Cardiovascular disease candidate genes, including genes previously associated with type 2 diabetes and diabetic nephropathy, were not associated with diabetic retinopathy, although a limited number of variants merit further investigation in larger cohorts.
Abstract
Purpose.
To investigate whether variants in cardiovascular candidate genes, some of which have been previously associated with type 2 diabetes (T2D), diabetic retinopathy (DR), and diabetic nephropathy (DN), are associated with DR in the Candidate gene Association Resource (CARe).
Methods.
Persons with T2D who were enrolled in the study (n = 2691) had fundus photography and genotyping of single nucleotide polymorphisms (SNPs) in 2000 candidate genes. Two case definitions were investigated: Early Treatment Diabetic Retinopathy Study (ETDRS) grades ≥14 and ≥30. The χ2 analyses for each CARe cohort were combined by Cochran-Mantel-Haenszel (CMH) pooling of odds ratios (ORs) and corrected for multiple hypothesis testing. Logistic regression was performed with adjustment for other DR risk factors. Results from replication in independent cohorts were analyzed with CMH meta-analysis methods.
Results.
Among 39 genes previously associated with DR, DN, or T2D, three SNPs in P-selectin (SELP) were associated with DR. The strongest association was to rs6128 (OR = 0.43, P = 0.0001, after Bonferroni correction). These associations remained significant after adjustment for DR risk factors. Among other genes examined, several variants were associated with DR with significant P values, including rs6856425 tagging α-l-iduronidase (IDUA) (P = 2.1 × 10−5, after Bonferroni correction). However, replication in independent cohorts did not reveal study-wide significant effects. The P values after replication were 0.55 and 0.10 for rs6128 and rs6856425, respectively.
Conclusions.
Genes associated with DN, T2D, and vascular diseases do not appear to be consistently associated with DR. A few genetic variants associated with DR, particularly those in SELP and near IDUA, should be investigated in additional DR cohorts.
Diabetic retinopathy (DR) is the leading cause of blindness in working-age Americans1,2 and is increasing in prevalence as rates of type 2 diabetes (T2D) soar worldwide.3,4 The frequency and severity of DR are heterogeneous within and across ethnic groups,5 even with adjustment for risk factors such as duration of diabetes and glycemic control.2,6,7 There are people who have a long duration of diabetes without DR and those who have severe DR despite relatively good glycemic control. For these reasons, genetic risk factors are thought to play a role in DR. Heritability has been estimated to be as high as 27% for DR and 52% for proliferative diabetic retinopathy (PDR).8–10 However, genetic association studies for DR have been thus far limited mostly to studies of one or a modest number of candidate genes.11,12 Most reported associations have not been consistently reproduced.11,13,14
In contrast to DR, genetic association studies for T2D have revealed many consistently associated genes. Genes that increase T2D risk may also predispose to development of retinopathy. In the case of diabetic nephropathy (DN), a TCF7L2 variant increases the risk of developing DN beyond the risk of diabetes.15 Because there is evidence that DR shares risk factors and pathophysiological mechanisms with DN and macrovascular diabetic complications,6,16–21 genes associated with DN and atherosclerotic vascular disease may also be associated with DR.
The Candidate gene Association Resource (CARe) is a collaboration for association analyses of genotypes and cardiovascular disease phenotypes.22 It comprises >40,000 participants from nine cohorts who have been genotyped for 49,320 single nucleotide polymorphisms (SNPs) from approximately 2,000 candidate genes postulated or known to increase risk of cardiovascular, metabolic, and inflammatory diseases.23 It includes 2691 T2D participants with fundus photographs of multiple ethnicities. Thus, the CARe framework provides an opportunity to investigate genetic associations for DR with a candidate gene approach. CARe genotyped many genes previously associated with DR,24,25 DN,25–27 and T2D.28–34 The first purpose of this study was to investigate whether these genes are also associated with the presence of DR in CARe. The second purpose was to determine whether the remaining genes included in the CARe genotyping platform, which were also chosen as potential cardiovascular disease genes, are associated with DR.
Methods
Study Population and Fundus Photography Procedures
Four CARe cohorts have fundus photographs of T2D participants: Atherosclerosis Risk in Communities (ARIC) Study, Cardiovascular Health Study (CHS), Jackson Heart Study (JHS), and Multiethnic Study of Atherosclerosis (MESA).35–38 T2D was defined according to the American Diabetes Association 2003 Criteria.39 The fundus photography protocol for each cohort is described in Table 1.40–42 In all studies, except for the JHS, fundus photographs were graded by masked readers at the University of Wisconsin Ocular Epidemiology Reading Center according to the modified Airlie House Classification system.43 Fundus photographs for the JHS were graded by masked JHS ophthalmologist investigators according to the same criteria.
Table 1.
CARe Participants with T2D and DR Grading by Cohort and Ethnicity
| Cohort/Population | Eyes Photographed per Participant (n) | Fields Photographed per Eye (n) | Size of Each Field Photographed (deg) | Participants with T2D, DR Grading and IBC Chip Genotyping (n) | Participants with ETDRS Grade <14 (n) | Participants with ETDRS Grade ≥14(n) | Participants with ETDRS Grade ≥30 (n) |
|---|---|---|---|---|---|---|---|
| ARIC | |||||||
| EA | One | One | 45 | 885 | 732 | 153 | 91 |
| AfrA | 439 | 315 | 124 | 95 | |||
| CHS | |||||||
| EA | One | One | 45 | 193 | 160 | 33 | 20 |
| AfrA | 54 | 35 | 19 | 15 | |||
| JHS | |||||||
| AfrA | Two | Seven | 30 | 55 | 26 | 29 | 22 |
| MESA | |||||||
| EA | Two | Two | 45 | 176 | 140 | 36 | 11 |
| AfrA | 275 | 176 | 99 | 57 | |||
| AsA | 79 | 54 | 25 | 14 | |||
| HA | 231 | 151 | 80 | 46 | |||
| All | |||||||
| EA | 1254 | 1032 | 222 | 122 | |||
| AfrA | 823 | 552 | 271 | 189 | |||
| AsA | 79 | 54 | 25 | 14 | |||
| HA | 231 | 151 | 80 | 46 |
EA, European American; AfrA, African American; AsA, Asian American; HA, Hispanic American.
Definition of Diabetic Retinopathy
We examined two DR phenotypes. First we defined cases as participants with an Early Treatment Diabetic Retinopathy Study (ETDRS) grade ≥14 in the eye with the higher ETDRS grade or in the only eye photographed, depending on the study's protocol. These analyses were designed to detect associations with the presence of any DR. Our second phenotype defined cases as participants with ETDRS grade ≥30. The latter was intended to reduce misclassification of patients with minimal signs of DR, which may be seen even in persons without diabetes.44–46 For all analyses, controls were defined as T2D participants with an ETDRS grade <14 (no DR).
Measurement of Other Variables
Data on DR risk factors were obtained from the study examination at which fundus photography was performed. These included duration of diabetes, fasting blood glucose, systolic and diastolic blood pressures, and fasting total cholesterol.47–50 The procedures for measuring these variables are described in online documentation (www.cscc.unc.edu/aric/, www.chs-nhlbi.org, and www.mesa-nhlbi.org, provided by the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD; and www.jsums.edu/jhs, JHS, Jackson State University, Jackson, MS). Some participants were unaware of a diabetes diagnosis and received the diagnosis based on their laboratory values at the study visit at which they also had fundus photography. For these patients, the duration of diabetes was calculated by halving the number of years between their prior study visit (when they did not meet criteria for T2D) and the visit at which they met criteria. If data on a risk factor were not measured at the fundus photography visit, the information was obtained from the visit closest in time to the fundus photography visit.
Genotyping
CARe participant DNA samples were interrogated on a custom genotyping array (iSelect ITMAT-Broad-CARe [IBC] Chip; Illumina, San Diego, CA). Its design is described elsewhere.23 SNP selection criteria and genotyping quality control (QC) procedures are explained in the Supplementary Methods (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental).23
Statistical Analysis
We first investigated genes that have been previously associated with DR, DN, and T2D. For DR, we chose the genes that had the most robust evidence of association from a comprehensive review of the literature24 and a subsequent strong association with the erythropoietin (EPO) gene promoter.25 For DN, we chose genes that have shown nominal associations (P < 0.05) with DN or a related quantitative trait.25–27 For T2D, we chose genes with SNPs that had met genome-wide significance.28–31,33 Supplementary Table S1 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental) lists the 39 genes included on the IBC chip that met these criteria. In the second phase, we investigated the remaining genes on the IBC chip which were primarily cardiovascular candidate genes.23
For genetic association testing, we used χ2 analysis comparing European-American cases (participants with T2D and DR) to controls (participants with T2D and no DR) in each CARe cohort. Results were combined by Cochran-Mantel-Haenszel (CMH) pooling of the odds ratios (ORs).51,52 This CMH method is a robust way of maintaining consistency with individual study ORs while estimating a single fixed-effects OR across all cohorts. We report correction for the multiple association tests performed with per gene Bonferroni correction and permutation testing. Per gene Bonferroni correction was performed because it is an intuitive, easily understood correction. This Bonferroni correction is not as conservative as the one that corrects for the total number of unique genetic loci tested, as would be represented by correcting for the total number of tag SNPs that are not in strong LD. For this reason, we also present the empiric permutation testing correction, which, although less intuitive, does account for this LD between SNPs. We defined statistical significance as P < 0.05 after per gene Bonferroni correction. We chose this threshold for the discovery phase of the experiment, although it is a less stringent threshold than one based on a correction that could completely account for LD between SNPs, to minimize type II error (false negatives) in this initial phase. We apply a more stringent threshold for the replication phase (see below), and the final decision of whether an SNP is truly associated is based on its final P value after replication. For SNPs that were statistically significant in the discovery phase, we performed haplotype analyses using the omnibus test and further examined the associations with logistic regression models that included other DR risk factors. Age and duration of diabetes were defined as continuous variables in years. Fasting glucose and total cholesterol were incorporated as continuous variables in milligrams per deciliter. Systolic and diastolic blood pressures were evaluated as continuous variables (in mm Hg). If a participant was taking antihypertension medication, 15 and 10 mm Hg were added to the systolic and diastolic blood pressure values, respectively.53 Sex and study site were also incorporated. All statistical analyses were performed in PLINK.54
Replication
Top significant findings were pursued in the non-European American populations in CARe and in independent Caucasian cohorts with genome-wide genotyping results: Age, Gene/Environment Susceptibility (AGES) study55; Blue Mountains Eye Study (BMES)56; Genetics of Diabetes Audit and Research Tayside Study (Go-DARTS)57; Finnish Diabetic Nephropathy (FinnDiane) Study9; Family Investigation of Nephropathy and Diabetes-Eye (FIND-Eye) Study10; Singapore Malay Eye Study (SiMES)58; and the Singapore Prospective Study Program (SP2).59 The Medical University of Lublin T2D cohort60 performed de novo genotyping for replication. The phenotyping protocols for these studies are described in the Supplementary Methods and Supplementary Table S2 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). Because ETDRS grading was not used by all cohorts, phenotype data were harmonized into two categories that were analogous to ETDRS grade ≥14 and ETDRS grade ≥30. Meta-analysis of the discovery cohort and replication cohorts results was performed by CMH pooling of ORs. Statistical heterogeneity was assessed with Cochran's Q statistic.61,62 The Q statistic calculates a weighted sum of the square distances of the observed effects from the null hypothesis of equality of the effects. The weight for each study is the inverse of the variance of the effect estimator so that larger and more accurate studies are weighted more heavily. Statistical significance after replication was defined as a meta-analysis P < 1 × 10−6. This threshold was determined empirically on the basis of the number of genes tested, is analogous to the threshold for genome-wide significance of 5.0 × 10−8 for GWAS,63 and corresponds approximately to a P < 0.05, after Bonferroni correction.
This research adhered to the tenets of the Declaration of Helsinki and was approved by the institutional review boards of the Massachusetts Eye and Ear Infirmary and the primary cohorts. Informed consent was obtained from all participants.
Results
Table 1 shows the number of T2D CARe participants by cohort and ethnicity. The prevalence of DR is similar among the cohorts, although it is higher in the JHS African-American population when compared to the other African-American populations. This discrepancy is probably secondary to the more precise phenotyping performed in the JHS with seven-field photography. Duration of diabetes and fasting glucose levels were not significantly different between the JHS cohort's and the other cohorts' African-American populations.
Table 2 shows the most significant associations for the analysis of the DR, DN, and T2D genes with any DR in European Americans. Only the associations to three SNPs in the P-selectin gene (SELP) were significant (P < 0.05, after Bonferroni correction). The three associated SNPs tagged the only associated haplotype (Supplementary Table S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). In logistic regression including other DR risk factors, the associations to rs6128, rs6133, and rs3917779 remained significant (P = 0.026, 0.022, and 0.026, respectively). The mean values for covariates are presented in Supplementary Table S4 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental).
Table 2.
Top Association Results in European-American Samples with Cases Defined as ETDRS Grade ≥14 for Variants within Genes Previously Associated with DR, DN, or T2D
| SNP | Chr | Gene | Position | Minor Allele | ARIC |
CHS |
MESA |
CMH Combined Analysis |
P Correction |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
OR | P | MAF |
OR | P | MAF |
OR | P | MAF |
OR | L95 | U95 | P | Bonferroni | Permutation | |||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |||||||||||||||||
| rs6128 | 1 | SELP | 167829528 | T | 0.08 | 0.16 | 0.48 | 0.0007 | 0.06 | 0.18 | 0.30 | 0.02 | 0.08 | 0.20 | 0.36 | 0.02 | 0.08 | 0.17 | 0.43 | 0.31 | 0.62 | 3.1 × 10−6 | 0.0001 | 0.0001 |
| rs6133 | 1 | SELP | 167831970 | A | 0.05 | 0.12 | 0.39 | 0.0003 | 0.03 | 0.13 | 0.22 | 0.02 | 0.07 | 0.13 | 0.50 | 0.16 | 0.05 | 0.12 | 0.38 | 0.25 | 0.59 | 1.1 × 10−5 | 0.0004 | 0.0001 |
| rs3917779 | 1 | SELP | 167837472 | A | 0.05 | 0.12 | 0.40 | 0.0004 | 0.03 | 0.12 | 0.23 | 0.03 | 0.07 | 0.13 | 0.50 | 0.16 | 0.05 | 0.12 | 0.39 | 0.26 | 0.60 | 1.6 × 10−5 | 0.0006 | 0.0001 |
| rs12262390 | 10 | HHEX | 94436103 | C | 0.12 | 0.09 | 1.48 | 0.05 | 0.15 | 0.10 | 1.61 | 0.22 | 0.18 | 0.09 | 2.27 | 0.03 | 0.14 | 0.09 | 1.61 | 1.18 | 2.20 | 0.002 | 0.10 | 0.004 |
| rs9356754 | 6 | CDKAL1 | 20916721 | G | 0.51 | 0.43 | 1.37 | 0.01 | 0.53 | 0.44 | 1.43 | 0.18 | 0.40 | 0.38 | 1.09 | 0.74 | 0.49 | 0.42 | 1.32 | 1.08 | 1.63 | 0.007 | 0.29 | 0.008 |
| rs9465904 | 6 | CDKAL1 | 20929641 | C | 0.37 | 0.30 | 1.38 | 0.01 | 0.42 | 0.33 | 1.49 | 0.15 | 0.28 | 0.26 | 1.08 | 0.79 | 0.36 | 0.30 | 1.34 | 1.08 | 1.65 | 0.008 | 0.33 | 0.01 |
| rs11466493 | 3 | TGFBR2 | 30661786 | G | 0.01 | 0.04 | 0.14 | 0.002 | 0.06 | 0.05 | 1.23 | 0.72 | 0.04 | 0.07 | 0.59 | 0.41 | 0.02 | 0.05 | 0.41 | 0.21 | 0.80 | 0.009 | 0.35 | 0.01 |
| rs17025862 | 3 | TGFBR2 | 30660132 | G | 0.01 | 0.04 | 0.14 | 0.002 | 0.06 | 0.05 | 1.23 | 0.72 | 0.04 | 0.07 | 0.59 | 0.41 | 0.02 | 0.05 | 0.41 | 0.21 | 0.80 | 0.009 | 0.35 | 0.01 |
| rs7747989 | 6 | CDKAL1 | 20921354 | C | 0.37 | 0.30 | 1.35 | 0.02 | 0.44 | 0.33 | 1.60 | 0.09 | 0.28 | 0.26 | 1.08 | 0.79 | 0.36 | 0.30 | 1.33 | 1.07 | 1.65 | 0.009 | 0.36 | 0.01 |
| rs2328572 | 6 | CDKAL1 | 21323815 | A | 0.003 | 0.02 | 0.15 | 0.03 | 0 | 0.03 | NA | NA | 0.01 | 0.03 | 0.48 | 0.48 | 0.005 | 0.02 | 0.19 | 0.05 | 0.69 | 0.01 | 0.44 | 0.01 |
| rs10214694 | 6 | CDKAL1 | 21321533 | T | 0.003 | 0.02 | 0.15 | 0.03 | 0 | 0.03 | NA | NA | 0.01 | 0.03 | 0.48 | 0.48 | 0.005 | 0.02 | 0.19 | 0.05 | 0.69 | 0.01 | 0.45 | 0.01 |
| rs12190631 | 6 | CDKAL1 | 21020651 | G | 0.05 | 0.03 | 1.38 | 0.30 | 0.12 | 0.02 | 6.17 | 0.0001 | 0.01 | 0.01 | 1.29 | 0.83 | 0.05 | 0.03 | 1.85 | 1.14 | 3.01 | 0.01 | 0.51 | 0.02 |
| rs2275729 | 10 | HHEX | 94442410 | G | 0.15 | 0.12 | 1.32 | 0.13 | 0.15 | 0.12 | 1.29 | 0.51 | 0.21 | 0.10 | 2.26 | 0.02 | 0.16 | 0.12 | 1.44 | 1.08 | 1.92 | 0.01 | 0.52 | 0.02 |
| rs1511024 | 4 | FABP2 | 120459629 | T | 0.05 | 0.03 | 1.74 | 0.07 | 0.05 | 0.02 | 3.00 | 0.12 | 0.06 | 0.03 | 1.76 | 0.35 | 0.05 | 0.03 | 1.86 | 1.13 | 3.06 | 0.01 | 0.55 | 0.01 |
| rs9350294 | 6 | CDKAL1 | 20978072 | T | 0.38 | 0.32 | 1.26 | 0.07 | 0.44 | 0.32 | 1.65 | 0.07 | 0.33 | 0.29 | 1.24 | 0.45 | 0.38 | 0.32 | 1.31 | 1.05 | 1.61 | 0.01 | 0.55 | 0.01 |
The minor allele is the effect allele for the ORs. Chr, chromosome; L95, lower 95% CI boundary; U95, upper 95% CI boundary; NA, not available.
Table 3 shows the most significant associations from the analysis of the DR, DN, and T2D candidate genes with DR defined as an ETDRS grade ≥30 in European Americans. The three SELP SNPs, along with five SNPs in the fat mass and obesity-associated (FTO) gene, were significantly associated (P < 0.05, after Bonferroni correction). Variants rs12708942, rs9806929, and rs4783824 tagged the only associated haplotype (Supplementary Table S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). Because the effect of FTO on T2D risk is mediated by its effect on obesity, we executed a logistic regression model including age, sex, and body mass index (BMI). BMI was incorporated as a continuous variable (weight in kilograms divided by height in meters squared). Three FTO SNPs continued to be associated with DR (P = 0.002, 0.002, and 0.001 for rs9926180, rs7500562, and rs12149433, respectively). Of note, an EPO variant, rs551238, was associated with this more stringent definition of DR and had a P < 0.05 after permutation correction but not after Bonferroni correction.
Table 3.
Top Association Results in European-American Samples with Cases Defined as ETDRS Grade ≥30 for Variants in Genes Previously Associated with DR, DN, or T2D
| SNP | Chr | Gene | BP | Minor Allele | ARIC |
CHS |
MESA |
CMH Combined Analysis |
P Correction |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
OR | P | MAF |
OR | P | MAF |
OR | P | MAF |
OR | L95 | U95 | P | Bonferroni | Permutation | |||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |||||||||||||||||
| rs6133 | 1 | SELP | 167831970 | A | 0.04 | 0.12 | 0.34 | 0.002 | 0 | 0.12 | NA | NA | 0.09 | 0.12 | 0.74 | 0.69 | 0.05 | 0.12 | 0.32 | 0.17 | 0.58 | 2.3 × 10−4 | 0.009 | 0.0002 |
| rs3917779 | 1 | SELP | 167837472 | A | 0.04 | 0.12 | 0.35 | 0.003 | 0 | 0.12 | NA | NA | 0.09 | 0.12 | 0.74 | 0.69 | 0.05 | 0.12 | 0.32 | 0.17 | 0.59 | 2.9 × 10−4 | 0.01 | 0.0001 |
| rs9926180 | 16 | FTO | 52486108 | T | 0.33 | 0.25 | 1.51 | 0.01 | 0.38 | 0.25 | 1.81 | 0.08 | 0.32 | 0.17 | 2.32 | 0.07 | 0.34 | 0.24 | 1.65 | 1.24 | 2.18 | 5.0 × 10−4 | 0.02 | 0.001 |
| rs7500562 | 16 | FTO | 52488391 | C | 0.33 | 0.25 | 1.51 | 0.01 | 0.38 | 0.25 | 1.81 | 0.08 | 0.32 | 0.17 | 2.27 | 0.08 | 0.34 | 0.24 | 1.64 | 1.24 | 2.17 | 5.3 × 10−4 | 0.02 | 0.001 |
| rs12149433 | 16 | FTO | 52485580 | G | 0.15 | 0.09 | 1.75 | 0.01 | 0.20 | 0.08 | 2.84 | 0.01 | 0.05 | 0.04 | 1.07 | 0.95 | 0.15 | 0.09 | 1.93 | 1.33 | 2.81 | 5.5 × 10−4 | 0.02 | 0.001 |
| rs6128 | 1 | SELP | 167829528 | T | 0.08 | 0.16 | 0.49 | 0.009 | 0.05 | 0.17 | 0.26 | 0.05 | 0.09 | 0.18 | 0.45 | 0.27 | 0.08 | 0.16 | 0.44 | 0.27 | 0.70 | 5.6 × 10−4 | 0.02 | 0.001 |
| rs13335343 | 16 | FTO | 52488941 | A | 0.15 | 0.09 | 1.74 | 0.01 | 0.20 | 0.08 | 2.84 | 0.01 | 0.05 | 0.04 | 1.07 | 0.95 | 0.15 | 0.09 | 1.92 | 1.32 | 2.80 | 6.1 × 10−4 | 0.02 | 0.001 |
| rs12935710 | 16 | FTO | 52500306 | T | 0.32 | 0.23 | 1.55 | 0.01 | 0.33 | 0.25 | 1.48 | 0.28 | 0.27 | 0.15 | 2.09 | 0.14 | 0.32 | 0.22 | 1.61 | 1.21 | 2.14 | 0.001 | 0.04 | 0.002 |
| rs10852525 | 16 | FTO | 52496582 | A | 0.17 | 0.12 | 1.52 | 0.05 | 0.20 | 0.10 | 2.29 | 0.05 | 0.09 | 0.05 | 2.09 | 0.34 | 0.17 | 0.11 | 1.71 | 1.20 | 2.45 | 0.003 | 0.12 | 0.005 |
| rs9929152 | 16 | FTO | 52496904 | G | 0.33 | 0.25 | 1.45 | 0.03 | 0.35 | 0.26 | 1.53 | 0.23 | 0.32 | 0.20 | 1.89 | 0.18 | 0.33 | 0.25 | 1.52 | 1.15 | 2.02 | 0.004 | 0.14 | 0.006 |
| rs4783824 | 16 | FTO | 52509162 | T | 0.13 | 0.09 | 1.50 | 0.09 | 0.20 | 0.08 | 2.95 | 0.01 | 0.05 | 0.03 | 1.69 | 0.62 | 0.14 | 0.08 | 1.78 | 1.21 | 2.64 | 0.004 | 0.15 | 0.005 |
| rs1362570 | 16 | FTO | 52491048 | C | 0.14 | 0.10 | 1.52 | 0.07 | 0.20 | 0.09 | 2.45 | 0.03 | 0.05 | 0.04 | 1.07 | 0.95 | 0.14 | 0.09 | 1.70 | 1.16 | 2.48 | 0.007 | 0.26 | 0.009 |
| rs4611524 | 17 | FTO | 58945384 | T | 0.31 | 0.41 | 0.67 | 0.02 | 0.33 | 0.42 | 0.68 | 0.27 | 0.36 | 0.40 | 0.86 | 0.74 | 0.32 | 0.41 | 0.69 | 0.52 | 0.91 | 0.009 | 0.34 | 0.009 |
| rs12708942 | 16 | FTO | 52503705 | A | 0.14 | 0.10 | 1.46 | 0.1 | 0.20 | 0.09 | 2.63 | 0.02 | 0.05 | 0.04 | 1.15 | 0.89 | 0.14 | 0.09 | 1.68 | 1.14 | 2.47 | 0.009 | 0.35 | 0.01 |
| rs9806929 | 16 | FTO | 52507417 | A | 0.14 | 0.10 | 1.46 | 0.1 | 0.20 | 0.09 | 2.63 | 0.02 | 0.05 | 0.04 | 1.15 | 0.89 | 0.14 | 0.09 | 1.68 | 1.14 | 2.47 | 0.009 | 0.35 | 0.01 |
| rs551238 | 7 | EPO | 100159464 | G | 0.34 | 0.40 | 0.75 | 0.08 | 0.20 | 0.40 | 0.37 | 0.01 | 0.32 | 0.36 | 0.82 | 0.67 | 0.31 | 0.40 | 0.69 | 0.52 | 0.91 | 0.009 | 0.37 | 0.01 |
The minor allele is the effect allele for the ORs. Chr, chromosome; L95, lower 95% CI boundary; U95, upper 95% CI boundary; NA, not available.
We then examined the strength of these associations in the CARe non-European populations. For any DR, the associations with the SELP SNPs were not replicated in the African-, Hispanic-, or Asian-American populations (Table 4). When cases were defined as ETDRS grade ≥30, the SNPs in SELP and FTO were not associated, and in logistic regression models including age, sex, and BMI, no association with the FTO SNPs was detected (data not shown).
Table 4.
CHM Association Results for SELP SNPs in non-European-American CARe Populations, with Cases Defined as ETDRS Grade ≥14
| SNP | Minor Allele | MAF |
OR | L95 | U95 | P | |
|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||
| African American | |||||||
| rs6128 | T | 0.5 | 0.46 | 1.17 | 0.95 | 1.44 | 0.14 |
| rs6133 | C | 0.42 | 0.45 | 0.89 | 0.72 | 1.09 | 0.26 |
| rs3917779 | G | 0.48 | 0.50 | 0.94 | 0.76 | 1.15 | 0.55 |
| Hispanic American | |||||||
| rs6128 | T | 0.29 | 0.26 | 1.14 | 0.74 | 1.75 | 0.55 |
| rs6133 | A | 0.16 | 0.18 | 0.91 | 0.55 | 1.52 | 0.72 |
| rs3917779 | A | 0.14 | 0.16 | 0.89 | 0.52 | 1.52 | 0.67 |
| Asian American | |||||||
| rs6128 | C | 0.24 | 0.27 | 0.86 | 0.40 | 1.87 | 0.70 |
| rs6133 | A | 0 | 0 | NA | NA | NA | NA |
| rs3917779 | A | 0 | 0 | NA | NA | NA | NA |
L95, lower 95% CI boundary; U95, upper 95% CI boundary; NA, not available.
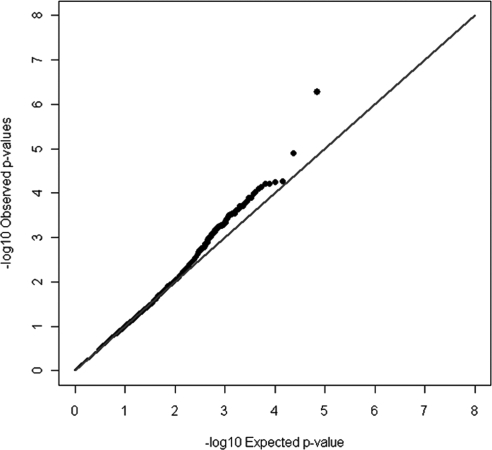
In the second phase of the analysis, we examined the remaining genes on the IBC chip in European Americans. The top association results for DR defined as ETDRS grade ≥14 and ETDRS grade ≥30 are shown in Tables 5 and 6, respectively. The lambdas for the quantile–quantile (Q–Q) plots were 1.01 and 1.00, respectively; Fig. 1). Supplementary Table S5 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental) shows the results from the principal components analysis. There was no evidence of population stratification. Several variants in the two analyses had associations that were significant after Bonferroni correction. One variant, rs6856425, was significantly associated with DR in all three CARe cohorts for both definitions of DR with P = 2.1 × 10−5 after Bonferroni correction in the ETDRS grade ≥30 analysis. This association could not be replicated in the CARe African American cohorts (MAF 18%, OR = 0.94, P = 0.69).
Table 5.
Top Association Results in European-American Samples with Cases Defined as ETDRS Score ≥14 for Variants in Genes on the IBC Chip
| SNP | Chr | Gene | Position | Minor Allele | ARIC |
CHS |
MESA |
CMH Combined Analysis |
P correction |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
OR | P | MAF |
OR | P | MAF |
OR | P | MAF |
OR | L95 | U95 | P | Bonferroni | Permutation | |||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |||||||||||||||||
| rs9332570 | 1 | F5 | 167794699 | G | 0.09 | 0.18 | 0.44 | 8.8 × 10−5 | 0.08 | 0.19 | 0.35 | 0.02 | 0.11 | 0.21 | 0.46 | 0.05 | 0.09 | 0.19 | 0.43 | 0.31 | 0.61 | 1.1 × 10−6 | 4.3 × 10−5 | 0.0001 |
| rs7545236 | 1 | F5 | 167796694 | C | 0.09 | 0.18 | 0.44 | 9.4 × 10−5 | 0.08 | 0.19 | 0.35 | 0.02 | 0.11 | 0.21 | 0.46 | 0.05 | 0.09 | 0.19 | 0.44 | 0.31 | 0.61 | 1.2 × 10−6 | 4.6 × 10−5 | 0.0001 |
| rs2298908 | 1 | F5 | 167805168 | T | 0.09 | 0.18 | 0.45 | 0.0001 | 0.08 | 0.19 | 0.35 | 0.02 | 0.11 | 0.21 | 0.46 | 0.05 | 0.09 | 0.19 | 0.44 | 0.31 | 0.61 | 1.3 × 10−6 | 4.9 × 10−5 | 0.0001 |
| rs1894701 | 1 | F5 | 167797210 | C | 0.09 | 0.18 | 0.44 | 8.8 × 10−5 | 0.08 | 0.19 | 0.36 | 0.03 | 0.11 | 0.21 | 0.46 | 0.05 | 0.09 | 0.19 | 0.44 | 0.31 | 0.61 | 1.3 × 10−6 | 5.0 × 10−5 | 0.0001 |
| rs6427203 | 1 | F5 | 167795655 | C | 0.09 | 0.18 | 0.45 | 0.0001 | 0.08 | 0.19 | 0.36 | 0.03 | 0.11 | 0.21 | 0.46 | 0.05 | 0.09 | 0.18 | 0.44 | 0.32 | 0.62 | 1.8 × 10−6 | 7.1 × 10−5 | 0.0001 |
| rs35260 | 5 | PDE4D | 58668304 | A | 0.38 | 0.47 | 0.68 | 0.0003 | 0.36 | 0.48 | 0.61 | 0.07 | 0.24 | 0.50 | 0.31 | 7.4 × 10−5 | 0.35 | 0.48 | 0.60 | 0.48 | 0.74 | 2.0 × 10−6 | 7.6 × 10−5 | 0.0001 |
| rs6128 | 1 | SELP | 167829528 | T | 0.08 | 0.16 | 0.48 | 0.0007 | 0.06 | 0.18 | 0.30 | 0.02 | 0.08 | 0.20 | 0.36 | 0.02 | 0.08 | 0.17 | 0.43 | 0.31 | 0.62 | 3.1 × 10−6 | 0.0001 | 0.0001 |
| rs7168655 | 15 | PCSK6 | 99782444 | A | 0.43 | 0.32 | 1.60 | 0.0002 | 0.47 | 0.33 | 1.76 | 0.04 | 0.43 | 0.33 | 1.50 | 0.13 | 0.44 | 0.33 | 1.61 | 1.31 | 1.98 | 7.6 × 10−6 | 0.0003 | 0.0001 |
| rs35259 | 5 | PDE4D | 58669348 | T | 0.39 | 0.47 | 0.70 | 0.005 | 0.36 | 0.48 | 0.61 | 0.07 | 0.26 | 0.50 | 0.36 | 0.0004 | 0.36 | 0.48 | 0.62 | 0.50 | 0.77 | 8.6 × 10−6 | 0.0003 | 0.0001 |
| rs6133 | 1 | SELP | 167831970 | A | 0.05 | 0.12 | 0.39 | 0.0003 | 0.03 | 0.13 | 0.22 | 0.02 | 0.07 | 0.13 | 0.50 | 0.16 | 0.05 | 0.12 | 0.38 | 0.25 | 0.59 | 1.1 × 10−5 | 0.0004 | 0.0001 |
| rs9332579 | 1 | F5 | 167791936 | T | 0.06 | 0.14 | 0.37 | 8.9 × 10−5 | 0.05 | 0.13 | 0.32 | 0.05 | 0.10 | 0.14 | 0.66 | 0.34 | 0.06 | 0.14 | 0.41 | 0.28 | 0.61 | 1.3 × 10−5 | 0.0005 | 0.0001 |
| rs3917779 | 1 | SELP | 167837472 | A | 0.05 | 0.12 | 0.40 | 0.0004 | 0.03 | 0.12 | 0.23 | 0.03 | 0.07 | 0.13 | 0.50 | 0.16 | 0.05 | 0.12 | 0.39 | 0.26 | 0.60 | 1.6 × 10−5 | 0.0006 | 0.0002 |
| rs6856425 | 4 | Near IDUA | 966918 | C | 0.04 | 0.02 | 2.36 | 0.01 | 0.11 | 0.02 | 6.17 | 0.0004 | 0.06 | 0.03 | 2.28 | 0.19 | 0.05 | 0.02 | 2.88 | 1.75 | 4.73 | 2.9 × 10−5 | 0.001 | 0.0002 |
| rs1824159 | 5 | PDE4D | 58654401 | T | 0.15 | 0.09 | 1.87 | 0.0006 | 0.11 | 0.09 | 1.15 | 0.76 | 0.19 | 0.08 | 2.81 | 0.004 | 0.15 | 0.09 | 1.88 | 1.39 | 2.53 | 3.5 × 10−5 | 0.001 | 0.0001 |
| rs1006273 | 15 | PCSK6 | 99782862 | A | 0.52 | 0.43 | 1.47 | 0.002 | 0.59 | 0.43 | 1.93 | 0.02 | 0.51 | 0.42 | 1.48 | 0.14 | 0.53 | 0.43 | 1.53 | 1.24 | 1.88 | 5.6 × 10−5 | 0.002 | 0.0001 |
| rs16879334 | 5 | MTRR | 7944506 | G | NA | NA | NA | NA | 0.11 | 0.03 | 3.33 | 0.01 | 0.10 | 0.02 | 5.88 | 0.001 | 0.10 | 0.03 | 4.11 | 2.05 | 8.21 | 6.4 × 10−5 | 0.002 | 0.0003 |
| rs10036406 | 5 | PDE4D | 58588070 | C | 0.16 | 0.10 | 1.82 | 0.0007 | 0.12 | 0.10 | 1.29 | 0.55 | 0.18 | 0.09 | 2.14 | 0.04 | 0.16 | 0.10 | 1.78 | 1.34 | 2.38 | 9.1 × 10−5 | 0.004 | 0.0001 |
| rs7105871 | 11 | CADM1 | 114717935 | C | 0.14 | 0.26 | 0.46 | 6.6 × 10−6 | 0.24 | 0.25 | 0.98 | 0.94 | 0.25 | 0.28 | 0.87 | 0.65 | 0.17 | 0.26 | 0.59 | 0.46 | 0.77 | 9.4 × 10−5 | 0.004 | 0.0002 |
The minor allele is the effect allele for the ORs. Chr, chromosome; L95, lower 95% CI boundary; U95, upper 95% CI boundary; NA, not available.
Table 6.
Top Association Results in European-American Samples with Cases Defined as ETDRS Grade ≥30 for Variants within All Genes on the IBC Chip
| SNP | Chr | Gene | Position | Minor Allele | ARIC |
CHS |
MESA |
CMH Combined Analysis |
P Correction |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
OR | P | MAF |
OR | P | MAF |
OR | P | MAF |
OR | L95 | U95 | P | Bonferroni | Permutation | |||||||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |||||||||||||||||
| rs6856425 | 4 | Near IDUA | 966918 | C | 0.05 | 0.02 | 3.01 | 0.002 | 0.15 | 0.02 | 8.50 | 1.8 × 10−5 | 0.09 | 0.03 | 3.54 | 0.10 | 0.07 | 0.02 | 3.83 | 2.26 | 6.46 | 5.3 × 10−7 | 2.1 × 10−5 | 0.0001 |
| rs1555179 | 6 | MTHFDIL | 151313049 | A | 0.19 | 0.35 | 0.45 | 3.0 × 10−5 | NA | NA | NA | NA | 0.18 | 0.32 | 0.47 | 0.17 | 0.19 | 0.34 | 0.45 | 0.32 | 0.65 | 1.3 × 10−5 | 0.0005 | 0.0002 |
| rs7078169 | 10 | CUBN | 17202320 | T | 0.09 | 0.03 | 2.86 | 0.0002 | 0.10 | 0.03 | 3.71 | 0.02 | 0.05 | 0.05 | 0.93 | 0.94 | 0.09 | 0.04 | 2.66 | 1.65 | 4.27 | 5.3 × 10−5 | 0.002 | 0.0001 |
| rs3901540 | 5 | PDE4D | 58465490 | T | 0.15 | 0.28 | 0.44 | 0.0001 | 0.18 | 0.29 | 0.52 | 0.13 | 0.32 | 0.32 | 0.98 | 0.96 | 0.17 | 0.29 | 0.50 | 0.35 | 0.70 | 5.6 × 10−5 | 0.002 | 0.0002 |
| rs6663966 | 1 | GLISI | 53842601 | C | 0.04 | 0.01 | 5.16 | 5.2 × 10−5 | 0.00 | 0.01 | NA | NA | 0.09 | 0.02 | 6.46 | 0.01 | 0.04 | 0.01 | 4.15 | 2.07 | 8.34 | 6.2 × 10−5 | 0.002 | 0.0009 |
| rs4697843 | 4 | HS3ST1 | 10704856 | A | 0.21 | 0.33 | 0.52 | 0.0005 | 0.20 | 0.33 | 0.51 | 0.10 | 0.18 | 0.31 | 0.50 | 0.21 | 0.20 | 0.33 | 0.52 | 0.38 | 0.72 | 6.3 × 10−5 | 0.002 | 0.0001 |
| rs4521758 | 8 | PRKDC | 48904123 | T | 0.17 | 0.11 | 1.59 | 0.03 | 0.30 | 0.11 | 3.47 | 0.0007 | 0.23 | 0.09 | 2.92 | 0.04 | 0.20 | 0.11 | 1.98 | 1.41 | 2.77 | 7.3 × 10−5 | 0.003 | 0.0006 |
| rs10752067 | 10 | CUBN | 17188898 | C | 0.09 | 0.04 | 2.76 | 0.0003 | 0.10 | 0.03 | 3.73 | 0.02 | 0.05 | 0.05 | 0.93 | 0.94 | 0.09 | 0.04 | 2.59 | 1.62 | 4.16 | 7.9 × 10−5 | 0.003 | 0.0001 |
| rs3917462 | 1 | Near SELE | 167958291 | C | 0.03 | 0.01 | 4.03 | 0.006 | 0.05 | 0.01 | 9.05 | 0.009 | 0.05 | 0.01 | 5.16 | 0.12 | 0.03 | 0.01 | 4.75 | 2.17 | 10.36 | 9.3 × 10−5 | 0.004 | 0.001 |
| rs2508450 | 11 | IL10RA | 117369039 | T | 0.34 | 0.44 | 0.65 | 0.008 | 0.28 | 0.49 | 0.40 | 0.01 | 0.23 | 0.44 | 0.38 | 0.06 | 0.32 | 0.45 | 0.58 | 0.44 | 0.76 | 0.0001 | 0.004 | 0.0001 |
| rs2195611 | 2 | PDEIA | 182964812 | C | 0.59 | 0.44 | 1.83 | 0.0001 | 0.58 | 0.45 | 1.69 | 0.12 | 0.45 | 0.48 | 0.91 | 0.83 | 0.58 | 0.45 | 1.68 | 1.29 | 2.19 | 0.0001 | 0.005 | 0.0001 |
| rs11163323 | 1 | Near PRKACB | 81759046 | T | 0.33 | 0.19 | 2.05 | 1.7 × 10−5 | 0.23 | 0.23 | 0.98 | 0.96 | 0.23 | 0.19 | 1.24 | 0.69 | 0.30 | 0.20 | 1.76 | 1.32 | 2.35 | 0.0001 | 0.005 | 0.0002 |
| rs11155762 | 6 | MTHFDIL | 151314489 | T | 0.19 | 0.35 | 0.45 | 3.0 × 10−5 | 0.38 | 0.34 | 1.17 | 0.64 | 0.18 | 0.32 | 0.47 | 0.17 | 0.22 | 0.34 | 0.55 | 0.40 | 0.75 | 0.0001 | 0.006 | 0.0005 |
| rs17186372 | 1 | NOTCH2 | 120302474 | G | NA | NA | NA | NA | 0.25 | 0.07 | 4.28 | 0.0002 | 0.14 | 0.07 | 2.08 | 0.26 | 0.21 | 0.07 | 3.45 | 1.81 | 6.56 | 0.0002 | 0.006 | 0.001 |
| rs6583305 | 3 | UBXN7 | 197603018 | T | 0.47 | 0.36 | 1.63 | 0.002 | 0.55 | 0.34 | 2.39 | 0.008 | 0.36 | 0.36 | 1.00 | 0.99 | 0.48 | 0.35 | 1.66 | 1.27 | 2.16 | 0.0002 | 0.007 | 0.0003 |
| rs6764533 | 3 | UBXN7 | 197572861 | A | 0.47 | 0.36 | 1.63 | 0.002 | 0.55 | 0.34 | 2.36 | 0.009 | 0.36 | 0.36 | 1.00 | 0.99 | 0.48 | 0.35 | 1.65 | 1.27 | 2.15 | 0.0002 | 0.007 | 0.0004 |
| rs11545078 | 8 | GGH | 64101318 | A | 0.02 | 0.10 | 0.15 | 0.0003 | 0.08 | 0.11 | 0.64 | 0.47 | 0.05 | 0.11 | 0.37 | 0.33 | 0.03 | 0.10 | 0.26 | 0.13 | 0.53 | 0.0002 | 0.008 | 0.0002 |
The minor allele is the effect allele for the ORs; Chr, chromosome; L95, lower 95% CI boundary, U95, upper 95% CI boundary, NA, not available.
Figure 1.
Quantile–quantile plot of all single SNPs examined on the IBC chip and their CMH association analysis to diabetic retinopathy, defined as an ETDRS score ≥30.
We pursued replication of top findings from Tables 5 and 6 in independent cohorts of European ancestry with a fixed-effects meta-analysis model adjusted for age and sex (Table 7, Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). None of the variants achieved significance in replication (P < 1 × 10−6). The smallest P value was for rs35260 (P = 0.03). For all SNPs examined in replication, there was a significant amount of heterogeneity (P < 0.05 for Q test). We performed a sensitivity analysis by removing the FinnDiane and Go-DARTS cohorts—the former because it had type 1 diabetes participants exclusively and both because they did not use ETDRS grading consistently for phenotyping (Supplementary Table S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). However, none of the associations were statistically significant in this analysis either, and significant heterogeneity remained for all SNPs with the exceptions of rs3917779 (Q test P = 0.06) and rs6856425 for the ETDRS grade ≥14 analysis (Q test P = 0.26). Given the significant residual heterogeneity, we then used a random effects model but found no significant difference in the results. We also performed meta-analyses without the CARe cohorts; there was no statistically significant result or any significant heterogeneity in these analyses. Of note, meta-analyses of the CARe cohorts alone also showed no significant heterogeneity.
Table 7.
Replication Results in European Samples
| ARIC |
CHS |
MESA |
AGES |
BMES |
FIND-Eye |
FinnDiane |
Go-DARTS |
Lublin |
Meta-analysis (Fixed Effects) |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls, n |
732 |
160 |
140 |
249 |
175 |
627 |
570 |
774 |
620 |
||||||||||||||||||||
| Cases ETDRS ≥14, n |
153 |
33 |
36 |
92 |
67 |
105 |
2009 |
923 |
576 |
||||||||||||||||||||
| Cases ETDRS ≥30, n |
91 |
20 |
11 |
37 |
26 |
NA |
1399 |
923 |
138 |
||||||||||||||||||||
| SNP | Minor Allele | Definition of Cases | OR | P | OR | P | OR | P | MAF | OR | P | MAF | OR | P | MAF | OR | P | MAF | OR | P | MAF | OR | P | MAF | OR | P | z Score | OR | P |
| rs9332570 | G | ETDRS ≥14 | 0.43 | 0.0001 | 0.33 | 0.02 | 0.49 | 0.06 | NA | 0.75 | 0.26 | 0.21 | 1.24 | 0.42 | 0.19 | 0.79 | 0.26 | 0.27 | 1.16 | 0.05 | NA | NA | NA | NA | NA | NA | 0.002 | 1 | 0.99 |
| rs35260 | A | ETDRS ≥14 | 0.68 | 0.002 | 0.59 | 0.08 | 0.29 | 0.0001 | NA | 1.01 | 0.98 | 0.49 | 1.23 | 0.32 | 0.48 | 0.9 | 0.45 | 0.5 | 0.96 | 0.59 | 0.44 | 0.98 | 0.77 | NA | NA | NA | −2.24 | 0.91 | 0.03 |
| rs6128 | T | ETDRS ≥14 | 0.48 | 0.0007 | 0.27 | 0.02 | 0.39 | 0.03 | 0.09 | 1.27 | 0.32 | 0.21 | 1.27 | 0.37 | 0.17 | 0.73 | 0.14 | 0.24 | 1.16 | 0.07 | NA | 1.13 | 0.21 | 0.16 | 1.01 | 0.96 | 0.6 | 1.03 | 0.55 |
| rs7168655 | A | ETDRS ≥14 | 1.59 | 0.0004 | 1.78 | 0.04 | 1.48 | 0.15 | NA | 0.91 | 0.57 | 0.34 | 0.96 | 0.83 | 0.36 | 1.08 | 0.63 | 0.34 | 1.02 | 0.82 | 0.34 | 0.96 | 0.6 | NA | NA | NA | 1.55 | 1.07 | 0.12 |
| rs6133 | A | ETDRS ≥14 | 0.39 | 0.0005 | 0.19 | 0.03 | 0.51 | 0.16 | 0.15 | 0.73 | 0.32 | 0.14 | 1.37 | 0.28 | 0.13 | 0.72 | 0.17 | 0.09 | 1.13 | 0.31 | NA | 1.08 | 0.45 | 0.08 | 1.03 | 0.84 | −0.42 | 0.98 | 0.68 |
| rs3917779 | A | ETDRS ≥14 | 0.40 | 0.0007 | 0.20 | 0.03 | 0.51 | 0.16 | 0.09 | 0.73 | 0.32 | 0.14 | 1.25 | 0.45 | 0.12 | 0.72 | 0.19 | 0.09 | 1.14 | 0.29 | NA | NA | NA | NA | NA | NA | −1.39 | 0.88 | 0.17 |
| rs6856425 | C | ETDRS ≥14 | 2.48 | 0.01 | 5.21 | 0.004 | 2.63 | 0.16 | 0.04 | 0.89 | 0.86 | 0.02 | 1.8 | 0.45 | 0.02 | 1.18 | 0.75 | 0.03 | 0.97 | 0.87 | 0.02 | 1.34 | 0.27 | 0.05 | 0.7 | 0.07 | 1.1 | 1.13 | 0.27 |
| rs7105871 | C | ETDRS ≥14 | 0.47 | 1.7 × 10−5 | 0.97 | 0.91 | 0.86 | 0.62 | NA | 0.81 | 0.2 | 0.25 | 1.16 | 0.54 | 0.22 | 0.88 | 0.47 | 0.31 | 1 | 0.96 | NA | NA | NA | NA | NA | NA | −1.85 | 0.9 | 0.07 |
| rs6856425 | C | ETDRS ≥30 | 3.19 | 0.003 | 6.8 | 0.002 | 3.25 | 0.19 | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0.03 | 0.79 | 0.39 | 0.02 | 1.34 | 0.27 | 0.05 | 0.46 | 0.06 | 1.65 | 1.28 | 0.1 |
All results are adjusted for age and sex. NA, not available.
In addition to the above replication efforts in European cohorts, we pursued replication of the same findings in two Asian cohorts, SiMES and SP2. None of the SNPs was statistically significant in these populations. We also investigated the FTO association in Go-DARTS; neither rs9926180 nor rs12935710 was significantly associated (P = 0.84 and 0.23, respectively).
Discussion
In this large international collaborative study, genes previously linked with T2D, DR, and DN and vascular diseases were not generally associated with DR. In the CARe European American population, among genes that have been previously associated with DR, DN, and T2D, three SNPs in SELP were associated with DR, even after adjustment for DR risk factors. However, we were unable to replicate this finding in other ethnic groups in CARe or in independent Caucasian cohorts. The SELP SNPs associated with DR in the present study were not in LD with rs6131, the SNP initially associated with diabetic albuminuria (Supplementary Fig. S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental).64 P-selectin plays a role in leukocyte adhesion to endothelium during inflammation, and thus there is a biological rationale for its role in both diabetic microalbuminuria and retinopathy.65 With regards to FTO, the SNPs associated with DR in CARe were not in significant LD with rs9939609, the SNP associated with T2D (Supplementary Fig. S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental).66
Importantly, we were unable to confirm an association to most genes that have previously been associated with DR and were included on the IBC chip. We note that for several of these genes, the IBC chip did not include SNPs in LD to the previously associated variants because the selection of tag SNPs may not have densely covered those genes (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). For some genes, most notably EPO, we did have excellent proxies to the initially reported variants. The chip included two perfect proxies (rs551238 and rs1734907) for the EPO SNP originally associated to PDR, rs1617640.25 When we defined cases as having an ETDRS grade ≥30, rs551238 had a significant effect consistent with that found in the previous study, where the minor allele is protective (OR = 0.69, P = 0.009); the P value remains significant after correction by permutation (P = 0.01) but not by the Bonferroni method (P = 0.37). It is possible that with a larger sample size, the association would withstand the Bonferroni correction. We were also unable to detect an association to DR in other genes previously associated with DN and T2D. For the DN genes, again the IBC chip may not have included SNPs in LD with the previously reported variants. For T2D, however, the IBC chip variants were specifically those previously associated at genome-wide significant levels.
Another explanation for the inability to replicate previous DR associations lies in the heterogeneity among studies regarding DR definitions and participants' mean duration of diabetes. We attempted to mitigate the heterogeneity of DR definitions by examining two different definitions. However, this may not be sufficient to account for all the possible phenotype heterogeneity. The studies from which we selected genes deemed to be previously associated with DR all used controls that were diabetic patients without DR, as we did; however, some of them were performed in type 1 diabetic patients, which is another potential source of heterogeneity. There was also great variability in the duration of T2D among cases and controls in CARe cohorts. In particular, there were participants with short durations of diabetes who were included. There is the potential for misclassification of controls if these participants did not have DR at the time of study inclusion but are at risk for significant DR with longer duration of diabetes. We attempted to correct for this by including duration of disease as a covariate in logistic regression, but these issues could still bias the results toward the null. However, our ability to detect an association with EPO indicates that the amount of control misclassification in CARe is not significant enough to prevent detection of associations of this effect size.25,67 Finally, while the current investigation is the largest candidate gene study for DR to date, it still has limited power to detect genetic associations of modest or small effects (Supplementary Table S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). In particular, CARe includes a modest number of European-American cases (122) defined as ETDRS grade ≥30. Examining milder degrees of DR as outcomes may have further decreased our ability to detect associations as the development of early DR has a lower heritability. Of note, the ARIC cohort is larger than the other cohorts, and the top findings in the analyses were often driven by the results in ARIC.
In the second phase of the analysis, we took an unbiased approach at the remaining genes available on the IBC chip. Although we found several strong associations in our discovery cohort, replication in independent samples did not yield variants with consistent effects nor any variants that met the replication significance threshold (P = 1 × 10−6). The rs6856425 association was initially compelling because it was consistent within each CARe cohort. Furthermore, the strength of the association was greater when DR was defined as ETDRS grade ≥30 vs. ETDRS grade ≥14, which is in line with the expected greater heritability of more advanced DR phenotypes. The rs35260 variant had the lowest P value in the replication meta-analyses (P = 0.03), but this was still far below the replication threshold for significance (P = 1 × 10−6).
Failure to replicate a genetic association can be explained broadly, either as a false positive in the discovery cohort or a false negative in the replication cohort. For rs6856425, a rare variant, the initial estimate was based on limited instances of the minor allele: 9 in cases and 20 in controls. Small numbers of observations can lead to unstable effect estimates that represent chance statistical fluctuations rather than true associations. This underscores the importance of large sample sizes in both discovery and replication cohorts, particularly for rare variants. Another possible reason for false positives is population stratification, but there was no significant population stratification in this study.
False negatives in the replication cohort can be due to a lack of power, genotyping/imputation imprecision or heterogeneity between cohorts. Our aggregate replication cohort sample was well powered (Supplementary Table S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental). Of note, imputation was used by most replication cohorts for at least one of the SNPs. Imputation quality scores were greater than 0.90 for all SNPs with the exception of rs6856425 in FIND-Eye where the imputation quality score was 0.67. Errors in imputed genotype calls could lead to false-negative results, particularly for rare SNPs, which are more susceptible to genotyping artifacts. In addition, there was significant heterogeneity among samples that could not be explained by excluding type 1 diabetes participants and cohorts that did not use ETDRS grading consistently. Because the heterogeneity was not present when meta-analysis was restricted to the replication cohorts alone or the CARe cohorts alone, the “winner's curse” effect of large effect sizes in the discovery cohorts likely explains most of this heterogeneity.68 Some heterogeneity might also derive from the different DR ascertainment methods and case–control definitions. Cohorts differed in their photography protocols, with some cohorts having one field of only one eye for phenotype determination. This introduces misclassification bias for participants for whom the DR grade in the one or two fields photographed may not accurately represent the DR grade in other fields or the contralateral eye. It is therefore possible that some of the variants associated with DR in CARe may eventually be replicated in larger studies with direct genotyping and better phenotype harmonization.
In summary, in this candidate gene analysis of DR with data from the CARe consortium, with replications in a several large, well-powered samples, we found little evidence of a major DR gene. This is the largest number of candidate genes studied for DR to date. Although no association could be confirmed with a high threshold for significance, the results are hypothesis generating and the genes associated with DR in CARe could be prioritized in studies. The importance of well-powered replication and phenotype harmonization are highlighted by our study. These issues will continue to be important as results from genome-wide association studies for DR become increasingly available.
Supplementary Material
Acknowledgments
The authors thank Joan W. Miller and David M. Altshuler for their support of the study.
Footnotes
Supported by National Eye Institute Grant K12-EY16335; a Research to Prevent Blindness Career Development Award; the Massachusetts Lions Eye Research Fund; the Sara Elizabeth O'Brien Trust; and Harvard Catalyst/The Harvard Clinical and Translational Science Center, National Institutes of Health Award Grant UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers. The National Heart, Lung Blood Institute (NHLBI) Grant N01-HC-65226 has supported genotyping and has created a genotype–phenotype database with data and samples from nine cohorts as part of Candidate Gene Association Resource (CARe). Please see the Supplementary Material (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental), as well as http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx for detailed information about the grants supporting the four cohorts in this study: Atherosclerosis Risk in Communities (ARIC) Study, Cardiovascular Health Study (CHS), Jackson Heart Study (JHS), and Multi-ethnic Study of Atherosclerosis (MESA). Please see the Supplementary Material (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-7510/-/DCSupplemental) for detailed information about the grants supporting the replication cohorts.
Disclosure: L. Sobrin, None; T. Green, None; X. Sim, None; R.A. Jensen, None; E.S. Tai, None; W.T. Tay, None; J.J. Wang, None; P. Mitchell, None; N. Sandholm, None; Y. Liu, None; K. Hietala, None; S.K. Iyengar, None; M. Brooks, None; M. Buraczynska, None; N. Van Zuydam, None; A.V. Smith, None; V. Gudnason, None; A.S.F. Doney, None; A.D. Morris, None; G.P. Leese, None; C.N.A. Palmer, None; A. Swaroop, None; H.A. Taylor, Jr, None; J.G. Wilson, None; A. Penman, None; C.J. Chen, None; P.-H. Groop, None; S.-M. Saw, None; T. Aung, None; B.E. Klein, None; J.I. Rotter, None; D.S. Siscovick, None; M.F. Cotch, None; R. Klein, None; M.J. Daly, None; T.Y. Wong, None
References
- 1. National Institute of Diabetes and Digestive and Kidney Diseases National diabetes statistics fact sheet: general information and national estimates on diabetes in the United States. 2000. Bethesda, MD: U.S. Department of Health and Human Services, National Institute of Health, Publication No. 02-3892 [Google Scholar]
- 2. Klein R, Klein BE, Moss SE, Cruickshanks KJ. The Wisconsin Epidemiologic Study of Diabetic Retinopathy, XVII: The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998;105:1801–1815 [DOI] [PubMed] [Google Scholar]
- 3. Klein BE. Overview of epidemiologic studies of diabetic retinopathy. Ophthalmic Epidemiol. 2007;14:179–183 [DOI] [PubMed] [Google Scholar]
- 4. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124–136 [DOI] [PubMed] [Google Scholar]
- 5. Zhang X, Saaddine JB, Chou CF, et al. Prevalence of diabetic retinopathy in the United States, 2005–2008. JAMA. 2010;304:649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nathan DM. Long-term complications of diabetes mellitus. N Engl J Med. 1993;328:1676–1685 [DOI] [PubMed] [Google Scholar]
- 7. Klein R, Klein BE, Moss SE, Cruickshanks KJ. Relationship of hyperglycemia to the long-term incidence and progression of diabetic retinopathy. Arch Intern Med. 1994;154:2169–2178 [PubMed] [Google Scholar]
- 8. Looker HC, Nelson RG, Chew E, et al. Genome-wide linkage analyses to identify loci for diabetic retinopathy. Diabetes. 2007;56:1160–1166 [DOI] [PubMed] [Google Scholar]
- 9. Hietala K, Forsblom C, Summanen P, Groop PH. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arar NH, Freedman BI, Adler SG, et al. Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest Ophthalmol Vis Sci. 2008;49:3839–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abhary S, Hewitt AW, Burdon KP, Craig JE. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009;58:2137–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liew G, Klein R, Wong TY. The role of genetics in susceptibility to diabetic retinopathy. Int Ophthalmol Clin. 2009;49:35–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182 [DOI] [PubMed] [Google Scholar]
- 14. Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61 [DOI] [PubMed] [Google Scholar]
- 15. Kottgen A, Hwang SJ, Rampersaud E, et al. TCF7L2 variants associate with CKD progression and renal function in population-based cohorts. J Am Soc Nephrol. 2008;19:1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krentz AJ, Clough G, Byrne CD. Interactions between microvascular and macrovascular disease in diabetes: pathophysiology and therapeutic implications. Diabetes Obes Metab. 2007;9:781–791 [DOI] [PubMed] [Google Scholar]
- 17. Ohno T, Kinoshita O, Fujita H, et al. Detecting occult coronary artery disease followed by early coronary artery bypass surgery in patients with diabetic retinopathy: report from a diabetic retinocoronary clinic. J Thorac Cardiovasc Surg. 2010;139:92–97 [DOI] [PubMed] [Google Scholar]
- 18. Kofler B, Mueller EE, Eder W, et al. Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet. 2009;10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008;359:1577–1589 [DOI] [PubMed] [Google Scholar]
- 20. Cheung N, Wong TY. Diabetic retinopathy and systemic vascular complications. Prog Retin Eye Res. 2008;27:161–176 [DOI] [PubMed] [Google Scholar]
- 21. Kawasaki R, Cheung N, Islam FM, et al. Is diabetic retinopathy related to subclinical cardiovascular disease? Ophthalmology. 2011;118:860–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Musunuru K, Lettre G, Young T, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keating BJ, Tischfield S, Murray SS, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhlmann K, Kovacs P, Boettcher Y, Hammes HP, Paschke R. Genetics of diabetic retinopathy. Exp Clin Endocrinol Diabetes. 2006;114:275–294 [DOI] [PubMed] [Google Scholar]
- 25. Tong Z, Yang Z, Patel S, et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A. 2008;105:6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng DP, Tai BC, Lim XL. Is the presence of retinopathy of practical value in defining cases of diabetic nephropathy in genetic association studies?—the experience with the ACE insertion/deletion polymorphism in 53 studies comprising 17,791 subjects. Diabetes. 2008;57:2541–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic factors in diabetic nephropathy. Clin J Am Soc Nephrol. 2007;2:1306–1316 [DOI] [PubMed] [Google Scholar]
- 28. Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662 [DOI] [PubMed] [Google Scholar]
- 29. Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 30. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 31. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sandhu MS, Weedon MN, Fawcett KA, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39:951–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56:685–693 [DOI] [PubMed] [Google Scholar]
- 35. Bild DE, Bluemke DA, Burke GL, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881 [DOI] [PubMed] [Google Scholar]
- 36. The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702 [PubMed] [Google Scholar]
- 37. Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276 [DOI] [PubMed] [Google Scholar]
- 38. Wilson JG, Rotimi CN, Ekunwe L, et al. Study design for genetic analysis in the Jackson Heart Study. Ethn Dis. 2005;15:S6–30- 37 [PubMed] [Google Scholar]
- 39. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 2003;26(suppl 1):S5–S20 [DOI] [PubMed] [Google Scholar]
- 40. Wong TY, Klein R, Islam FM, et al. Diabetic retinopathy in a multi-ethnic cohort in the United States. Am J Ophthalmol. 2006;141:446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheung N, Wang JJ, Rogers SL, et al. Diabetic retinopathy and risk of heart failure. J Am Coll Cardiol. 2008;51:1573–1578 [DOI] [PubMed] [Google Scholar]
- 42. Klein R, Marino EK, Kuller LH, et al. The relation of atherosclerotic cardiovascular disease to retinopathy in people with diabetes in the Cardiovascular Health Study. Br J Ophthalmol. 2002;86:84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diabetic retinopathy study. Report Number 6. Design, methods, and baseline results. Report Number 7. A modification of the Airlie House classification of diabetic retinopathy. Prepared by the Diabetic Retinopathy. Invest Ophthalmol Vis Sci 1981;21:1–226 [PubMed] [Google Scholar]
- 44. Ojaimi E, Nguyen TT, Klein R, et al. Retinopathy signs in people without diabetes the multi-ethnic study of atherosclerosis. Ophthalmology. 2011;118:656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colagiuri S, Lee CM, Wong TY, Balkau B, Shaw JE, Borch-Johnsen K. Glycemic thresholds for diabetes-specific retinopathy: implications for diagnostic criteria for diabetes. Diabetes Care. 2011;34:145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wong TY, Liew G, Tapp RJ, et al. Relation between fasting glucose and retinopathy for diagnosis of diabetes: three population-based cross-sectional studies. Lancet. 2008;371:736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guillausseau PJ, Massin P, Charles MA, et al. Glycaemic control and development of retinopathy in type 2 diabetes mellitus: a longitudinal study. Diabet Med. 1998;15:151–155 [DOI] [PubMed] [Google Scholar]
- 48. Stratton IM, Kohner EM, Aldington SJ, et al. UKPDS 50: risk factors for incidence and progression of retinopathy in type II diabetes over 6 years from diagnosis. Diabetologia. 2001;44:156–163 [DOI] [PubMed] [Google Scholar]
- 49. Tight blood pressure control and risk of macrovascular and microvascular complications in type 2 diabetes: UKPDS 38. UK Prospective Diabetes Study Group. BMJ. 1998;317:703–713 [PMC free article] [PubMed] [Google Scholar]
- 50. Ferris FL, 3rd, Chew EY, Hoogwerf BJ. Serum lipids and diabetic retinopathy. Early Treatment Diabetic Retinopathy Study Research Group. Diabetes Care. 1996;19:1291–1293 [DOI] [PubMed] [Google Scholar]
- 51. Cochran WG. Some methods for strengthening the common X2 tests. Biometrics. 1954;10:417–451 [Google Scholar]
- 52. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748 [PubMed] [Google Scholar]
- 53. Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935 [DOI] [PubMed] [Google Scholar]
- 54. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qiu C, Cotch MF, Sigurdsson S, et al. Retinal and cerebral microvascular signs and diabetes: the age, gene/environment susceptibility-Reykjavik study. Diabetes. 2008;57:1645–1650 [DOI] [PubMed] [Google Scholar]
- 56. Mitchell P, Smith W, Wang JJ, Attebo K. Prevalence of diabetic retinopathy in an older community. The Blue Mountains Eye Study. Ophthalmology. 1998;105:406–411 [DOI] [PubMed] [Google Scholar]
- 57. Doney AS, Leese GP, Olson J, Morris AD, Palmer CN. The Y402H variant of complement factor H is associated with age-related macular degeneration but not with diabetic retinal disease in the Go-DARTS study. Diabet Med. 2009;26:460–465 [DOI] [PubMed] [Google Scholar]
- 58. Wong TY, Cheung N, Tay WT, et al. Prevalence and risk factors for diabetic retinopathy: the Singapore Malay Eye Study. Ophthalmology. 2008;115:1869–1875 [DOI] [PubMed] [Google Scholar]
- 59. Yim-Lui Cheung C, Wong TY, Lamoureux EL, et al. C-reactive protein and retinal microvascular caliber in a multiethnic Asian population. Am J Epidemiol. 2010;171:206–213 [DOI] [PubMed] [Google Scholar]
- 60. Buraczynska M, Baranowicz-Gaszczyk I, Tarach J, Ksiazek A. Toll-like receptor 4 gene polymorphism and early onset of diabetic retinopathy in patients with type 2 diabetes. Hum Immunol. 2009;70:121–124 [DOI] [PubMed] [Google Scholar]
- 61. Cochran WG. Problems arising in the analysis of a series of similar experiments. J R Stat Soc. 1937;4:102–118 [Google Scholar]
- 62. Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129 [Google Scholar]
- 63. Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385 [DOI] [PubMed] [Google Scholar]
- 64. Liu Y, Burdon KP, Langefeld CD, et al. P-selectin gene haplotype associations with albuminuria in the Diabetes Heart Study. Kidney Int. 2005;68:741–746 [DOI] [PubMed] [Google Scholar]
- 65. Tedder TF, Steeber DA, Chen A, Engel P. The selectins: vascular adhesion molecules. FASEB J. 1995;9:866–873 [PubMed] [Google Scholar]
- 66. Freathy RM, Timpson NJ, Lawlor DA, et al. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abhary S, Burdon KP, Casson RJ, Goggin M, Petrovsky NP, Craig JE. Association between erythropoietin gene polymorphisms and diabetic retinopathy. Arch Ophthalmol. 2010;128:102–106 [DOI] [PubMed] [Google Scholar]
- 68. Kraft P. Curses—winner's and otherwise—in genetic epidemiology. Epidemiology. 19:649–651, 2008; discussion 57–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.