These data suggest that there are simplified, quantitative volume estimators of NVAMD lesion components that are accessible for use by clinicians without the need for specialized software or time-consuming manual segmentation.
Abstract
Purpose.
To evaluate simple methods of estimating the volume of clinically relevant features in neovascular age-related macular degeneration (NVAMD) using spectral domain–optical coherence tomography (SD-OCT).
Methods.
Using a database of NVAMD cases imaged with macular cube (512 A-scans × 128 B-scans) SD-OCT scans, the authors retrospectively selected visits where cystoid macular edema (CME), subretinal fluid (SRF), or pigment epithelial detachments (PEDs) were evident. Patients with single visits were analyzed in the cross-sectional analysis (CSA) and those with a baseline visit and three or more follow-up visits in the longitudinal analysis (LA). The volume of each feature was measured by manual grading using validated grading software. Simplified measurements for each feature included: number of B-scans or A-scans involved and maximum height. Automated measurements of total macular volume and foveal central subfield were also collected from each machine. Correlations were performed between the volumes measured with 3D-OCTOR, automated measurements, and the simplified measures.
Results.
Forty-five visits for 25 patients were included in this study: 26 cube scans from 26 eyes of 25 patients in the CSA and 24 scans from 5 eyes of 5 patients in the LA. The simplified measures that correlated best with manual grading in the CSA group were maximum lesion height for CME (r2 value = 0.96) and B-scan count for SRF and PED volume (r2 values of 0.88 and 0.70). In the LA group, intervisit differences were correlated. Change in B-scan count correlated well with change in SRF volume (r2 = 0.97), whereas change in maximum height correlated with change in CME and PED volume (r2 = 0.98 and 0.43, respectively).
Conclusions.
These data suggest that simplified estimators of some NVAMD lesion volumes exist and are accessible by clinicians without the need for specialized software or time-consuming manual segmentation. These simple approaches could enhance quantitative disease monitoring strategies in clinical trials and clinical practice.
Imaging data have been an integral part of both ophthalmic clinical practice and clinical trials for decades. Despite the enthusiastic adoption of optical coherence tomography (OCT) into clinical practices over the last decade, the use of OCT in clinical trials has generally lagged behind its clinical use. For example, time domain OCT took years to become commonplace in neovascular age-related macular degeneration (NVAMD) clinical trials, in part because of inconsistencies in measurements due to software segmentation and registration irregularities in this disease.1–3 In the recent era of spectral domain OCT (SD-OCT), large data sets, a multitude of device manufacturers, and segmentation errors may again have hampered rapid integration of SD-OCT data into multicenter studies. Given the widespread adoption of SD-OCT into practices, continued failure to use this same technology in clinical trials maintains a disconnect between trials and practice that may interfere with the evidence-based management and therapy that often constitute the goal of these trials. Instead, it seems that a desirable goal for clinical trials should be to use and study techniques that can be readily adapted to clinical practice to guide patient management based on the conclusions from those trials.
One OCT-based measure that has been used in both clinical trials and clinical practices is the automated report produced by each commercial OCT instrument.4–7 Despite the differences in measurements made by different machines, these data, such as macular volume and Early Treatment Diabetic Retinopathy Study grid retinal thickness measurements, have been useful in clinical trials because they provide a simple, objective, numerical distillation of the imaging data contained in the SD-OCT data set. However, these measurements are still prone to segmentation errors and fail to extract much of the rich data concerning specific lesions that are contained in OCT images.8–10
One common tool that is used in clinical trials but not in clinical practice is a reading center assessment. These evaluations extract data, such as the presence of cystoid macular edema (CME), subretinal fluid (SRF), or pigment epithelial detachments (PEDs), that may be overlooked by automated measurements. Based on the expertise of human graders, reading center assessments can detect and sometimes correct segmentation errors. However, the standardized training, protocols, and time involved in these assessments make them impractical for most clinicians.
The goal of this study was to investigate simplified, quantitative methodologies that approach the accuracy of a detailed, time-consuming reading center assessment but can be completed in a short enough time to make it practical for most clinicians.
Methods
Data Collection
Using a database of cases collected at our institute over a 3-year period of patients with NVAMD imaged with macular cube (512 A-scans × 128 B-scans) volume SD-OCT scans (either a 3D-OCT-1000, Topcon, Inc., Tokyo, Japan; or Cirrus HD-OCT, Carl Zeiss Meditec, Inc., Dublin, CA), we retrospectively selected single visits where the SD-OCT scans demonstrated CME, SRF, and/or PEDs. Data from these single visits were analyzed as one group and called the cross-sectional analysis (CSA). Patients with one baseline visit and three or more follow-up visits of sufficient quality for analysis were also selected and grouped into a separate longitudinal analysis (LA) group. Change analyses were conducted on the longitudinal data in the LA group by subtracting measurements at one visit from the same measurements at the next visit. These measurements were referred to as “delta ” measurements. All research herein adhered to the tenets set forth in the Declaration of Helsinki.
Volumetric Grading
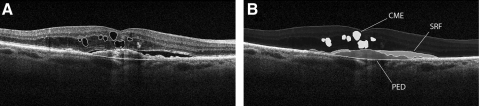
Gold-standard volumetric data were generated by exporting the raw OCT data from the SD-OCT devices and importing the data into previously described validated grading software (titled “3D-OCTOR”) for manual grading.11,12 This software allows the user to navigate through all available scans and manually draw structure boundaries for all or a subset of scans. Based on these boundaries the software then calculates measurements such as thickness, area, and volume for each designated space (Fig. 1). Volumetric grading was performed on every B-scan of each SD-OCT volume scan in accordance with our Doheny Image Reading Center protocols. The reproducibility of these grading methods has been previously described.11 In addition, automated measurements from the OCT devices, specifically macular volume (MV) and foveal central subfield (FCS) thickness, were collected for each case for comparison with the manual grading.
Figure 1.
(A, B) Images show the same B-scan from an SD-OCT data set after manual grading within a validated reading center tool (3D-OCTOR). CME, SRF, and PED were manually graded on every B-scan (A) and the validated reading tool (3D-OCTOR) measured the corresponding spaces (B) to give detailed volumetric results.
Simplified Grading
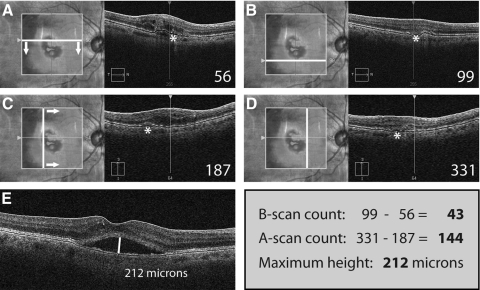
In a second step, simplified measurements were made for each feature (CME, SRF, or PED) including: number of B-scans demonstrating the feature, number of A-scans including the feature, and the maximum height of the feature across the macular cube. With the exception of A-scan counts (on 3D-OCT-1000 data), these measurements were obtained directly from the aforementioned SD-OCT devices using the preinstalled software (Cirrus HD-OCT, version 5.0.0.326, Carl Zeiss Meditec; or Topcon Data Viewer, version 4.13.002.02; Analyze.dll v 2.01.51, respectively). B-scan count and A-scan count were determined by identifying the first scan containing the feature and then moving forward through the volume scan until the feature was no longer visible. The difference between the numbers of the first and last scans represented the number of scans involved (Fig. 2). For the purpose of this study, a PED was defined as the single largest, contiguous detachment of the retinal pigment epithelium (RPE) across the macula excluding satellite PED lesions that appeared to be disconnected from the main PED. Since our version of the data viewer (Topcon Data Viewer) did not allow visualization of A-scans as cross-sections, we vertically reconstructed the 3D-OCT-1000 data set with custom software and registered adjacent scans for display within 3D-OCTOR. This was done only for the purpose of simplified grading of A-scan counts but not for manual segmentation. Simplified measures were also combined in the following way: (1) A-scan count × maximum height; (2) B-scan count × maximum height; and (3) A-scan count × B-scan count × maximum height.
Figure 2.
Simplified grading of SRF in a sample case from the Cirrus HD-OCT. The first B-scan of the cube scan that shows the feature to be graded (SRF marked with an asterisk) is identified and the respective B-scan number noted (A). Then, progressing through the lesion (arrows), the number of the B-scan representing the lower boundary of the lesion feature is also noted (B). In the same way, the A-scans representing the left and right boundaries of the lesion feature are identified and their respective numbers noted. The Cirrus HD-OCT natively supports the A-scan view of a cube scan. Regarding the 3D-OCT-1000 used in our study, we engineered custom software to facilitate an A-scan view similar to that shown for the Cirrus HD-OCT. Finally, the maximum height of the feature is measured with a caliper tool found in virtually any OCT software. The final values for the simplified grading of SRF in this case are: number of B-scans: 99 − 56 = 43; number of A-scans = 331 − 187 = 144; maximum height = 212 μm. Grading of these three parameters will take an average time of approximately 30 seconds per feature.
All grading was done independently in a dual-grader process by two reading center certified graders (FMH, ACW). Discrepancies between the two graders were adjudicated and the average of the final results was used for subsequent statistical analysis. Both volumetric grading and simplified grading were timed for a subset of cases to estimate the total time required to complete each task.
Statistical Methods
Pearson correlation coefficients were calculated for the CSA and LA groups between manual volumetric measurements and the simplified measurements, the derived parameters from the simplified measurements such as lesion area, and the automated results from each OCT device. Delta volume measurements were compared with delta versions of these simplified parameters. Nonlinear comparisons, such as between volume and thickness or volume and area, were linearized by calculating the cube root of volumetric measurements and the square root of area measurements before calculating the Pearson correlation.
To determine the predictive power of a model derived from these data, the cross-sectional data were split randomly into two groups (split sample methodology). Linear regression models developed using one group were tested against the other for each of the two halves. The average percentage error for each simplified parameter was calculated by comparing its predicted volume with the gold standard from volumetric grading.
Custom software was written to perform a Monte Carlo permutation correlation analysis on the entire data set. In our approach, correlation coefficients were calculated for billions of random permutations of data subsets from the full correlation analysis. For example, given only 8 data pairs in a data set, a Monte Carlo permutation analysis also calculates correlation coefficients for 8 permutations of 7 data pairs, 28 permutations of 6 pairs, 56 permutations of 5 pairs, and 70 permutations of 4 pairs in addition to the single permutation of 8 data pairs. In addition to standard P values, these data can be used to calculate 95% confidence intervals (CIs) for correlations derived from subsets of data. Small CIs suggest that these data are exchangeable and that future data points will correlate in the same way. In contrast, large CIs suggest uncertainty about the significance of the conclusion since a different conclusion might have been reached using only a subset of the same data. The greatest degree of uncertainty regarding the correlation of two data sets occurs when the CI value dips below significance after leaving out only 1 or 2 data pairs.
To test the potential utility of these parameters in treatment versus observation clinical decisions, the sensitivity and specificity of simplified and automated measurements for detection of increases or decreases in manually measured lesion volumes were also computed. To accomplish this, changes in measurements for CME, SRF, and PED were subcategorized into one of three groups: decreased, no change, or increased. To account for inaccuracies in manual measurements, such as those arising from subjective boundary choices or limitations of image resolution, any change ≤2 B-scans, 8 A-scans, 7 μm maximum height/retinal thickness, 0.1 mm3 macular volume, or 0.019-mm cube-root volume was categorized as “no change. ” These thresholds were derived from the assumption that the OCT volume scans have an axial resolution of ≤7 μm, a horizontal resolution of ≤20 μm, and a vertical resolution of ≤48 μm.
Finally, interrater agreement for the simplified grading was assessed with weighted Cohen's kappa (κ) and Bland–Altman plots. The weights of the individual groups for the κ-analysis are shown in Table 1.
Table 1.
Groups and Weights for Analysis of Cohen's kappa
| κ Analysis |
Measurement Parameters* |
|||
|---|---|---|---|---|
| Group | Weight | B-Scan Count (n) | A-Scan Count (n) | Maximum Height (μm) |
| 1 | 1.00 | 0–2 | 0–8 | 0–7 |
| 2 | 0.70 | 3–6 | 9–24 | 8–15 |
| 3 | 0.00 | >6 | >24 | >15 |
For analysis of Cohen's kappa, the difference between the two graders was weighted according to its absolute value. Measurement errors of more than 3 B-scans, more than 9 A-scans, or more than 8 μm in maximum height were considered negligible.
Measurement parameters are given as the difference between the two graders.
Results
A total of 45 visits for 25 patients were included in this study. Twenty-six cube scans from 26 eyes of 25 patients with a diagnosis of NVAMD were included in the CSA. Of these cube scans, 20 were taken on the 3D-OCT-1000 (Topcon), whereas 6 were taken on the Cirrus HD-OCT. Twenty-four visits from 5 eyes of 5 patients were included in the LA, and 19 of the total 24 cube scans were taken on the 3D-OCT-1000 (Topcon), whereas 5 scans were taken on the Cirrus HD-OCT. The OCT device used at baseline did not change during the follow-up for all 5 patients in the LA. Sixteen patients were female. The age range of the subjects was 73–94 years (mean, 82.5 years).
For the CSA group, nearly all simplified parameters correlated with the manual volumetric measurements (Table 2). The range of R2 values for each feature was 0.7–0.96 for CME, 0.87–0.96 for SRF, and 0.32–0.81 for PED. Automated measurements from the SD-OCT devices did not correlate as well with gold-standard manual measurements. The range of R2 values for automated measurements was 0.13–0.44 for CME, 0.34–0.54 for SRF, and 0.11–0.17 for PED.
Table 2.
Summary of Correlation Analysis
| Measurement Parameters | CSA Group |
LA Group* |
||||
|---|---|---|---|---|---|---|
| CME | SRF | PED | CME | SRF | PED | |
| Correlation of simplified measurements (R2) | (n = 26) | (n = 26) | (n = 26) | (n = 19) | (n = 19) | (n = 19) |
| B-scan count | 0.80 | 0.88 | 0.70 | 0.97 | 0.97 | 0.39 |
| A-scan count | 0.77 | 0.88 | 0.70 | 0.95 | 0.88 | 0.07 |
| Maximum height | 0.96 | 0.87 | 0.32 | 0.98 | 0.89 | 0.43 |
| B-scan count × maximum height | 0.95 | 0.95 | 0.67 | 0.99 | 0.97 | 0.53 |
| A-scan count × maximum height | 0.96 | 0.96 | 0.70 | 0.99 | 0.98 | 0.49 |
| Estimated area × maximum height | 0.93 | 0.96 | 0.81 | 0.98 | 0.99 | 0.52 |
| Correlation of automated measurements (R2) | ||||||
| Cirrus HD-OCT | (n = 6) | (n = 6) | (n = 6) | (n = 0) | (n = 4) | (n = 4) |
| FCS | 0.47 | 0.01 | 0.10 | — | 0.72 | 0.42 |
| Macular volume | 0.08 | 0.38 | 0.08 | — | 0.63 | 0.34 |
| 3D-OCT-1000 | (n = 20) | (n = 20) | (n = 20) | (n = 15) | (n = 15) | (n = 15) |
| FCS | 0.19 | 0.24 | 0.17 | 0.33 | 0.81 | 0.26 |
| Macular volume | 0.60 | 0.62 | 0.16 | 0.27 | 0.70 | 0.72 |
The coefficient of determination (R2) for each parameter is shown for the CSA group and the LA group separately. The simplified measurements are listed in the upper half, whereas the automated measurements collected from the OCT devices are shown in the lower half.
For the longitudinal group measurements are given as change from the previous visit.
Results for the LA group were similar when analyzed as single visits. However, analysis of changes in measurements between visits (delta analysis) demonstrated better correlation with automated MV (range of R2 values: 0.27 to 0.72) and FCS measurements (range of R2 values: 0.26 to 0.81). Delta analysis for the simplified parameters was still superior to automated measurements for CME and SRF lesions, with maximum R2 values of 0.99 each. Interestingly, delta analysis of PEDs yielded worse correlations with manual measurements than automated analyses (R2 = 0.53, Table 2). There were obvious differences in the R2 values of automated measurements between the two devices used in this study (Table 2). Assessment of the segmentation algorithm performance revealed only minor errors on both devices, and thus we attribute this observed difference in correlation to the small case number in the Cirrus cohort (n = 6 and n = 4, for the CSA and LA groups, respectively).
Sensitivity–specificity analysis revealed that the simplified parameters had a higher sensitivity and specificity for predicting intervisit changes in CME, SRF, and PED volumes than changes in automated macular volume or FCS (Table 3). FCS thickness was more specific than MV in detecting fluid changes, but it also produced a number of false-negative results, despite the use of a relatively sensitive cutoff of approximately 7 μm. When the clinical trial standard of a 100-μm change in FCS was applied, the sensitivity of FCS for detection of changes in SRF and CME (12.5%) was inferior to the simplified measures (100%).
Table 3.
Sensitivity and Specificity Analysis Results
| Feature | Measurement Parameters |
|||||
|---|---|---|---|---|---|---|
| B-Scan Count | A-Scan Count | Maximum Height | FCS Thickness (>7 μm) | FCS Thickness (>100 μm) | Macular Volume | |
| CME | ||||||
| Sensitivity, % | 100.0 | 100.0 | 100.0 | 75.0 | 0.0 | 100.0 |
| Specificity, % | 100.0 | 100.0 | 100.0 | 18.2 | 81.8 | 0.0 |
| SRF | ||||||
| Sensitivity, % | 100.0 | 91.7 | 100.0 | 91.7 | 16.7 | 100.0 |
| Specificity, % | 85.7 | 66.7 | 71.4 | 42.9 | 100.0 | 0.0 |
| PED | ||||||
| Sensitivity, % | 66.7 | 100.0 | 66.7 | 66.7 | 0.0 | 66.7 |
| Specificity, % | 56.3 | 31.3 | 18.8 | 25.0 | 87.5 | 0.0 |
| CME and SRF* | ||||||
| Sensitivity, % | 100.0 | 100.0 | 100.0 | 81.3 | 12.5 | 100.0 |
| Specificity, % | 66.7 | 33.3 | 33.3 | 33.3 | 100.0 | 0.0 |
The results show that the simplified parameters are both sensitive and specific for a detection of change in CME or SRF volume. FCS thickness, a parameter used in clinical trials, has a sensitivity of only 81.3% for detecting a change in either intra- or subretinal fluid (CME and SRF), although a cutoff of only 7 μm was used in this analysis.
CME and SRF were treated as a single feature in this group.
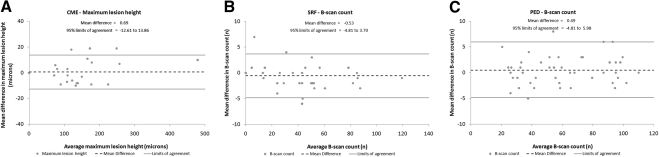
Interrater agreement for both B-scan count and maximum lesion height measurements was good with Cohen's kappa values of 0.95 and 0.87 for CME, 0.92 and 0.88 for SRF, and 0.87 and 0.70 for PEDs, respectively. A-scan count had lower interrater agreement, with an average κ = 0.69 for all features (range, 0.60 to 0.78). Bland–Altman plots confirmed the good interrater agreement of B-scan count and maximum lesion height (Fig. 3).
Figure 3.
Bland–Altman plots showing the interrater agreement for the one single parameter from simplified grading of each CME (A), SRF (B), and PED (C) that correlated best with exact volumes. The mean difference between the graders is shown as a dashed line; the boundaries of the 95% limits of agreement are marked by the upper and lower gray lines.
The best linear regression models used to predict feature volumes in opposite halves of the CSA data set produced an average relative error of 17.1% for CME based on maximum lesion height, and 15.2% and 20.1% for SRF and PED, respectively, when based on B-scan count × maximum height (Table 4). A-scan count was not used in this analysis due to unsatisfactory interrater agreement.
Table 4.
Relative Errors of Estimated Volume versus Gold-Standard Volume
| Feature | Relative Errors from Models |
||
|---|---|---|---|
| B-Scan Count | Maximum Height | B-Scan Count × Maximum Height | |
| CME | |||
| Group 1 (n = 13) | 49.4% | 20.4% | 23.7% |
| Group 2 (n = 13) | 35.7% | 13.7% | 16.6% |
| SRF | |||
| Group 1 (n = 13) | 22.9% | 21.9% | 11.8% |
| Group 2 (n = 13) | 22.3% | 29.8% | 18.6% |
| PED | |||
| Group 1 (n = 13) | 29.2% | 47.5% | 28.4% |
| Group 2 (n = 13) | 15.6% | 18.7% | 11.8% |
| Average (n = 26) | |||
| CME | 42.6% | 17.1% | 20.2% |
| SRF | 22.6% | 25.9% | 15.2% |
| PED | 22.4% | 33.1% | 20.1% |
To determine the predictive power of our analysis, we used a random split sample approach.
Models were built on one group and applied to the other. The absolute errors of each model were averaged and compared with the averaged gold-standard volumes of the respective features (CME, SRF, PED). The resulting relative errors are shown individually for both groups and as the averaged value of the two.
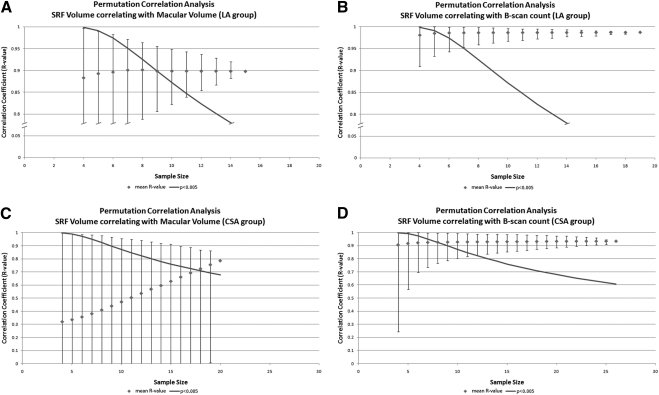
Monte Carlo permutation correlation analysis revealed that all simplified grading parameters exceeded statistical significance of P = 0.005 at one third of our total sample size. In comparison, the correlation of automated measurements with gold-standard volumes failed to reach a significant level (Fig. 4) in many cases. When they did, the CIs sometimes suggested that a different conclusion might have been reached had a few cases been excluded. This is particularly true for the correlation of automated MV with SRF volume, where the R value dropped from 0.79 to 0.00 after exclusion of only a single case (Fig. 4).
Figure 4.
Plots show the correlation coefficients (R values) obtained by permutation correlation analysis, with the error bars indicating a range of 2SDs around the mean on either side. The continuous line represents the minimum Pearson correlation coefficient, with a statistical significance of P ≤ 0.005 for two-tailed tests. Graph (A) shows the results when correlating automated macular volume and gold-standard volumetric grading of SRF in the LA group. Graph (B) is equivalent but compares the simplified parameter maximum height with the gold-standard volume. In both graphs (A and B) the y-axis is broken, as indicated for better readability. Graphs (C and D) compare the same parameters in the CSA group, respectively.
On average, the time required to perform manual volumetric grading of CME, SRF, and PED in a single SD-OCT volume scan using 3D-OCTOR was 92 ± 16 minutes. In contrast, simplified grading was >50 times faster, with an average time to completion of 1.4 ± 0.2 minutes for all features. Simplified grading of only B-scan counts or maximum lesion height from a single SD-OCT volume scan took an average of 13 and 9 seconds, respectively.
Discussion
In this study, we report on comparisons between a detailed volumetric analysis of clinically relevant OCT features associated with NVAMD and simplified quantification methods that are quick and practical for both clinicians and reading centers to perform. OCT-driven monitoring of NVAMD, based at least in part on changes in CME, SRF, and PEDs, is becoming commonplace in both clinical practice and in clinical trials.6,7,13,14
In the absence of software capable of measuring these clinically relevant parameters accurately and automatically, clinicians are left either to assess changes qualitatively or use automated measurements such as MV or FCS. In the interest of time and efficiency, many clinicians choose the latter option since these automated machine–derived measurements are objective, quantitative, somewhat consistent, and can be calculated quickly. On the down side, automated measurements have higher rates of errors in complex retinal diseases,8–10,15 they do not measure subcomponents such as CME or SRF independently, they may not reflect sub-RPE changes reliably, and they may be insensitive to small changes in lesion size. Although the data in this study suggest a highly significant correlation between automated MV and SRF volumes (R2 = 0.62), this disappears entirely (R2 = 0.00) when only a single data pair of 20 is removed. This highlights that this seemingly significant conclusion is dependent on a single data point and may not be found in other data sets.
In contrast, reading centers have developed customized methods to extract more data from SD-OCT scans than are available to the typical clinician. For instance, some reading centers perform manual measurements at the foveal center point, some provide protocol-driven qualitative assessments of abnormal features, whereas others provide exhaustive manual grading of every feature in every B-scan. In general, these processes are either quick with limited data extraction or too time-consuming to be implemented in clinical practices where busy clinicians need a quick and reliable measure of progression.
In this context, we aimed to find a compromise solution that was accurate enough to be used by reading centers in clinical trials yet quick enough to be used by busy clinicians. Based on the assumption that fluid-based features have similar morphologies between patients (e.g., CME is spherical, whereas SRF and PEDs are dome-shaped), our hypothesis was that we could use a single dimension of these lesions to predict the lesion volume with a high degree of accuracy. We did not include subretinal tissue (SRT) in this study since we felt that this premise applied only to fluid-filled lesions and would not extend to SRT since solid tissue spaces retain their own shape instead of obeying common laws of fluid dynamics.
By reducing the grading to three basic parameters—A-scan count, B-scan count, and maximum height—we demonstrated excellent correlations with our gold-standard lesion volumes, producing R2 values of up to 0.96 (Table 2). B-scan count and maximum lesion height are measures that can be easily estimated using software of most manufacturers. In contrast, A-scan count is more difficult to obtain. In our study, the only instrument that natively supported A-scan display of SD-OCT volume data was the Cirrus HD-OCT. Furthermore, the decreased transverse resolution and undulating baseline due to axial motion during acquisition led to decreased reproducibility and greater time investment in subjective A-scan grading of these features (κ = 0.69). Although PED volumes did not correlate as well as SRF and CME volumes with the simplified measures, we attribute this difference to the heterogeneous morphology of PEDs. Some PEDs were high and dome-shaped, whereas most showed a highly variable contour with several peaks. In contrast, both SRF and CME seem to spread out in a more symmetric regular pattern, thereby enabling more reliable modeling of actual volumes. Following this logic, it is possible that simple measures may correlate better with serous PED volumes than fibrovascular PED volumes, although this was not studied in this analysis.
Combinations of at least two parameters such as A-scan count × B-scan count × Maximum lesion height produced slightly better correlations. However, the fractional improvements in correlation with these combinations were outweighed by a >100% increase in time required to arrive at these estimations. In addition, incorporation of A-scan counts with low reproducibility may increase the chance of a spurious conclusion. Therefore, we do not recommend including A-scans as a valid parameter for simplified grading at the time being. Ultimately, split sample analysis demonstrated that models derived from simple parameters can predict absolute volumes with average relative errors of 20% or less for all tested features (Table 4).
In practice, clinicians may be less interested in calculating absolute lesion volumes than they are in determining improvement or worsening in a single patient. Therefore, analysis of the LA data in this study may be more applicable to clinical practice since these cases were followed over time. The commonly used OCT retreatment criteria that attempt to predict worsening or improvement were proposed by the PrONTO trial.3,6 In particular, changes in FCS thickness ≥ 100 μm and visible fluid on OCT in combination with a loss of visual acuity have been adopted by some clinicians in their practices. In the 24 volume scans of five patients in our study, FCS thickness correlated with changes only in some features over time. This is not surprising since significant fluctuations in lesion size may still be seen well below FCS changes of 100 μm. Furthermore, changes in FCS are not specific for the underlying reasons for this change.
In contrast, changes in our simplified grading correlated better with changes in lesion volumes over time than changes in MV or FCS. Sensitivity–specificity analysis for categorical assessments of increased or decreased lesion size also was superior to MV and FCS measurements for CME and SRF. In this study, macular volume had a high sensitivity and lower specificity, whereas FCS was more specific in detection of changes measured by manual grading. With the exception of PED, changes in simplified measures of SRF and CME were very accurate at assessing increases or decreases in lesion volumes.
From a reading center perspective, one crucial advantage of simplified grading is that the time required to arrive at a quantitative result is close to manual foveal center measurements, yet achieves accuracies that approach the laborious methods that take 50 times longer. In fact, the time for simplified grading may be reduced even further when focusing only on those parameters with the highest volume-correlation per feature. Although previous reports have suggested significant time savings without compromising the measurement accuracy by using only a subset of B-scans in a 128 B-scan volume set, this was established for retinal thickness only and may not be applicable to the small, more focal features seen in NVAMD.16
Despite the promising conclusions from this study, it has several important limitations. The number of visits included in the analysis is limited due to the tremendous amount of time required for manual multilayered segmentation of full SD-OCT volume scans. Therefore, despite achieving statistical significance in the study end points, it can be argued that the data set lacked enough power to prevent a type II error. Permutation correlation analysis suggests that these data were sufficient since the same conclusion would have been reached with a fraction of the data set.
Retrospective selection of cases focusing on scans with more pathologic features may have introduced a selection bias into these data. This could be especially problematic if the morphology of the individual features measured in this study differs with disease stage. For instance, if SRF has a certain typical shape on initial presentation that changes as the disease progresses (and CME or larger PEDs develop), then a selection bias might result in poor predictions of early SRF volumes. To our knowledge this difference in morphology has not been reported in the literature nor has it been noticed by the authors.
Another potential weakness is that we did not differentiate between treated and untreated eyes nor by the type of treatment being received. It could be argued that the morphology of these features might differ between eyes undergoing primary ranibizumab therapy versus eyes that failed preceding bevacizumab treatment. However, the similarity in results in both the longitudinal and cross-sectional groups of our randomly selected sample pool would suggest that our findings may be more universally applicable without a noticeable bias toward individual treatment status.
Finally, only two different SD-OCT devices and one scan protocol were evaluated in this study. Even though the scan protocol is the most commonly used protocol in clinical practice and clinical trials, it still cannot be assumed that these conclusions apply to other machines, especially those that use radically different scan sequences. Similarly, none of the devices evaluated used an eye-tracking mechanism. It is plausible that the correlation of automated measures in both cross-sectional and longitudinal settings may be improved by eye tracking, although we do not expect these differences to significantly change the correlation values we found.
In conclusion, simplified estimates for fluid volumes in NVAMD in this study were excellent predictors of actual fluid volumes as well as longitudinal changes in fluid volumes. Since these methods are quick and accessible to users of most SD-OCT devices, they may be viable alternatives for clinicians to use to obtain basic quantitative information without the need for manual segmentation or specialized grading software. Moreover, since these same methods could readily be adopted by reading centers, it is also possible that these measures could help bridge the methodologic gap that may exist between clinical trials and clinical practice.
Footnotes
Supported in part by National Eye Institute Grants EY03040 and R01 EY014375, and Deutsche Forschungsgemeinschaft Grant He 6094/1-1.
Disclosure: F.M. Heussen, None; Y. Ouyang, None; S.R. Sadda, Carl Zeiss Meditec AG (F), Optovue Inc. (F), Heidelberg Engineering (C), P; A.C. Walsh, P
References
- 1. Sadda SR, Wu Z, Walsh AC, et al. Errors in retinal thickness measurements obtained by optical coherence tomography. Ophthalmology. 2006;113:285–293 [DOI] [PubMed] [Google Scholar]
- 2. Patel PJ, Chen FK, Ikeji F, et al. Repeatability of stratus optical coherence tomography measures in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:1084–1088 [DOI] [PubMed] [Google Scholar]
- 3. Fung AE, Lalwani GA, Rosenfeld PJ, et al. An optical coherence tomography-guided, variable dosing regimen with intravitreal ranibizumab (Lucentis) for neovascular age-related macular degeneration. Am J Ophthalmol. 2007;143:566–583 [DOI] [PubMed] [Google Scholar]
- 4. Kaiser PK, Blodi BA, Shapiro H, Acharya NR. MARINA Study Group Angiographic and optical coherence tomographic results of the MARINA study of ranibizumab in neovascular age-related macular degeneration. Ophthalmology. 2007;114:1868–1875 [DOI] [PubMed] [Google Scholar]
- 5. Sadda SR, Stoller G, Boyer DS, et al. Anatomical benefit from ranibizumab treatment of predominantly classic neovascular age-related macular degeneration in the 2-year anchor study. Retina. 2010;30:1390–1399 [DOI] [PubMed] [Google Scholar]
- 6. Lalwani GA, Rosenfeld PJ, Fung AE, et al. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: year 2 of the PrONTO Study. Am J Ophthalmol. 2009;148:43–58 [DOI] [PubMed] [Google Scholar]
- 7. Regillo CD, Brown DM, Abraham P, et al. Randomized, double-masked, sham-controlled trial of ranibizumab for neovascular age-related macular degeneration: PIER Study year 1. Am J Ophthalmol. 2008;145:239–248 [DOI] [PubMed] [Google Scholar]
- 8. Krebs I, Falkner-Radler C, Hagen S, et al. Quality of the threshold algorithm in age-related macular degeneration: Stratus versus Cirrus OCT. Invest Ophthalmol Vis Sci. 2009;50:995–1000 [DOI] [PubMed] [Google Scholar]
- 9. Mylonas G, Ahlers C, Malamos P, et al. Comparison of retinal thickness measurements and segmentation performance of four different spectral and time domain OCT devices in neovascular age-related macular degeneration. Br J Ophthalmol. 2009;93:1453–1460 [DOI] [PubMed] [Google Scholar]
- 10. Keane PA, Mand PS, Liakopoulos S, Walsh AC, Sadda SR. Accuracy of retinal thickness measurements obtained with Cirrus optical coherence tomography. Br J Ophthalmol. 2009;93:1461–1467 [DOI] [PubMed] [Google Scholar]
- 11. Joeres S, Tsong JW, Updike PG, et al. Reproducibility of quantitative optical coherence tomography subanalysis in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2007;48:4300–4307 [DOI] [PubMed] [Google Scholar]
- 12. Sadda SR, Joeres S, Wu Z, et al. Error correction and quantitative subanalysis of optical coherence tomography data using computer-assisted grading. Invest Ophthalmol Vis Sci. 2007;48:839–848 [DOI] [PubMed] [Google Scholar]
- 13. Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431 [DOI] [PubMed] [Google Scholar]
- 14. Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–1444 [DOI] [PubMed] [Google Scholar]
- 15. Han IC, Jaffe GJ. Comparison of spectral- and time-domain optical coherence tomography for retinal thickness measurements in healthy and diseased eyes. Am J Ophthalmol. 2009;147:847–858 [DOI] [PubMed] [Google Scholar]
- 16. Sadda SR, Keane PA, Ouyang Y, Updike JF, Walsh AC. Impact of scanning density on measurements from spectral domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2010;51:1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]