Abstract
Human-induced alteration of natural habitats has the potential to impact on the genetic structuring of remnant populations at multiple spatial scales. Species from higher trophic levels, such as snakes, are expected to be particularly susceptible to land-use changes. We examined fine-scale population structure and looked for evidence of sex-biased dispersal in smooth snakes (Coronella austriaca), sampled from 10 heathland localities situated within a managed coniferous forest in Dorset, United Kingdom. Despite the limited distances between heathland areas (maximum <6 km), there was a small but significant structuring of populations based on eight microsatellite loci. This followed an isolation-by-distance model using both straight line and ‘biological' distances between sampling sites, suggesting C. austriaca's low vagility as the causal factor, rather than closed canopy conifer forest exerting an effect as a barrier to dispersal. Within population comparisons of male and female snakes showed evidence for sex-biased dispersal, with three of four analyses finding significantly higher dispersal in males than in females. We suggest that the fine-scale spatial genetic structuring and sex-biased dispersal have important implications for the conservation of C. austriaca, and highlight the value of heathland areas within commercial conifer plantations with regards to their future management.
Keywords: colubridae, forestry, genetic diversity, isolation-by-distance, microsatellites, habitat loss
Introduction
Anthropogenic pressures, through processes such as habitat alteration, have been attributed to the decline and disappearance of many populations (Pimm and Raven, 2000). However, such processes also have significant impacts on the genetic diversity, and ultimately, evolutionary potential of those populations that remain (Templeton et al., 2001). For a given species, the genetic structuring of populations reflects the range within that individuals are more closely related to one another than to those randomly selected from the general population (Repaci et al., 2007). Genetic structuring in contiguous habitats is typically expected to follow an isolation-by-distance model, whereby the geographical distance between two populations is the single biggest determinant of their genetic differentiation (Slatkin, 1993). In contrast, the population genetic structure of organisms inhabiting habitat mosaics can be facilitated or restricted by the specific habitat features an organism encounters (Adriaensen et al., 2003).
Limits to gene flow between populations are typically the result of a species reproductive mode and vagility (Lowe et al., 2005). However, habitat change can result in the creation of dispersal barriers (Hitchings and Beebee, 1998), which can alter patterns of gene flow and lead to the isolation of remnant populations (Gerlach and Musolf, 2000). Such small and isolated populations may continue to lose genetic variation as a consequence of genetic drift (Reed and Frankham, 2003), and become inbred (Madsen et al., 1996). Those populations that are able to persist and even expand in size following such perturbations are likely to be less genetically diverse because of bottleneck effects, which may result in reduced population fitness (Hoelzel et al., 2002).
Species at higher trophic levels have been shown both theoretically (Holt et al., 1999) and empirically (Komonen et al., 2000) to be particularly sensitive to habitat alteration. As snakes typically occur in the middle to higher levels of food webs, habitat changes are likely to have strong effects on their population dynamics (Lind et al., 2005) and have been linked to widespread population declines (Reading et al., 2010). Changes in the population status of snakes could also result in cascade effects, as has been seen with garter snakes (Thamnophis spp.) and their amphibian prey (Matthews et al., 2002). In addition to their high trophic level, snakes are typically limited in their dispersal ability, even in continuous habitats (Luiselli and Capizzi, 1997), which could have profound effects on their population genetics (Keogh et al., 2007). As a result, genetic structuring is likely to increase in populations that are subjected to habitat fragmentation.
Although theoretical evidence supports the idea that dispersal ability is a critical factor in determining the persistence of species in a fragmented habitat (Hanski and Thomas, 1994), it remains one of the least understood factors in conservation biology (Macdonald and Johnson, 2001). This is largely a result of difficulties associated with its direct study (Templeton et al., 1990; Prugnolle and de Meeus, 2002), particularly for inconspicuous species such as snakes (Parker and Plummer, 1987; Gibbs and Weatherhead, 2001). In this respect genotypic data have become an invaluable tool to indirectly measure species dispersal capabilities, while simultaneously estimating differentiation between sampled populations (DeYoung and Honeycutt, 2005).
Coronella austriaca is an ovoviviparous, colubrid species that primarily feeds on lizards and small mammals using ambush tactics (Spellerberg and Phelps, 1977). Radio-tracking studies suggest C. austriaca to be sedentary in nature (Gent and Spellerberg, 1993), with adults becoming sexually mature at 4 years of age and potentially living in excess of 17 years (Reading, 2004b). Although widely distributed throughout continental Europe and areas of western Asia, C. austriaca populations in the United Kingdom are limited to lowland heathland habitats in Dorset, Hampshire and Surrey (Braithwaite et al., 1989). A plagioclimax vegetation community dominated by ericaceous dwarf shrubs (Chapman et al., 1989), lowland heathland has undergone significant loss and fragmentation as a result of agricultural intensification, shrub encroachment, urbanisation and commercial afforestation (Bakker and Berendse, 1999; Rose et al., 2000). This has been implicated as the determining factor in historical population declines of C. austriaca in the United Kingdom (Spellerberg and Phelps, 1977). At present some of the largest patches of heathland that remain are contained within the boundaries of commercial forests. Silvicultural practices within forest sites have resulted in a heterogeneous landscape, comprising stands of Pinus spp. of varying ages interspersed with patches of heathland. These heathland patches represent important remnant habitats for C. austriaca populations (Reading, 2004a, 2004b); whereas forest stands have been suggested as ‘effective barriers' to dispersal between populations (Phelps, 1978). This assumption has remained unstudied to date.
Given our limited knowledge of snake populations in altered landscapes and particularly the impacts of heathland alteration on United Kingdom populations of C. austriaca, this study was designed to examine (a) whether fine-scale population genetic structuring occurs within Wareham Forest (maximum distance between sampling sites <6 km), and to determine whether the observed structure was a result of isolation-by-distance effects or the limits to dispersal ability conferred by the presence of coniferous forest stands, and (b) to determine whether dispersal in this species is sex-biased.
Methods
Field surveys and sampling
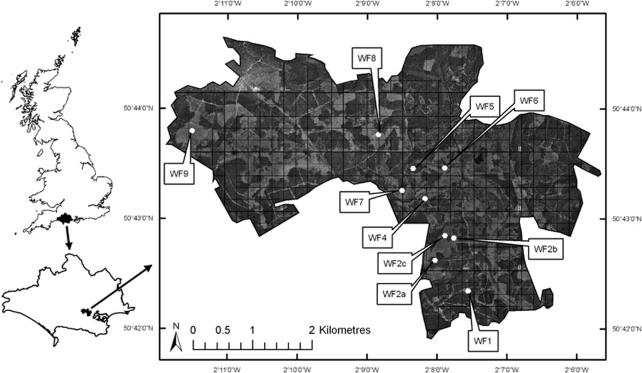
A total of 10 hexagonal arrays of 37 artificial refugia (sensu Reading, 1997) were located within the boundaries of Wareham Forest, Dorset (Figure 1). Arrays were located on homogeneous areas of lowland heathland between closed-canopy stands of Corsican pine (Pinus nigra var. maritima) and Scots pine (Pinus sylvestris). Straight-line distances (m) between the central refugia of each array at sampling sites were calculated in ArcGIS 9.1 using ortho-rectified aerial photographs. Habitat structure, in addition to geographical distance, is known to be a significant factor in determining population genetic structure for a number of species (Manel et al., 2003), and so a ‘biological distance' between sampling sites within Wareham Forest was also measured from ortho-rectified aerial photographs. The ‘biological distance' (m) was based on the assumption that snakes moving between pairs of heathland patches in Wareham Forest would preferentially use habitats that were primarily open canopied (for example, forest rides, clear-felled stands, young Pinus spp. stands, heath and moorland). C. austriaca is a lowland heathland specialist and, as such, is typically found in areas with few or no trees present (Spellerberg and Phelps, 1977). As a result the distance between the nearest edges of pairs of sampling sites, rather than the central refugia within arrays at sampling sites, was measured from the same aerial photographs, but based on a nonlinear path that avoided stands of trees that formed a closed canopy (Table 1).
Figure 1.
Localities of the 10 arrays of artificial refugia (white hexagons) within Wareham Forest, Dorset, from which individual Coronella austriaca were sampled. Darker coloured areas indicate stands of closed canopy Pinus spp., whereas lighter coloured areas include heathland, clearfelled stands and young Pinus spp. plantations.
Table 1. Matrix table of pairwise distances (m) between 10 arrays of artificial refugia used to collect DNA samples from populations of Coronella austriaca within Wareham Forest, Dorset.
| WF1 | WF2a | WF2b | WF2c | WF4 | WF5 | WF6 | WF7 | WF8 | WF9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| WF1 | — | 844.7 | 926.7 | 975.6 | 1720.4 | 2223.9 | 2295.6 | 2292.7 | 3092.1 | 5452.2 |
| WF2a | 1025.0 | — | 217.9 | 128.8 | 878.8 | 1385.6 | 1496.9 | 1450.2 | 2251.8 | 4724.0 |
| WF2b | 1124.6 | 206.7 | — | 211.0 | 831.2 | 1308.4 | 1368.6 | 1141.2 | 2204.6 | 4789.7 |
| WF2c | 1128.4 | 110.3 | 149.8 | — | 741.0 | 1255.3 | 1369.4 | 1318.3 | 2114.1 | 4614.3 |
| WF4 | 1963.5 | 965.9 | 853.0 | 822.6 | — | 523.3 | 733.5 | 625.0 | 1376.1 | 4056.3 |
| WF5 | 2468.1 | 1444.8 | 1338.9 | 1295.7 | 498.6 | — | 382.9 | 437.2 | 918.4 | 3806.0 |
| WF6 | 2620.6 | 1635.9 | 1585.0 | 1541.7 | 883.8 | 435.3 | — | 822.4 | 1081.3 | 4093.9 |
| WF7 | 2612.0 | 1595.2 | 1474.0 | 1460.6 | 633.9 | 494.9 | 873.5 | — | 832.4 | 3451.0 |
| WF8 | 3502.5 | 2481.0 | 2375.7 | 2338.0 | 1524.9 | 1079.1 | 1297.2 | 885.9 | — | 3062.3 |
| WF9 | 6046.4 | 5100.0 | 4940.8 | 4946.2 | 4103.4 | 3947.9 | 4355.4 | 3567.3 | 3299.4 | — |
Values above the dashed lines are the straight-line distances between the central refugia in each array. Values below the dashed lines are the ‘biological' pairwise distances between the nearest edges of the two heathland samplings sites, containing arrays based on the assumption that smooth snakes only move through habitats with open canopies.
Surveys for C. austriaca were completed between April and October of 2006 through 2008 (Pernetta, 2009). All snakes were captured by hand and sexed using tail length and shape (van Gelder et al., 1998; Reading, 2004a, 2004b). Individuals were permanently marked using Passive Integrated Transponders (UKID Systems Model: UKID 122 IJ, 12-mm long, 2.12-mm wide), and DNA samples were collected upon first capture to prevent sampling individuals more than once.
Samples comprised either blood spots collected on Whatman grade 2 filter paper strips (Whatman plc, Maidstone, UK), obtained by puncturing the caudal sinus below the cloaca, or shed skin samples in cases where animals were in the process of sloughing when captured. Both sample types were air dried at room temperature before being frozen (−20 °C) before analysis. All blood samples were collected under licence from the Home Office (No: PIL 80/9699) Natural England (No: 20070553).
Sample extraction and microsatellite analysis
DNA was extracted from the dried blood and/or skin samples using a modified version of Gemmell and Akiyama's (1996) method for vertebrate tissues. Samples were placed in 300 μl of digestion solution (100 m TRIS, 1%sodium dodecyl sulfate, 100 m NaCl) and 7 μl of protinase K (10 mg ml−1), and mixed overnight (≈12 h) on a rotating wheel at 37 °C. Following digestion, 300 μl of 10 LiCl and 600 μl of chloroform were added and the samples mixed on the rotating wheel for a further 30 min at room temperature. The samples were then centrifuged for 10 min at 13 000 rpm to separate the supernatant, which was transferred to a new eppendorf tube. DNA was then precipitated with ethanol, dried and dissolved in 50-μl sterile distiled water, and stored at −20 °C.
We used eight previously characterised microsatellite loci Ca19, Ca20, Ca26, Ca27, Ca30, Ca40, Ca47 and Ca66 (Bond et al, 2005), on the basis of their consistency in amplification and ease of scoring. Amplifications were carried out in 20 μl volumes containing 1 μl of the DNA extract. Reactions contained 10 m Tris-HCl pH 8.3, 50 m KCI, 1.5 m MgCl2, 0.2 μm of each oligonucleotide primer, 100 μm dGTP, dCTP and dTTP, 5-μm unlabelled dATP, 3.7 KBq [α-33P]dATP (37 TBq mmol−1) and 0.5–1.0 units Taq polymerase (New England Biolabs, Hitchin, Hertfordshire, UK). Conditions for each primer set used followed those of Beebee (2008) and comprised an initial 4-min denaturation period (94 °C), followed by 35 amplification cycles each with 1-min denaturation at 94 °C, 1-min annealing (Ca20, Ca30, Ca40=51 °C, Ca19, Ca26, Ca27, Ca47, Ca66=60 °C) and 1-min elongation at 72 °C. All reactions culminated with a 4-min elongation step at 72 °C. PCR products were electrophoresed alongside M13 sequence markers in standard sequencing gels (6% wv, polyacrylamide). Following electrophoresis, gels were visualised using autoradiography and genotypes scored from the X-ray film.
Statistical analyses
Loci were examined for null alleles and mis-scoring using (Van Oosterhout et al, 2004). All loci were tested, at 99.5% confidence intervals, for evidence of selection using L (Antao et al, 2008) with 50 000 simulations based on a stepwise mutation model. All retained loci were also tested for the presence of linkage disequilibria between loci using G v4.0 (Rousset, 2008), and for deviations from Hardy–Weinberg equilibrium (HWE) using v6.2 (Peakall and Smouse, 2006). In both cases the critical probability was adjusted for multiple comparisons using a sequential Bonferonni correction (Weir, 1990). Observed and expected heterozygosities (HO, HE) were calculated in , whereas Allelic richness (AR) and the inbreeding coefficient within populations (FIS) were calculated using F v2.9.3.2 (Goudet, 1995). The significance of FIS estimates was tested in F based on 1600 randomisations. Allelic richness and measures of genetic diversity were compared between populations using Kruskal–Wallis one-way analysis of variance tests implemented in Minitab v.15 (Minitab inc., Philadelphia, PA, USA).
Bayesian clustering methods of population assignment were implemented using S 2.3.3. as an initial attempt to determine whether the sampling regions represent genetically distinct populations (Pritchard et al., 2000). A correlated alleles model with admixture incorporating sampling locality information was run in triplicate for each potential real population size (K=1–10). An initial burn-in period of 305 Markov chain replicates, followed by 106 iterations was used for each run. The number of ‘true' populations was determined by calculating ΔK using the method as described by Evanno et al. (2005).
Genetic differentiation was assessed using FST analysis, which generally performs well when divergence among samples is expected to be low (Balloux and Goudet, 2002). Total genetic differentiation between populations was assessed using an analysis of molecular variance (AMOVA), in addition to pairwise estimates of FST between populations using The significance of these estimators was assessed using a non-parametric permutation approach (9999 permutations) with a subsequent Bonferonni correction applied to significance values.
Isolation by distance between populations was assessed using Mantel tests conducted in (9999 permutations). Both linear and ‘biological' distances (Ln transformed) between pairs of populations were compared with the genetic difference between population pairs (FST/(1−FST); a useful transformation for testing isolation-by-distance effects (Rousset, 1997)). Four tests for evidence of sex-biased dispersal were conducted using F for the samples collected within Wareham Forest, based on 10 000 permutations. The first test compared relatedness (r) between sexes, with the assumption that the dispersing sex should exhibit lower levels of within population relatedness than the philopatric sex. The second test compared among population FST values between sexes, based on the principle that allelic frequencies of the dispersing sex should exhibit higher levels of homogeneity across the populations than the philopatric sex. The third and fourth tests are based on the use of a corrected assignment index to determine the probability of a genotype originating from the population in which the individual was collected (Favre et al, 1997). The first of these two tests compared the mean of the corrected assignment index (mAIc). In this instance, immigrants to a population are expected to have lower AIc values than residents. Therefore, the dispersing sex is likely to have a significantly lower mAIc value. The second of these tests compared the difference in the variance of AIc (vAIc). In this instance, the dispersing sex is likely to have a larger vAIc value than the philopatric sex because of increased probability of both resident and immigrant individuals being present in the sampled populations (Dubey et al, 2008). Because of the inequality in the number of males and females sampled at each site, we used a random re-sampling procedure to create five new data sets (n=110) with an equal sex ratio (male=55 females=55) and calculated the mean statistic and probability values for each test (sensu Dubey and Shine, 2010)
Results
A total of 149 individual smooth snakes were captured within Wareham Forest. Samples from two individuals failed to amplify any of the eight loci and so were excluded from further analyses. Initial examination of the data with M-C showed no evidence of null alleles, and L analyses showed no evidence of any loci being under selection. There was also no evidence of linkage disequilibrium at the 5% level (Bonferonni adjustment P<0.000625). Tests for deviation from HWE found five significant results (Table 2). However, these were not limited to a single locus or sampling site and so all eight loci were retained for further analysis. The mean allelic richness, inbreeding coefficients, observed and expected heterozygosities are shown for each sample site in Table 2. Kruskall–Wallis one-way ANOVA tests showed no significant difference between sample sites in any of the calculated measures of genetic diversity (AR, H=0.960, P=0.384; HO, H=2.54, P=0.980; HE. H=15.21, P=0.085; FIS, H=2.11, P=0.990). Mean inbreeding coefficients were high within all 10 sampling sites (overall mean=0.250, s.e.±0.018) and strongly significant for two of them (Table 2).
Table 2. Total sample size (n), based on males (M) and females (F) sampled at each array, average number of individuals genotyped per locus (N), allelic richness (AR), observed heterozygosity (HO), expected heterozygosity (HE), inbreeding coefficients (FIS) and loci showing significant deviations from Hardy–Weinberg equilibrium (HWE) for 10 smooth snake-sample sites within Wareham Forest, Dorset.
| Array | N | M | F | N | AR | HO | HE | FIS | HWE |
|---|---|---|---|---|---|---|---|---|---|
| WF1 | 15 | 6 | 9 | 13.5 | 2.113 | 0.455 | 0.568 | 0.236 (0.0013) | Ca47 |
| WF2a | 17 | 12 | 5 | 13.38 | 2.295 | 0.476 | 0.591 | 0.249 (0.0019) | |
| WF2b | 24 | 11 | 13 | 18.63 | 2.236 | 0.393 | 0.526 | 0.280 (0.0006*) | Ca19, Ca20, Ca40 |
| WF2c | 8 | 3 | 5 | 7 | 2.492 | 0.414 | 0.555 | 0.336 (0.0056) | |
| WF4 | 21 | 11 | 10 | 18.25 | 2.603 | 0.507 | 0.582 | 0.156 (0.0025) | |
| WF5 | 10 | 6 | 4 | 7.75 | 2.536 | 0.388 | 0.507 | 0.305 (0.0013) | Ca40 |
| WF6 | 14 | 7 | 7 | 9.88 | 2.705 | 0.448 | 0.518 | 0.189 (0.0063) | |
| WF7 | 12 | 11 | 1 | 9 | 3.013 | 0.448 | 0.601 | 0.314 (0.0006*) | |
| WF8 | 12 | 7 | 5 | 7.88 | 2.768 | 0.394 | 0.455 | 0.212 (0.0331) | |
| WF9 | 14 | 9 | 5 | 9.5 | 2.807 | 0.357 | 0.418 | 0.220 (0.0038) |
Probabilities of FIS estimates are given in parentheses. *significant at adjusted nominal 5% significance level for multiple samples of 0.00063.
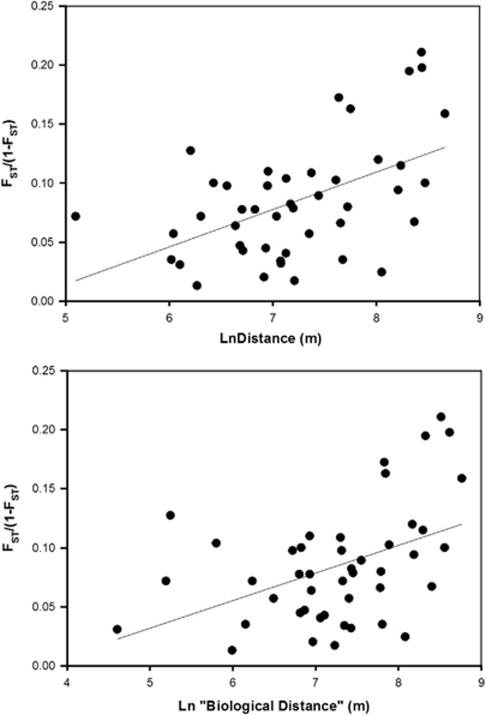
Bayesian clustering analysis using S identified two ‘true' populations, based on the samples obtained. However, the two identified ‘populations' did not correspond to the geographical locality of samples, suggesting the relatively low levels of variability for each loci resulted in a failure to correctly resolve discrete population clusters. Despite the close proximity of sample sites within Wareham Forest, AMOVA showed a small, but highly significant genetic differentiation between populations (FST=0.078, P<0.001). In addition, a total of 29 of the 45 pairwise FST values were significantly different following Bonferonni's correction (P<0.0011; Table 3). Although ‘biological' distance tended to be longer than the Euclidean distance between sampling sites, there was a highly significant correlation between both measures (Pearson's r=0.998, P>0.001). Mantel tests showed significant effects of isolation by distance using both straight-line (rS=0.511, P=0.003) and ‘biological' distances between sampling sites (rS=0.445, P=0.005; Figure 2).
Table 3. Pairwise estimates of FST among 10 smooth snake-sample sites within Wareham Forest.
| WF1 | WF2a | WF2b | WF2c | WF4 | WF5 | WF6 | WF7 | WF8 | WF9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| WF1 | — | |||||||||
| WF2a | 0.060 | — | ||||||||
| WF2b | 0.072 | 0.113 | — | |||||||
| WF2c | 0.020 | 0.030 | 0.067 | — | ||||||
| WF4 | 0.082 | 0.099 | 0.072 | 0.089 | — | |||||
| WF5 | 0.074 | 0.098 | 0.017 | 0.094 | 0.067 | — | ||||
| WF6 | 0.062 | 0.054 | 0.031 | 0.067 | 0.091 | 0.013 | — | |||
| WF7 | 0.093 | 0.076 | 0.033 | 0.089 | 0.054 | 0.034 | 0.045 | — | ||
| WF8 | 0.107 | 0.140 | 0.034 | 0.147 | 0.073 | 0.041 | 0.039 | 0.043 | — | |
| WF9 | 0.137 | 0.165 | 0.091 | 0.174 | 0.163 | 0.103 | 0.063 | 0.086 | 0.024 | — |
Pairwise FST values that are significantly different from zero following Bonferonni's correction are highlighted in bold.
Figure 2.
Relationship between pairwise FST values and (a) the straight-line distance (rS=0.511, P=0.003), and (b) the ‘biological' distance (rS=0.445, P=0.005) between smooth snake-sampling sites in Wareham Forest.
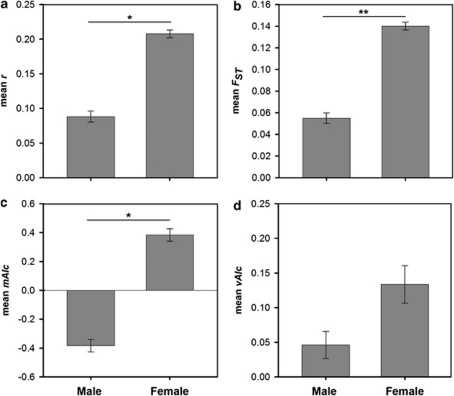
Analysis of sampling sites for sex-biased dispersal showed significant evidence for males being the dispersing sex. Female smooth snakes had significantly higher levels of both within-population relatedness (Figure 3a, mean P=0.011) and between population genetic differentiation (Figure 3b, mean P=0.0072). In addition, females showed significantly higher mAIc values (Figure 3c, mean P=0.046), as would be expected from the more philopatric sex. Finally, the difference in mean vAIc values was not statistically significant (mean P=0.134), the higher mean value for females is not consistent with the other three results in indicating males to be the dispersing sex (Figure 3d).
Figure 3.
Mean±s.e. (a) relatedness, (b) FST, (c) mAIc and (d) vAIc values for male and female smooth snakes from Wareham Forest, Dorset. Significant differences between the sexes are indicated by asterisks (*P<0.05, ** P<0.01).
Discussion
As far as we are aware, the results presented in this study provide the first detailed microsatellite-based analysis of the population genetic structure of C. austriaca at any spatial scale. Importantly, these results have shown that there is significant genetic structuring of populations at a fine-scale, within Wareham Forest. Comparable results have been found in North American species, notably eastern massasauga rattlesnakes (Sistrurus catenatus catenatus; Gibbs et al., 1997), black rat snakes (Elaphe obsoleta obsoleta; Prior et al., 1997), timber rattlesnakes (Crotalus horridus; Clark et al., 2008) and eastern foxsnakes (Mintonius gloydi; DiLeo et al., 2010). Typically, these patterns result from the presence of anthropogenic barriers or a lack of permeable habitats between sub-populations. North American snakes associated with water bodies also have significant population genetic structuring at fine geographical scales (Nerodia sipedon, Prosser et al., 1999; Thamnophis sirtalis and Thamnophis elegans, Manier and Arnold, 2005), likely a result of their habitat specificity (Manier and Arnold, 2005). Genetic differentiation of snake populations has also been linked to body size (King and Lawson, 2001), which may be a reflection of absolute dispersal capacity (DiLeo et al., 2010).
Unlike these previous studies (but see Clark et al., 2008), the fine-scale population genetic structuring observed in this study is consistent with an isolation-by-distance model. This result suggests that the observed differentiation may be because of the previously observed low vagility of the species (Gent and Spellerberg, 1993). Given the highly significant correlation relating straight-line and biological distances between sampling sites, it remains difficult to explicitly state that stands of mature Pinus spp. do not represent an ‘effective barrier' to C. austriaca dispersal (sensu Phelps, 1978). However, it does seem that in this particular site smooth snakes are either capable of using these stands, or other non-heathland habitats within Wareham forest, such as rides bordering such stands, as routes for dispersal between populations.
In contrast to Gent and Spellerberg (1993), whose telemetry-based study found no evidence of sex-biased dispersal, the microsatellite analyses showed clear evidence for male-biased dispersal in smooth snakes. More recently, long-term mark-recapture studies on the ranging behaviour of smooth snakes at a site in Wareham Forest have shown that, although the mean range of male and female smooth snakes is similar (≈0.5–0.6 ha), males typically exhibit increased ranging movements once sexual maturity is reached (Reading, 2005). The biased dispersal observed in this study may therefore be a result of sexually mature males dispersing to find suitable mates, which can be an adaptive trait to reduce inbreeding (Dubey et al., 2008). Coronella austriaca effectively discriminates kin in a foraging context (Pernetta et al., 2009), although further work is required to understand whether this has any functional role in mate selection and male dispersal. The second possible explanation for the observed sex-biased dispersal could be a result differences in dispersal between sexes following parturition. Male neonatal slatey-grey snakes (Stegonotus cucullatus) typically disperse further from natal sites than female litter mates (Dubey et al., 2008). However, it is unknown whether this also occurs in C. austriaca because of the very-low incidence of neonate snake captures (Pernetta, 2009). Positive FIS values were recorded from all of the sampling localities within Wareham Forest, suggesting that relatively high levels of inbreeding may still be occurring. Although positive values of FIS may also be indicative of the Wahlund effect (the sampling of an assumed single population that is actually composed of two or more populations; Sinnock, 1975), in this instance this seems extremely unlikely, because of the restricted area covered by each array of artificial refugia (≈0.284 ha).
To date only four published studies have used genetic data to investigate sex-biased dispersal in snakes, all of which have found a male-bias (Rivera et al., 2006; Keogh et al., 2007; Clark et al., 2008; Dubey et al., 2008). Male-biased dispersal in mammalian species has been attributed to a female-defence polygyny mating system (Greenwood, 1980), and while polygyny has been regarded as typical for snakes (Duvall et al., 1993), more recent research suggests that this may not always be the case (Rivas and Burghardt, 2005). As a result, further work is required to determine the reproductive system of C. austriaca, through paternity analysis of litters, thereby establishing whether the observed bias in dispersal can be explained by their reproductive strategy.
From a conservation perspective our data emphasise the sedentary nature of C. austriaca on United Kingdom lowland heathland sites within commercial coniferous forest plantations; a land-use change that has been suggested as the cause of significant declines in heathland reptiles (Spellerberg, 1988). However, this study has shown that C. austriaca is able to use areas of open heath and areas containing young stands of Pinus spp. before complete canopy closure. In addition, although areas containing mature stands of trees may not represent suitable permanent habitats, the presence of isolation-by-distance effects in population genetic structuring suggests that such stands, under current management regimes, do not represent significant barriers to dispersal. Wareham forest represents a heterogeneous landscape, despite the presence of large stands of Pinus spp., as it also contains areas of forest rides, clearfelled trees and moor land, which could exert an effect as corridors for C. austriaca. As a result, movements of individuals between heathland patches are more likely to be limited by their dispersal capabilities. Inevitably, the commercial nature of such sites results in conflict between maximising profits from forestry and the conservation benefits of land for biodiversity. However, this research suggests that maintaining a mosaic of habitats within managed commercial forests could benefit smooth snake populations. Although plantation forests are not able to support the biodiversity of primary habitats, they can have a role in complementary conservation services (Barlow et al., 2007). Further research concentrating on determining the precise stage at which young forestry stands may become unsuitable habitat for smooth snakes should be seen as a priority for forest management practices.
Acknowledgments
We are grateful to the Forestry Commission and in particular Mark Warn for permission to work within Wareham Forest. Financial support for this study was provided by a Natural Environment Research Council studentship to APP (NER/S/A/2005/13642) with additional funding from the Centre for Ecology & Hydrology. APP is grateful to Inga Zeisset and Shoshanna May for laboratory advice and support. Three anonymous reviewers provided comments that greatly improved the manuscript.
The authors declare no conflict of interest.
References
- Adriaensen F, Chardon JP, De Blust G, Swinnen E, Villalba S, Gulinck H, et al. The application of ‘least-cost' modelling as a functional landscape model. Landscape Urban Plann. 2003;64:233–247. [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart K. Lositan: a workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker JP, Berendse F. Constraints in the restoration of ecological diversity in grassland and heathland communities. Trends Ecol Evol. 1999;14:63–68. doi: 10.1016/s0169-5347(98)01544-4. [DOI] [PubMed] [Google Scholar]
- Balloux F, Goudet F. Statistical properties of population differentiation estimators under stepwise mutation in a finite island model. Mol Ecol. 2002;11:771–783. doi: 10.1046/j.1365-294x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Barlow J, Gardner TA, Araujo IS, vila-Pires TC, Bonaldo AB, Costa JE, et al. Quantifying the biodiversity value of tropical primary, secondary, and plantation forests. Proc Natl Acad Sci USA. 2007;104:18555–18560. doi: 10.1073/pnas.0703333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebee TJC. Buccal swabbing as a source of DNA from squamate reptiles. Conserv Genet. 2008;9:1087–1088. [Google Scholar]
- Bond JM, Porteus R, Hughes S, Mogg RJ, Gardner MG, Reading CJ. Polymorphic microsatellite markers, isolated using a simple enrichment procedure, in the threatened smooth snake (Coronella austriaca) Mol Ecol Notes. 2005;5:42–44. [Google Scholar]
- Braithwaite AC, Buckley J, Corbett KF, Edgar PW, Haslewood ES, Haslewood GAD, et al. The distribution in England of the smooth snake (Coronella austriaca Laurenti), results of the British Herpetological society survey, 1984–7. Herpetol J. 1989;1:370–376. [Google Scholar]
- Chapman SB, Clarke RT, Webb NR. The survey and assessment of heathland in Dorset, England, for conservation. Biol Conserv. 1989;47:137–152. [Google Scholar]
- Clark RW, Brown WS, Stechert R, Zamudio KR. Integrating individual behaviour and landscape genetics: the population structure of timber rattlesnake hibernacula. Mol Ecol. 2008;17:719–730. doi: 10.1111/j.1365-294X.2007.03594.x. [DOI] [PubMed] [Google Scholar]
- DeYoung RW, Honeycutt RS. The molecular toolbox: genetic techniques in wildlife ecology and management. J Wildl Manage. 2005;69:1362–1384. [Google Scholar]
- DiLeo MF, Row JR, Lougheed SC. Discordant patterns of population structure for two co-distributed snake species across a fragmented Ontario landscape. Diversity Distrib. 2010;16:571–581. [Google Scholar]
- Dubey S, Brown GP, Madsen T, Shine R. Male-biased dispersal in a tropical Australian snake (Stegonotus cucullatus, Colubridae) Mol Ecol. 2008;17:3506–3514. doi: 10.1111/j.1365-294x.2008.03859.x. [DOI] [PubMed] [Google Scholar]
- Dubey S, Shine R. Restricted dispersal and genetic diversity in populations of an endangered montane lizard (Eulamprus leuraensis, Scincidae) Mol Ecol. 2010;19:886–897. doi: 10.1111/j.1365-294X.2010.04539.x. [DOI] [PubMed] [Google Scholar]
- Duvall D, Schuett GW, Arnold SJ.1993Ecology and evolution of mating systemsIn: Seigel, RA, Collins, JT (eds).Snakes: Ecology and Behaviour McGraw-Hill: New York; 165–200. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Favre L, Balloux F, Goudet J, Perrin N. Female-biased dispersal in the monogamous mammal Crocidura russula: evidence from field and microsatellite patterns. Proc R Soc Lond [Biol] 1997;269:127–132. doi: 10.1098/rspb.1997.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Akiyama S. An efficient method for the extraction of DNA from vertebrate tissues. Trends Genet. 1996;12:338–339. doi: 10.1016/s0168-9525(96)80005-9. [DOI] [PubMed] [Google Scholar]
- Gent AH, Spellerberg IF. Movement rates of the smooth snake Coronella austriaca (Colubridae): a radio-telemetric study. Herpetol J. 1993;3:140–146. [Google Scholar]
- Gerlach G, Musolf K. Fragmentation of landscape as a cause for genetic subdivision in bank voles. Conserv Biol. 2000;14:1066–1074. [Google Scholar]
- Gibbs HL, Prior KA, Weatherhead PJ, Johnson G. Genetic structure of populations of the threatened eastern massasauga rattlesnake, Sisturus c. catenatus: evidence from microsatellite DNA markers. Mol Ecol. 1997;6:1123–1132. doi: 10.1046/j.1365-294x.1997.00284.x. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Weatherhead PJ. Insights into population ecology and sexual selection in snakes through the application of DNA-based genetic markers. J Hered. 2001;92:173–179. doi: 10.1093/jhered/92.2.173. [DOI] [PubMed] [Google Scholar]
- Goudet J. Fstat (version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Anim Behav. 1980;28:1140–1162. [Google Scholar]
- Hanski I, Thomas CD. Metapopulation dynamics and conservation—a spatially explicit model applied to butterflies. Biol Conserv. 1994;68:167–180. [Google Scholar]
- Hitchings SP, Beebee TJC. Loss of genetic diversity and fitness in common toad (Bufo bufo) populations isolated by inimical habitat. J Evol Biol. 1998;11:269–283. [Google Scholar]
- Hoelzel AR, Fleischer RC, Campagna C, Le Boeuf BJ, Alvord G. Impact of a population bottleneck on symmetry and genetic diversity in the northern elephant seal. J Evol Biol. 2002;15:567–575. [Google Scholar]
- Holt RD, Lawton JH, Polis GA, Martinez ND. Trophic rank and species-area relationship. Ecology. 1999;80:1495–1504. [Google Scholar]
- Keogh JS, Webb JK, Shine R. Spatial genetic analysis and long-term mark recapture data demonstrate male-biased dispersal in a snake. Biol Lett. 2007;3:33–35. doi: 10.1098/rsbl.2006.0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RB, Lawson R. Patterns of population sub-division and geneflow in three sympatric natricine snakes. Copeia. 2001;3:602–614. [Google Scholar]
- Komonen A, Pentillä R, Lindgren M, Hanksi I. Forest fragmentation truncates a food chain based on an old-growth forest bracket fungus. Oikos. 2000;90:119–126. [Google Scholar]
- Lind AJ, Welsh HH, Tallmon DA. Garter snake population dynamics from a 16 year study: considerations for ecological monitoring. Ecol Appl. 2005;15:294–303. [Google Scholar]
- Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity. 2005;95:255–273. doi: 10.1038/sj.hdy.6800725. [DOI] [PubMed] [Google Scholar]
- Luiselli L, Capizzi D. Influences of area, isolation and habitat features on distribution of snakes in Mediterranean fragmented woodlands. Biodiversity Conserv. 1997;6:1339–1351. [Google Scholar]
- Macdonald DW, Johnson DDP.2001Dispersal in theory and practice: consequences for conservation biologyIn: Clobert, J, Danchin, E, Dhondt, AA, Nichols, JD (eds).Dispersal Oxford University Press: Oxford; 358–372. [Google Scholar]
- Madsen T, Stille B, Shine R. Inbreeding depression in an isolated population of adders, Vipera berus. Biol Conserv. 1996;75:113–118. [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- Manier MK, Arnold SJ. Population genetic analysis identifies source-sink dynamics for two sympatric garter snake species (Thamnophis elegans and Thamnophis sirtalis) Mol Ecol. 2005;14:3965–3976. doi: 10.1111/j.1365-294X.2005.02734.x. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Knapp RA, Pope KL. Garter snake distributions in high elevation aquatic ecosystems: is there a link with declining amphibians populations and non-native trout introductions. J Herpetol. 2002;36:16–22. [Google Scholar]
- Parker WS, Plummer MV.1987Population EcologyIn: Seigel RA, Collins JT, Novak SS (eds).Snakes: Ecology and Evolutionary Biology Macmillan: New York; 253–301. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernetta AP.2009Population ecology and conservation genetics of the smooth snake (Coronella austriaca) in a fragmented heath landscapePhD Thesis, University of Southampton.
- Pernetta AP, Reading CJ, Allen JA. Chemoreception and kin discrimination by neonate smooth snakes, Coronella austriaca. Anim Behav. 2009;77:363–368. [Google Scholar]
- Phelps TE. Seasonal movement of the snakes Coronella austriaca, Vipera berus and Natrix natrix in southern England. Brit J Herpetol. 1978;5:775–761. [Google Scholar]
- Pimm SL, Raven P. Extinction by numbers. Nature. 2000;403:843–845. doi: 10.1038/35002708. [DOI] [PubMed] [Google Scholar]
- Prior KA, Gibbs HL, Weatherhead PJ. Population genetic structure in the black rat snake: implications for management. Conserv Biol. 1997;11:1147–1158. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser MR, Gibbs HL, Weatherhead PJ. Microgeographic population genetic structure in the northern water snake, Nerodia sipedon sipedon detected using microsatellite DNA loci. Mol Ecol. 1999;8:329–333. doi: 10.1046/j.1365-294x.1999.00530.x. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, de Meeus T. Inferring sex-biased dispersal from population genetic tools: a review. Heredity. 2002;88:161–165. doi: 10.1038/sj.hdy.6800060. [DOI] [PubMed] [Google Scholar]
- Reading CJ. A proposed standard method for surveying reptiles on dry lowland heath. J Appl Ecol. 1997;34:1057–1069. [Google Scholar]
- Reading CJ. Age, growth and sex determination in a population of smooth snakes, Coronella austriaca in southern England. Amphibia-Reptilia. 2004a;25:137–150. [Google Scholar]
- Reading CJ. The influence of body condition and prey availability on female breeding success in the smooth snake (Coronella austriaca Laurenti) J Zool. 2004b;264:61–67. [Google Scholar]
- Reading CJ.2005Ranging behaviour in the smooth snake Coronella austriaca Laurenti Proceedings 5th World Congress of Herpetology. Stellenbosch, p85.
- Reading CJ, Luiselli LM, Akani GC, Bonnet X, Amori G, Ballouard JM, et al. Are snake populations in widespread decline. Bio Lett. 2010;6:777–780. doi: 10.1098/rsbl.2010.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D, Frankham R. Correlation between fitness and genetic diversity. Conserv Biol. 2003;17:230–237. [Google Scholar]
- Repaci V, Stow AJ, Briscoe DA. Fine-scale genetic structure, co-founding and multiple mating in the Australian allodapine bee Ramphocinclus brachyurus. J Zool. 2007;270:687–691. [Google Scholar]
- Rivas JA, Burghardt GM. Snake mating systems, behaviour, and evolution: the revisionary implications of recent findings. J Comp Psychol. 2005;119:447–454. doi: 10.1037/0735-7036.119.4.447. [DOI] [PubMed] [Google Scholar]
- Rivera PC, Gardenal CN, Chiaraviglio M. Sex-biased dispersal and high levels of gene flow among local populations in the argentine boa constrictor, Boa constrictor occidentalis. Austral Ecol. 2006;31:948–955. [Google Scholar]
- Rose RJ, Webb NR, Clarke RT, Traynor CH. Changes on the heathlands in Dorset, England between 1987 and 1996. Biol Conserv. 2000;93:117–125. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Sinnock P. The Wahlund effect for the two-locus model. Am Nat. 1975;109:565–570. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Spellerberg IF. Ecology and management of retile populations in forests. Q J Forest. 1988;82:99–109. [Google Scholar]
- Spellerberg IF, Phelps TE. Biology, general ecology and behaviour of the snake, Coronella austriaca Laurenti. Biol J Linn Soc. 1977;9:133–164. [Google Scholar]
- Templeton AR, Robertson RJ, Brisson J, Strasburg J. Disrupting evolutionary processes: the effects of habitat fragmentation on collared lizards in the Missouri Ozarks. Proc Natl Acad Sci USA. 2001;98:5426–5432. doi: 10.1073/pnas.091093098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Shaw K, Routman E, Davis SK. The genetic consequences of habitat fragmentation. Ann Missouri Bot Gard. 1990;77:13–27. [Google Scholar]
- van Gelder JJ, Olfers JHJ, Mertens LAJM, Kersten HLM. Field identification of the sex of the smooth snake (Coronella austriaca Laurenti) J Herpetol. 1988;22:53–60. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Weir BS. Sinauer Associates: Sunderland; 1990. Genetic data analysis: methods for discrete population genetic data. [DOI] [PubMed] [Google Scholar]