distograms were produced using Spatial Genetic Software v.1.0d (Degen, 2000; Degen et al., 2001), comparing genetic distances with geographic distances, using the number of alleles/haplotypes in common over all loci, between pairs of individuals belonging to a given spatial distance class (adapted from Surles et al. (1990) and Hamrick et al. (1993)). Values exceeding the reference value indicate positive spatial genetic structure (that is, individuals are more related than by random chance), and values below the reference value show negative spatial structure (that is, individuals are less related than by random chance) (Degen, 2000). Permutation tests were used to evaluate the statistical significance of all measures, to assess deviations from a spatially random distribution. A total of 1000 permutations were performed to obtain 95% confidence intervals under the hypothesis of no spatial correlation. The data set consisted of all genetically surveyed adults and seedlings (Table 1).
Results
We found no native California cordgrass, S. foliosa, at either Alameda in the north or Robert's Landing in the south, based on the molecular determinations in this study. Introduced S. alterniflora was extremely rare at Alameda and occurred only at the upper end of the tidal gradient among hybrid Spartina and other species of plants. No S. alterniflora was detected at the other two sites. The nine large plants that established upon the tidal flat at Alameda during the decade preceding this study were all hybrids; all of the seedlings recruiting around these tidal flat plants were hybrids. At Robert's Landing all adults and seedlings were hybrid Spartina; we found no plants of the parental species. The Hayward Spartina included 10 native S. foliosa plants growing along shore and 24 large adult Spartina hybrids growing on the tidal flats well below other vegetation. All of the tested 299 seedlings at all sites were hybrid. We found neither native S. foliosa nor S. alterniflora growing on tidal flats at any of the three sites.
Genetic variation
S. foliosa was genetically depauperate throughout its range with an average of only 1.6 (±0.9) alleles per locus. Only 5 of 17 loci were polymorphic in S. foliosa, mainly indicating distinctions between San Francisco Bay and the southern California/northern Mexico group. The Hayward S. foliosa population had only two polymorphic loci. S. alterniflora had genetic diversity range roughly five times more than S. foliosa, with an average of 8.7 (±3.0) alleles per locus. San Francisco Bay S. alterniflora at Alameda had lower average allelic diversity with 4.2 (±1.4) alleles per locus, compared to the range wide S. alterniflora value. This is consistent with the introduction history of S. alterniflora into San Francisco Bay from a single Virginia S. alterniflora source (Faber, 2000). We found hybrid Spartina allelic diversity to range from 3.65 to 5.65 alleles per locus.
All adult hybrid and S. alterniflora individuals had unique genetic fingerprints (differing at >3 alleles) confirming that we did not sample clones more than once. We found 21 unique S. foliosa multilocus genotypes (in 43 samples, differing at only one or two alleles); some S. foliosa plants with the same genotypes were present at more than one sampling site, whereas genetically indistinguishable plants were spatially discrete within sites, indicating that the low genetic variation detected in this species could not resolve all individuals' genotypes. All seedlings assayed in this study were hybrids. We found no S. alterniflora during sampling at either Robert's Landing or Hayward.
All seedling populations were inbred as assessed by the inbreeding coefficient FIS (Table 1). Adult tidal flat plants at Robert's Landing were the most inbred of established plants, followed by the tidal flat adults at Alameda. Inbreeding was not detected in Spartina in the shoreline meadows at Robert's Landing and Alameda, or among Hayward adults growing on open mud below the meadows. Adults of neither hybrids nor S. foliosa at Hayward were inbred.
H-W exact tests, examining the random union of gametes, showed more loci to be in H-W equilibrium (HWE) at Alameda (Alameda meadow=77% of loci in HWE, Alameda tidal flat=94% of loci in HWE, Alameda S. alterniflora=88% of loci in HWE), and in the meadow at Robert's Landing (Robert's Landing=65% of loci in HWE), than in the tidal flats at Robert's Landing (12% of loci in HWE), and Hayward (18%). Loci in S. foliosa populations were either monomorphic (65% of loci in HWE), or deviated from H-W (35% of loci in HWE). In a mix of S. alterniflora populations from the native range on the Atlantic and Gulf coasts of North America, only 12% of tested loci showed HWE, as expected for a combination of individuals from several geographically distinct source populations.
Bayesian cluster analysis
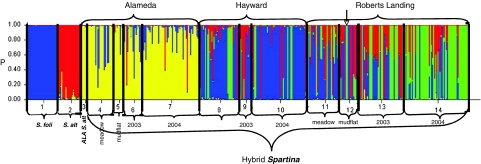
Bayesian cluster analysis clearly distinguished S. foliosa (blue color in Figure 2), native range S. alterniflora (red color in Figure 2) and the S. alterniflora plants at Alameda (yellow color in Figure 2). It also confirmed at least two distinct hybrid membership groups. The first group aligned Alameda hybrid plants (Figure 2, groups 4–7) with the Alameda S. alterniflora (Figure 2, group 3). A second group included the hybrid Spartina at Robert's Landing (Figure 2, groups 11–14) and Hayward (Figure 2, groups 8–10). At all tidal flat sites, seedling groups (Figure 2, groups 6, 7, 9, 10, 13 and 14) are most like adults at the same location, shown by similar color patterns (Figure 2, groups 4, 5, 8, 11 and 12).
Figure 2.
Bar plot estimated by STRUCTURE depicting group membership (hybrid ancestry), representing each individual as a line segment. Each segment is partitioned into K=4 shaded (colored) components, representing the individual's estimated membership coefficients in the K clusters (y axis). Numbers (x axis) represent the initial geographic/demographic groupings: 1=S. foliosa, 2=S. alterniflora, 3=Alameda S. alterniflora, 4=Alameda meadow, 5=Alameda mudflat, 6=Alameda 2003 seedlings, 7=Alameda 2004 seedlings, 8=Hayward shoreline mudflat, 9=Hayward 2003 seedlings, 10=Hayward 2004 seedlings, 11=Robert's Landing meadow, 12=Robert's Landing mudflat, 13=Robert's Landing 2003 seedlings, 14=Robert's Landing 2004 seedlings. Arrow indicates Robert's Landing plant 14 (only green in group 12). A full color version of this figure is available at the Heredity Journal online.
Analysis of molecular variance
We found a large regional genetic distinction between plants at Alameda and the Hayward/Robert's Landing populations: FST=0.21 (Table 2). Genetic distinction between neighboring Hayward and Robert's Landing adults was lower and ranged between FST=0.08 (across mudflats at both sites, Table 3), and FST=0.01 (Hayward mudflat versus Robert's Landing inland marsh, Table 3). Population structure among all three sites increased when both seedling generations from 2003 and 2004 were included: FCT=0.08 (Table 4) adults only among three populations, FCT=0.11 adults plus seedlings among three populations (Table 5). Generation differences (founders versus descendents versus seedlings) varied among populations (Table 6). There were small generational genetic differences at Alameda and Hayward (FST <0.03), whereas there were more pronounced generational differences at Robert's Landing between primary founders, and their descendents and successive seedling generations; that is, the FST between founders and 2004 seedlings was 0.26. This general trend was independent of the potential influence of unequal seedling sample sizes on FST results, as determined by a randomized resampling of the 2004 seedling data to equalize sample sizes.
Table 2. Analysis of molecular variance (average over 17 loci) among regions 1 and 2.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage variation |
|---|---|---|---|---|
| Among regions | 1 | 131.232 | 0.32044 | 16.63** |
| Among individuals within regions | 9 | 76.051 | 0.08898 | 4.62** |
| Within individuals | 895 | 1358.208 | 1.51755 | 78.75** |
| Total | 905 | 1565.491 | 1.92698 |
Region 1: North, Alameda (adult S. alterniflora and hybrid shoreline meadow, mudflat hybrid adults and 2003 and 2004 seedlings).
Region 2: South, Robert's Landing (adult hybrid shoreline meadow and mudflat, and 2003 and 2004 mudflat hybrid seedlings) and Hayward (adult S. foliosa and hybrid shoreline plants, and 2003 and 2004 mudflat hybrid adults and seedlings).
Average F-statistics over all loci. Fixation indices: FST, 0.21; FSC, 0.06; FCT, 0.17.
**Significance at P<0.0001.
Table 3. Population pairwise FST values.
| Adults RL mudflat | Adults RL meadow | Seedlings RL 2003 | Seedlings RL 2004 | Adults HAY mudflat | Adults HAY S. foliosa | Seedlings HAY 2003 | Seedlings HAY 2004 | Adults ALA mudflat | Adults ALA meadow | Adults ALA meadow S. alterniflora | Seedlings ALA 2003 | Seedlings ALA 2004 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adults RL mudflat | 0.00 | ||||||||||||
| Adults RL meadow | 0.06 | 0.00 | |||||||||||
| Seedlings RL 2003 | 0.17 | 0.04 | 0.00 | ||||||||||
| Seedlings RL 2004 | 0.26 | 0.11 | 0.03 | 0.00 | |||||||||
| Adults HAY mudflat | 0.08 | 0.01 | 0.04 | 0.11 | 0.00 | ||||||||
| Adults HAY S. foliosa | 0.17 | 0.27 | 0.41 | 0.50 | 0.28 | 0.00 | |||||||
| Seedlings HAY 2003 | 0.13 | 0.04 | 0.03 | 0.08 | 0.01 | 0.38 | 0.00 | ||||||
| Seedlings HAY 2004 | 0.14 | 0.04 | 0.05 | 0.10 | 0.03 | 0.35 | 0.01 | 0.00 | |||||
| Adults ALA mudflat | 0.27 | 0.20 | 0.22 | 0.27 | 0.18 | 0.51 | 0.21 | 0.24 | 0.00 | ||||
| Adults ALA meadow | 0.20 | 0.16 | 0.16 | 0.21 | 0.15 | 0.39 | 0.16 | 0.19 | 0.01 | 0.00 | |||
| Adults ALA meadow S. alterniflora | 0.31 | 0.25 | 0.27 | 0.31 | 0.24 | 0.61 | 0.25 | 0.28 | 0.02 | 0.04 | 0.00 | ||
| Seedlings ALA 2003 | 0.19 | 0.15 | 0.16 | 0.21 | 0.13 | 0.38 | 0.14 | 0.16 | 0.03 | 0.02 | 0.03 | 0.00 | |
| Seedlings ALA 2004 | 0.22 | 0.18 | 0.20 | 0.24 | 0.16 | 0.38 | 0.18 | 0.21 | 0.02 | 0.02 | 0.02 | 0.02 | 0.00 |
Abbreviations: ALA, Alameda; HAY, Hayward; RL, Robert's Landing.
Bold values indicate significance at P<0.05.
Table 4. AMOVA among sites: adults only.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage variation |
|---|---|---|---|---|
| Among sites | 2 | 51.500 | 0.16367 | 7.50** |
| Among adult populations within sites | 4 | 29.223 | 0.19799 | 9.08* |
| Among individuals within populations | 122 | 234.025 | 0.09865 | 4.52* |
| Within individuals | 129 | 222.000 | 1.72093 | 78.90 |
| Total | 257 | 536.748 | 2.18124 |
Site 1: Alameda, S. alterniflora and hybrid shoreline meadow adults, mudflat hybrid adults.
Site 2: Robert's Landing, adult hybrid shoreline meadow and mudflat.
Site 3: Hayward, adult S. foliosa and hybrid shoreline plants.
**Significance at P<0.0001. *Significance at P<0.05.
Average F-statistics over all loci. Fixation indices: FIS, 0.05; FSC, 0.09; FCT, 0.08; FIT, 0.21. FIS, among individuals within populations; FSC, among adult populations but within sites; FCT, adult populations among sites; FIT, among adult populations and among sites.
Table 5. AMOVA among three sites: across generations.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage variation |
|---|---|---|---|---|
| Among sites | 2 | 173.934 | 0.23677 | 11.12** |
| Among generations within sites | 10 | 94.474 | 0.11990 | 5.63** |
| Among individuals within generations | 452 | 898.340 | 0.21471 | 10.08** |
| Within individuals | 465 | 724.500 | 1.55806 | 73.17 |
| Total | 929 | 1891.248 | 2.12944 |
Site 1: Alameda, S. alterniflora and hybrid shoreline meadow adults, mudflat hybrid adults and 2003 and 2004 seedlings.
Site 2: Robert's Landing, adult hybrid shoreline meadow and mudflat, and 2003 and 2004 mudflat hybrid seedlings.
Site 3: Hayward, adult S. foliosa and hybrid shoreline plants, and 2003 and 2004 mudflat hybrid adults and seedlings.
**Significance at P<0.0001. *Significance at P<0.001.
Average F-statistics over all loci. Fixation indices: FIS, 0.12; FSC, 0.06; FCT, 0.11; FIT, 0.27. FIS, among individuals within generations; FSC, among generations but within sites; FCT, generations among sites; FIT, among generations and among sites.
Table 6. Temporal genetic structure among generations of individuals: primary founders, descendents and seedlings at three population sites of hybrid Spartina.
| Primary founders | Descendents | 2003 Seedlings | 2004 Seedlings |
|---|---|---|---|
| Alameda meadow | 0.01 | 0.02* | 0.02* |
| Robert's Landing mudflat | 0.06* | 0.17* | 0.26* |
| Hayward mudflat | NA | 0.01 | 0.03* |
*FST difference at P<0.05.
Spatial genetic analysis
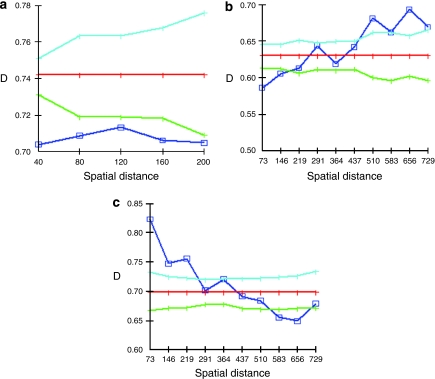
Spatial genetic analysis (Figure 3) showed significant positive spatial structure between adults and seedlings up to a distance of ca. 200 m at all three sites, and negative spatial structure above a distance of approximately 440 m at Hayward and above ∼530 m at Robert's Landing (Figure 3). At Alameda there were too few individuals in a distance class >200 m to include in the analysis (minimum value=30 individuals per distance class; Degen, 2000).
Figure 3.
Spatial genetic analysis using the number of alleles/haplotypes in common over all loci for hybrid adults and seedlings at the three population sites (a, Alameda; b, Hayward; c, Robert's Landing) performed with SGS (Degen et al., 2001). Values exceeding the reference value (95% confidence intervals—blue and green lines) indicate positive spatial genetic structure, and values below the reference line (red) show negative spatial structure (Degen, 2000). The spatial distance unit is meters. A full color version of this figure is available at the Heredity Journal online.
Discussion
Spatial genetic structure existed among and temporal genetic structure existed within Spartina hybrid populations in San Francisco Bay. We found genetically distinct hybrid populations at Alameda and the Robert's Landing/Hayward shorelines (FCT=0.21) that likely resulted from different founding histories: human plantings versus natural tidal establishment. We posit that at Alameda, S. alterniflora, F1 and early generation hybrids were planted together, interbred and the hybrids proliferated. Our Bayesian analysis supported the view that the preponderance of introgression occurred with a set of S. alterniflora ancestors (yellow color in Figure 2) at Alameda that was genetically distinct from the S. alterniflora ancestors of the southern populations. This may have been due to a founding effect where only a small number of the original S. alterniflora plants were transplanted or due to a second, unreported, introduction of S. alterniflora, from a different East Coast source population, at Alameda. To the detriment of both parental species, hybrids spread clonally, and interbred with neighboring hybrids (Sloop et al., 2009). This resulted in a majority of loci in HWE with low linkage disequilibrium across loci. S. foliosa introgressive hybrids were either less frequent there, or possibly lost out in competition. Presently, there are no S. foliosa plants at Alameda (Ayres et al., 2004).
We propose that the development of spatial genetic structure proceeded differently at Robert's Landing and Hayward. As neither hybrids nor S. alterniflora were planted at these sites, these shorelines were colonized by hybrid seed floating northward from invaded marshes to the south, the original plantation of S. alterniflora being 8 km to the south at New Alameda Creek (Faber, 2000). This colonization trajectory would have taken much more time than the human-aided hybrid establishment at Alameda allowing natural selection to act upon these later-generation hybrids. In previous work we have found that advanced generation hybrids, but not the parental species or early generation hybrids, are self-fertile (Sloop et al., 2009). One outcome of self-fertility was reflected by a relatively low number of loci in HWE along the southern shoreline compared to Alameda. As the presence of native S. foliosa at the Hayward shoreline predates the arrival of hybrids, colonizing hybrids backcrossed with native S. foliosa, as suggested by high levels of S. foliosa alleles (blue color) in Hayward plants in Figure 2. The lower level of genetic structure between populations at Robert's Landing and Hayward (FST=0.08, mudflat adults) suggests that seed movement on tidal currents may link these neighboring sites, 2 km apart. In comparison, our results imply that little if any gene flow has occurred between Alameda, 16 km northward, and the Robert's Landing and Hayward sites (FST=0.21).
Many hybrids contained unique alleles. As most of the unique hybrid alleles were at relatively low frequencies (0.02–0.13), this may point to the allelic composition(s) of one or more S. alterniflora ancestors not present in our samples. Alternatively, these unique alleles may be due to genomic changes, including chromosomal rearrangements, differential gene expression, and gene silencing as has been noted to occur in hybridizing species (Baack and Rieseberg, 2007).
Despite the potential for high rates of gene flow through seed dispersal in the dynamic San Francisco Bay tidal system, parentage results confirmed that high proportions of seedlings were the progeny of nearby plants (Sloop et al., 2009). Thus, predominately local seedling recruitment maintains the distinct genetic structure between Alameda and Robert's Landing/Hayward. As an extreme example of local parentage, a single plant at Robert's Landing produced, by selfing, 10% of all seedlings we genotyped there (Sloop et al., 2009, see arrow Figure 2). Selfing combined with local seed dispersal can lead to the development of fine-scale genetic structure, as has been shown in hybrid oaks by Valbuena-Carabaña et al. (2007).
Our finding of seedling spatial autocorrelation distances of <200 m (Figure 3) indicated that within a 200 m distance, seedlings are genetically more similar to each other than expected by chance, as perhaps a plant surrounded by its seedlings. This further confirmed the local dispersal of seeds suggested by our findings of fine-scale spatial genetic structure of seedlings, and paternity analyses in previous studies (Sloop et al., 2009). Plants >440 m apart, on the other hand, are more genetically dissimilar than expected by chance (Figure 3). This dissimilarity is likely above a certain geographic distance, where seeds of genetically and geographically distant plants have settled out of the tidal waters. The Alameda site was too small to make autocorrelations at >200 m (Figure 3).
We found that genetic structure changed over time among generations within populations (Table 6). At Alameda the primary founders were meadow plants growing high in the tidal range and among other plant species. Tidal flat descendents (growing alone on open mud) were similar to the meadow founders while seedlings from successive years showed increasing genetic divergence from the founding population (Table 6). At Robert's Landing FST values between founders, descendants and 2003 and 2004 generations of seedling increased from 0.06 to 0.17 to 0.26, respectively, showing a substantially growing genetic distinction from the founders over generations (Table 6). At Hayward the temporal dynamics were similar to, but weaker than those at Robert's Landing. Increased production of self-fertilized seedlings especially at Robert's Landing and Hayward from 2003 to 2004 (Table 1, Sloop et al., 2009) may explain these changes in genetic structure and are also reflected by increased inbreeding coefficients (Table 1). We found in previous work that self-seed set under experimental pollen exclusion treatments was much higher in the later-generation hybrids growing upon tidal flats than in early generation hybrids or the parental species (Sloop et al., 2009).
Increased self-compatibility of isolated tidal flat colonizers and their production of inbred recruits can result in lower levels of heterozygosity than expected under HWE, and higher levels of inbreeding (Sloop et al., 2009). In open San Francisco Bay tidal flats, colonizing hybrid family groups growing in isolation have set self-fertilized seeds, and have thus overcome the pollen limitation seen in invading, self-incompatible S. alterniflora in Willapa Bay, WA (Davis et al., 2004; Taylor et al., 2004). There, pollen limitation of isolated plants caused an Allee effect that greatly slowed the tidal flat invasion (Davis et al., 2004; Taylor et al., 2004). In San Francisco Bay, these few self-compatible hybrids, having adapted to environmentally challenging conditions, maximize their reproductive fitness by producing large numbers of self-fertilized seeds in isolation (Daehler, 1998; Sloop et al., 2009). These seeds then spread on the tide and established as vigorous seedlings throughout the tidal flat, over time dominating this environmentally harsh habitat. Producing offspring in this way may greatly increase the rate of the colonization of new and challenging environments by maximizing the effects of selective forces on highly adapted genotypes, and so avoiding both inbreeding and outbreeding depression during this difficult phase of invasion (Lynch, 1991; Waser and Price, 1994; Sloop et al., 2009).
The natural stepping-stone dispersal mechanism (Kimura and Weiss, 1964) and human movement of hybrid plants (for example, unintentional use of cryptic hybrid plants rather than natives in restoration efforts) have increased hybrid dispersal and influenced hybrid population genetic structure by introducing varying population founding trajectories at various sites and times around San Francisco Bay. Genetic structure in San Francisco Bay hybrid populations has been affected by a number of different founding events: site-specific environmental dynamics affecting colonization rates, and relative isolation from other populations over time. The proximity of a native or hybrid marsh, the distance between established adult populations, wind directions, tidal currents and environmental selection forces determined the rate and direction of local tidal flat seed and pollen dispersal/gene flow. Our findings therefore suggest that even though long-distance dispersal has been manifestly important in the very rapid spread of hybrid Spartina throughout San Francisco Bay over the past 35 years (Ayres et al., 2004), local recruitment and the evolution of self-fertility (Sloop et al., 2009) have also contributed, generating substantial regional, fine-scale and temporal genetic structure. Our findings provide a glimpse in the evolutionary dynamics that underlie the expansion of invasive hybrids and the colonization of new habitats as well as those occupied by the parent species.
Acknowledgments
We thank J Bando, H Davis, J Lambrinos, H McGray, A Lee and R Hall for their assistance. This work was supported by the California Coastal Conservancy (CalFed Grant No. 99-110), California Sea Grant (Project R/CZ-176) and the National Science Foundation Biocomplexity program (DEB No. 0083583).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott RJ. Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- Austerlitz F, Garnier-Géré PH. Modelling the impact of colonization on genetic diversity and differentiation of forest trees: interaction of life cycle, pollen flow and seed long-distance dispersal. Heredity. 2003;90:282–290. doi: 10.1038/sj.hdy.6800243. [DOI] [PubMed] [Google Scholar]
- Ayres DA, Zaremba K, Sloop CM, Strong DR. Sexual reproduction of cordgrass hybrids (Spartina foliosa × alterniflora) invading tidal marshes in San Francisco Bay. Divers Distrib. 2008;14:187–195. [Google Scholar]
- Ayres DR, Garcia-Rossi D, Davis HG, Strong DR. Extent and degree of hybridization between exotic (Spartina alterniflora) and native (S. foliosa) cordgrass (Poaceae) in California, USA determined by random amplified polymorphic DNA (RAPDs) Mol Ecol. 1999;8:1179–1186. doi: 10.1046/j.1365-294x.1999.00679.x. [DOI] [PubMed] [Google Scholar]
- Ayres DR, Smith DL, Zaremba K, Klohr S, Strong DR. Spread of exotic cordgrasses and hybrids (Spartina sp.) in the tidal marshes of San Francisco Bay. Biol Invasions. 2004;6:221–231. [Google Scholar]
- Baack EJ, Rieseberg LH. A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev. 2007;17:513–518. doi: 10.1016/j.gde.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, Laboratoire Génome, Populations. Université de Montpellier II: Montpellier, France; 1996–2004. [Google Scholar]
- Blum MJ, Sloop CM, Ayres DR, Strong DR. Characterization of microsatellite loci in Spartina species (Poaceae) Mol Ecol Notes. 2004;4:39–42. [Google Scholar]
- Buggs RJA. Empirical study of hybrid zone movement. Heredity. 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- Civille J, Sayce K, Smith SD, Strong DR. Reconstructing a century of Spartina invasion with historical record and contemporary remote sensing. Ecoscience. 2005;12:330–338. [Google Scholar]
- Daehler CC. Variation in self-fertility and the reproductive advantage of self-fertility for an invading plant. Evol Ecol. 1998;12:553–568. [Google Scholar]
- Daehler CC, Strong DR. Hybridization between introduced smooth cordgrass (Spartina alterniflora, Poaceae) and native California cordgrass (Spartina foliosa) in San Francisco Bay, California, USA. Am J Bot. 1997;85:607–611. [PubMed] [Google Scholar]
- Davis HG, Taylor CM, Lambrinos JG, Strong DR. Pollen limitation causes an Allee effect in a wind-pollinated invasive grass (Spartina alterniflora) Proc Natl Acad Sci USA. 2004;101:13804–13807. doi: 10.1073/pnas.0405230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen B, Petit R, Kremer A. SGS—Spatial Genetic Software: a computer program for analysis of spatial genetic and phenotypic structures of individuals and populations. J Hered. 2001;92:447–448. doi: 10.1093/jhered/92.5.447. [DOI] [PubMed] [Google Scholar]
- Degen B.2000SGS: Spatial Genetic Software. Computer program and user's manual . ftp://ghd.dnsalias.net/degen/software.html . [DOI] [PubMed]
- Donoghue K. Maternal determinants of seed dispersal in Cakile edentula: fruit, plant, and site traits. Ecology. 1998;79:2771–2788. [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants. Proc Natl Acad Sci USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson BK, Chung MG, Telewski FW. Spatial pattern of allozyme variation in a contact zone of Pinus ponderosa and P. arizonica (Pinaceae) Am J Bot. 2003;90:25–31. doi: 10.3732/ajb.90.1.25. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Faber P.2000Grass wars: good intensions gone awry. Why would anyone bring an alien cordgrass into S. F. BayIn: Rasa Gustaitis (ed). California Coast & Oceanvol. 16. California Coastal Conservancy: Oakland, CA, USA; , http://www.coastalconservancy.ca.gov/coast&ocean/summer2000/pages/pthr.htm . [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick JL, Murawski DA, Nason JD. The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio. 1993;107:281–297. [Google Scholar]
- Hamrick JL, Nason JD.1996Consequences of dispersal in plantsIn: Rhodes OE, Chesser RK, Smith MH (eds.).Population Dynamics in Ecological Space and Time University of Chicago Press: Chicago; 203–236. [Google Scholar]
- Higgins SI, Richardson DM. Predicting plant migration rates in a changing world: the role of long-distance dispersal. Am Nat. 1999;153:464–475. doi: 10.1086/303193. [DOI] [PubMed] [Google Scholar]
- Huiskes AHL, Koutstaal BP, Herman PMJ, Beeftink WG, Markusse MM, De Muck W. Seed dispersal of halophytes in tidal salt marshes. J Ecol. 1995;83:559–567. [Google Scholar]
- Husband BC, Schemske DW. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. [DOI] [PubMed] [Google Scholar]
- Kimura M, Weiss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. Dispersal versus gene flow in plants. Ann Mo Bot Gard. 1981;68:232–253. [Google Scholar]
- Lambrinos JG. How interactions between ecology and evolution influence contemporary invasion dynamics. Ecology. 2004;85:2061–2070. [Google Scholar]
- LeCorre VL, Machon N, Petit RJ, Kremer A. Colonization with long-distance dispersal and genetic structure of maternally inherited genes in forest trees: a simulation study. Genet Resour. 1997;69:117–125. [Google Scholar]
- Long JC. The genetic structure of admixed populations. Genetics. 1991;127:417–428. doi: 10.1093/genetics/127.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution. 1991;3:622–629. doi: 10.1111/j.1558-5646.1991.tb04333.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimate of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Bonvin G. Fine-scale genetic structure and marginal processes in an expanding population of Biscutella laevigata L. (Brassicaceae) Heredity. 2008;101:536–542. doi: 10.1038/hdy.2008.95. [DOI] [PubMed] [Google Scholar]
- Primack RB, Kang H. Measuring fitness and natural selection in wild populations. Ann Rev Ecol Syst. 1989;20:367–396. [Google Scholar]
- Pritchard JK, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen W.2004Documentation for structure softwareVersion 2, , http://pritch.bsd.uchicago.edu .
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests of ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation, and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Schnabel A, Nason JD, Hamrick JL. Understanding the population genetic structure of Gleditsia triacanthos L.: seed dispersal and variation in female reproductive success. Mol Ecol. 1998;7:819–832. [Google Scholar]
- Slatkin M. Gene flow and population structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Sloop CM, Ayres DR, Strong DR. The rapid evolution of self-fertility in Spartina hybrids (Spartina alterniflora × foliosa) invading San Francisco Bay, CA. Biol Invasions. 2009;11:1131–1144. [Google Scholar]
- Sloop CM, McGray HG, Blum MJ, Strong DR. Characterization of 24 additional microsatellite loci in Spartina species (Poaceae) Conserv Genet. 2005;6:1049–1052. [Google Scholar]
- Strong DR, Ayres DA.2009Spartina introductions and consequences in salt marshes: arrive, survive, thrive, and sometimes hybridizeIn: Silliman BR, Grosholz T, Bertness M (eds).Human Impacts on Salt Marshes University of California Press: Berkeley, CA, USA [Google Scholar]
- Surles SE, Arnold J, Schnabel A, Hamrick JL, Bongarten BC. Genetic relatedness in open-pollinated families of two leguminous tree species, Robinia pseudoacacia L. and Gleditsia triacanthos L. Theo Appl Genet. 1990;80:49–56. doi: 10.1007/BF00224015. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Davis HG, Civille JC, Grevstad FG, Hastings A. Consequences of an Allee effect in an invasive plant: Spartina alterniflora in Willapa Bay, Washington. Ecology. 2004;85:3254–3266. [Google Scholar]
- US Army Corps of Engineers . Pre-construction Report. Shoreline Erosion Control Demonstration Project: Alameda, CA; 1978. [Google Scholar]
- Valbuena-Carabaña M, Gonzalez-Martinez SC, Hardy OJ, Gil L. Fine-scale genetic structure in mixed oak stands with different levels of hybridization. Mol Ecol. 2007;16:1207–1219. doi: 10.1111/j.1365-294X.2007.03231.x. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Waser NM, Price MV. Crossing-distance effects in Delphinium nelsonii: outbreeding and inbreeding depression in progeny fitness. Evolution. 1994;3:842–852. doi: 10.1111/j.1558-5646.1994.tb01366.x. [DOI] [PubMed] [Google Scholar]
- Williams DA, Muchugu E, Overholt WA, Cuda JP. Colonization patterns of the invasive Brazilian peppertree, Schinus terebinthifolius, in Florida. Heredity. 2007;98:284–293. doi: 10.1038/sj.hdy.6800936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.