Abstract
Bioorthogonal reactions suitable for functionalization of genetically or metabolically encoded alkynes, e.g., copper-catalyzed azide-alkyne cycloaddition reaction (“click chemistry”), have provided chemical tools to study biomolecular dynamics and function in living systems. Despite its prominence in organic synthesis, copper-free Sonogashira cross-coupling reaction suitable for biological applications has not been reported. In this work, we report the discovery of a robust aminopyrimidine-palladium(II) complex for copper-free Sonogashira cross-coupling that enables selective functionalization of a homopropargylglycine (HPG)-encoded ubiquitin protein in aqueous medium. A wide range of aromatic groups including fluorophores and fluorinated aromatic compounds can be readily introduced into the HPG-containing ubiquitin under mild conditions with good to excellent yields. The suitability of this reaction for functionalization of HPG-encoded ubiquitin in E. coli was also demonstrated. The high efficiency of this new catalytic system should greatly enhance the utility of Sonogashira cross-coupling in bioorthogonal chemistry.
Because of their small sizes and ease in the genetic/metabolic incorporation, simple organic groups such as azide, aldehyde, terminal alkyne, and terminal alkenes are attractive bioorthogonal reporters for studying biomolecular dynamics and functions in their native environment.1 One of the key driving forces for this approach has been the emergence of a growing repertoire of bioorthogonal reactions2 through which designed small-molecule probes can be selectively conjugated to pre-tagged biomolecules. For tracking biological dynamics, bioorthogonal functionalization needs to be rapid, highly selective, and high yielding. Indeed, an aldehyde can be selectively functionalized with the hydrazide or alkoxyamine-based probes via nucleophilic addition;3 an azide can be functionalized with the alkyne probes via copper-catalyzed click chemistry,4 strained-promoted cycloaddition,5 or Staudinger ligation6; and terminal alkenes can be functionalized via either photoclick chemistry7 or ruthenium-catalyzed olefin metathesis.8 While many aqueous reactions involving terminal alkynes have been reported,9 the viable option for functionalization of terminal alkynes in biological systems remains to be click chemistry.10
In an effort to apply palladium-catalyzed reactions to modify proteins in vitro, Yokoyama and co-workers previously reported a Sonogashira cross-coupling reaction between N-(prop-2-yn-1-yl)biotinamide and an iodophenyl-containing Ras protein in an aqueous medium containing 18% DMSO.11 However, a low yield of 25% was achieved after 80-min incubation at 6 °C under N2 protection; in addition, copper(I) salt was used as a co-catalyst, which further limits its potential in cellular applications because of the known cytotoxicity associated with copper.12 Inspired by the recent report of a robust Suzuki-Miyaura cross-coupling with protein substrates in aqueous medium,13 we envisioned that with a suitable palladium catalyst, efficient copper-free Sonogashira cross-coupling with protein substrates could be realized. Here, we report the discovery of a new water-soluble palladium complex, and its utility in copper-free Sonogashira cross-coupling reactions in clean and rapid formation of new carbon-carbon bonds between a terminal alkyne-containing peptide/protein and a wide range of aryl iodides in aqueous medium. Furthermore, we demonstrate the suitability of this reaction for fluorescent labeling of an alkyne-encoded protein in E. coli cells.
Our initial study focused on the Sonogashira cross-coupling between fluorescein iodide 1a and a homopropargylglycine (HPG)-containing peptide 2a using a water-soluble 2-amino-4,6-dihydroxypyrimidine (ADHP) as the palladium ligand because ADHP has been previously used successfully in the copper-free Sonogashira reactions in organic solvents.14 However, no cross-coupling product was detected after 30 min (Table 1, entry 1); similar result was obtained when the reaction was extended to 3 hours. We attributed this lack of reactivity to the hydration of ADHP at either the two hydroxy groups or the amino group in aqueous medium. To alter the hydration sphere of ADHP and fine-tune the electron density of the pyrimidine ring, a series of pyrimidine-based ligands (L2–L7) were synthesized and their catalytic activities were examined in the model reaction in potassium phosphate buffer (Table 1). To our delight, we found that ligands L2 and L5 with the dimethylamino substituent gave the desired product 3a in 59% and 29% yield, respectively (entries 2 and 5). Blocking the two hydroxyl group led to trace amount of product (entries 3 and 4). Replacement of the amino group with bulkier morpholine or piperidine group resulted in substantially lower yields (entries 6 and 7). Using the ligand L2-palladium complex, we obtained the cross-coupled product 3a in 91% yield when palladium complex usage was increased to 30% along with an excess of 2a and a slightly longer reaction time (entry 8).
Table 1.
Ligand Screen and Reaction Condition Optimization a
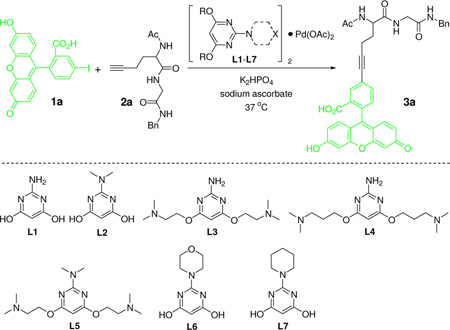 | |||||
|---|---|---|---|---|---|
| Entry | 1a : 2a | Pd • ligand | Ligand | Time | Yield (%) b |
| 1 | 1.05:1.00 | 20% | L1 | 30 min | NR |
| 2 | 1.05:1.00 | 20% | L2 | 30 min | 59 |
| 3 | 1.05:1.00 | 20% | L3 | 30 min | <5 |
| 4 | 1.05:1.00 | 20% | L4 | 30 min | <5 |
| 5 | 1.05:1.00 | 20% | L5 | 30 min | 29 |
| 6 | 1.05:1.00 | 20% | L6 | 30 min | <5 |
| 7 | 1.05:1.00 | 20% | L7 | 30 min | 13 |
| 8 | 1.00:2.40 | 30% | L2 | 40 min | 91 |
Reactions were carried out under ambient conditions without the need of insert gas protection.
Yields were derived by comparing the integration area of 3a in their respective HPLC traces to that of a standard curve. See Supporting Information for details.
With the optimal conditions in hand, we then investigated the generality of this protocol for various aryl iodide (Table 2). Both ortho- or para-substituted aryl iodides gave good conversions (entries 1, 3 and 5). Sterically hindered 2,4,6-trimethylphenyl iodide also participated in the reaction and gave a 72% conversion (entry 6). Heterocyclic aryl iodide, 2-iodothiophene, underwent successful reaction with 2a to afford the cross-coupled product in 95% conversion (entry 2). A fluorogenic 4-iodo-7-methoxy-coumarin was also a good substrate (entry 4). However, electron-deficient aryl iodides such as 4-nitrophenyl iodide gave poor conversions under these conditions.
Table 2.
Aryl Iodides in Aqueous Copper-Free Sonogashira Cross-Coupling Reactions a
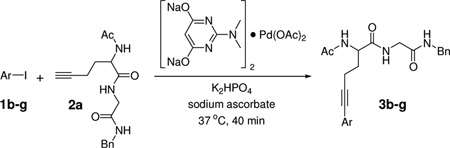 | |||
|---|---|---|---|
| Entry | Ar-I | Pd • L2 | Conversion (%)b |
| 1 | 30% | 84 | |
| 2 | 30% | 95 | |
| 3 | 30% | 77 | |
| 4 | 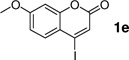 |
30% | 88 |
| 5 | 30% | 70 | |
| 6 | 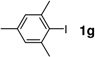 |
30% | 72 |
Reactions were carried out on a 4 µmol scale at 37 °C with 1:2a ratio of 1:2.4.
Conversions were calculated based on the disappearance of aryl iodides in the reaction mixtures in the HPLC. See Supporting Information for details.
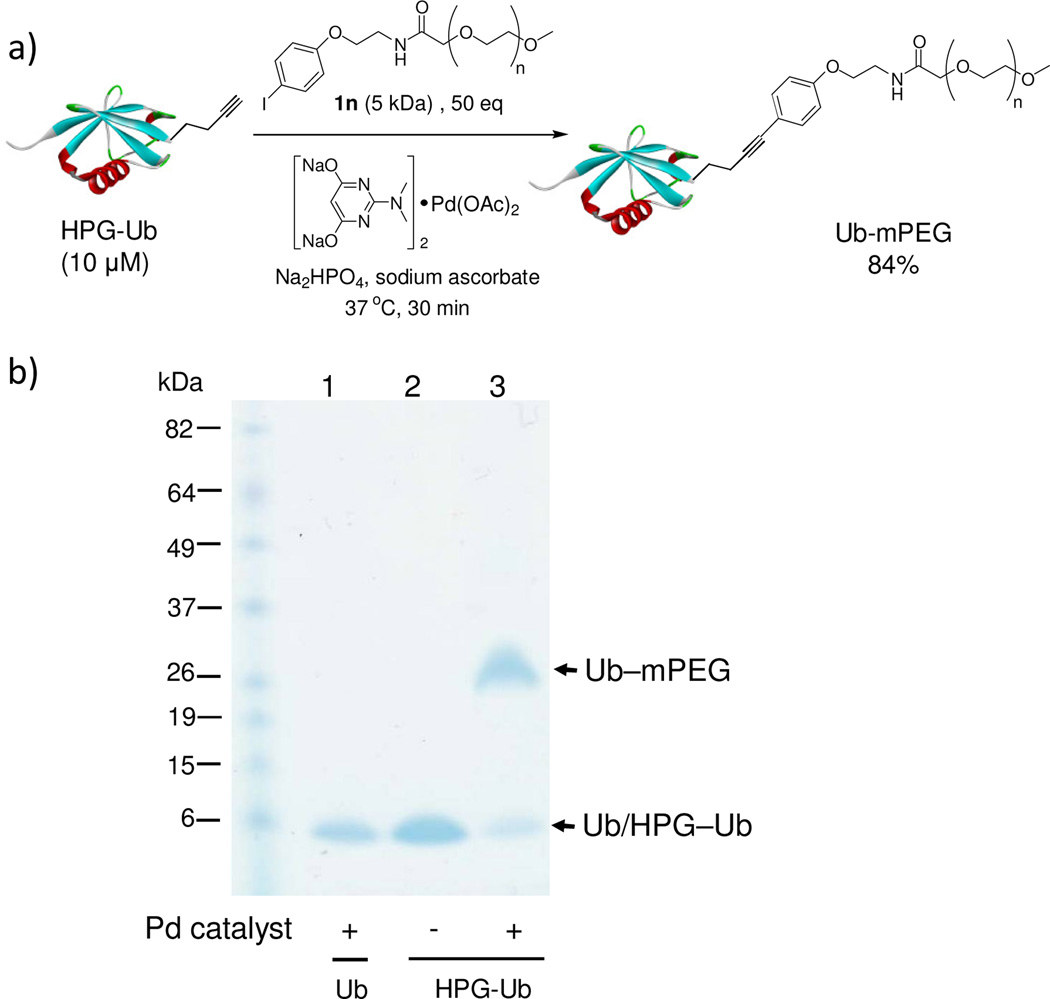
The encouraging results with peptide substrate 2a prompted us to examine whether these conditions can be employed to modify a HPG-encoded protein. We selected ubiquitin as a protein model because of its small size and robust fold.15 Since HPG has been shown in the literature to be an efficient methionine surrogate for metabolic incorporation,16 we appended a Met-Gly-Gly sequence to the C-terminus of ubiquitin on a pQE80 construct, and expressed the HAG-containing ubiquitin in M15A, a methionine auxotroph, in the presence of HPG. After Ni-NTA affinity chromatography, the N-terminal His-tag, together with the first Met or HPG in the sequence, was cleaved by treating the protein with PreScission protease. The resulting ubiquitin was purified by FPLC to afford either wild-type ubiquitin (Ub) or HPG-encoded ubiquitin (HPG–Ub) with one HPG at its C-terminus. The HPG occupancy was determined to be 92% based on the LC-MS analysis. To test Sonogashira reaction with the HPG-containing proteins, we incubated HPG-Ub with 50 equiv of 1a and 50 equiv of palladium complex using a two-step addition procedure.17 Gratifyingly, the reaction reached completion in 30 min (Table 3, entry 1) as monitored by LC-MS. Control experiments showed that the reaction did not proceed in the absence of palladium complex. By taking advantage of the intrinsic fluorescence of 1a, we subjected the reaction mixture to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by in-gel fluorescence analysis. A clear green fluorescent band was detected only for HPG–Ub but not for Ub (Figure S2). We then examined a range of aryl iodides containing various functional groups including hydroxy (1i), carboxylic acid (1h), fluorine (1j, 1k), nitro (1l), and thiophene (1c) groups (Table 3). In general, Sonogashira cross-coupling reactions with HPG–Ub gave the modified proteins in good yields (55–93%) independent of the electronic structures of aryl iodides. Sterically hindered substrates such as 1g and 1m gave lower yields (entries 7 and 13), even with extended reaction times. To verify the yield and selectivity of the reaction, HPG–Ub was incubated with 50 equiv of phenyl iodide covalently linked to monodisperse polyethylene glycol (1n, MW ≈ 5 kDa) in a sodium phosphate buffer at 37 °C for 30 min, and the mixture was then analyzed by SDS-PAGE (Figure 1). A distinct higher molecular weight band corresponding to Ub–mPEG with an estimated 84% yield was observed, matching closely the yield for analogous substrate, 1-iodo-4-methoxy benzene (1f; Table 3, entry 6).18 Importantly, no PEGylated products were detected in the absence of the palladium complex, and the reaction proceeded only with HPG–Ub and not with Ub (lane 1 in Figure 1), indicating that PEGylation occurs selectively with HPG via the Sonogashira cross-coupling.
Table 3.
Aryl Iodide Substrates for Cross-Coupling with HPG–Ub a
 | |||
|---|---|---|---|
| Entry | Ar-I | Time | Yield (%)b |
| 1 | 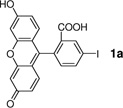 |
30 min | 93 |
| 2 | 30 min | 83 | |
| 3 | 30 min | 55 | |
| 4 | 30 min | 86 | |
| 5 | 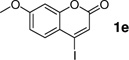 |
240 min | 73 c |
| 6 | 30 min | 83 | |
| 7 | 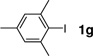 |
180 min | 73 |
| 8 | 30 min | 82 | |
| 9 | 30 min | 85 | |
| 10 | 30 min | 78 | |
| 11 | 30 min | 79 | |
| 12 | 180 min | 66 | |
| 13 | 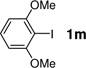 |
180 min | 60 |
Reactions were carried out with 5–10 µM of HPG-Ub and 50 equiv of aryl iodide and palladium complex at 37 °C under ambient conditions.
Yields were determined based on LC-MS analysis of the reaction mixtures based on the equation: yield % = Iproduct/(IHPG-Ub + Iproduct). When side products were detected, the yield was calculated using the equation: yield % = Iproduct/(IHPG-Ub + Iproduct + Iside product), where IHPG-Ub, Iproduct, and Iside product represent the ion counts of the remaining HPG-Ub, product, and side product, respectively. See Supporting Information for details.
Hydrolysis of the lactone ring was observed in mass spectrum.
Figure 1.
Selective PEGylation of HPG-Ub with an mPEG-linked phenyl iodide 1n via copper-free Sonogashira cross-coupling. (a) Reaction scheme. (b) Coomassie blue staining of SDS-PAGE gel showing selective formation of the PEGylated Ub (Ub-mPEG): lane 1, control reaction with wild-type ubiquitin; lane 2: control reaction with HPG-Ub in the presence of 50 equiv of 1n but absence of palladium complex; lane 3: reaction with HPG-Ub in the presence of 50 equiv of 1n and palladium catalyst.
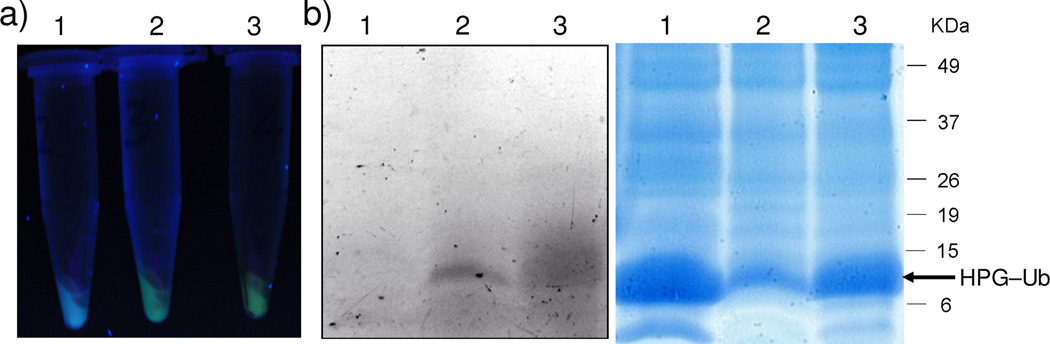
Since many enzyme-mediated biochemical transformations are facilitated by transition metals,19 selective palladium-catalyzed transformations inside living cells may offer an organometallic tool to manipulate biological processes. To assess whether the copper-free Sonogashira cross-coupling reaction can be employed to modify HPG–Ub in bacterial cells, we added 1 mM of palladium complex, 100 µM of fluorescein iodide 1a, and 5 mM of sodium ascorbate in sodium phosphate buffer to HPG–Ub-overexpressing M15A cells. After growing at 37 °C for 4 h, cells were collected and washed extensively with 0.9% NaCl solution to remove the excess reagents. The palladium complex-treated cells showed green fluorescence upon 365-nm excitation while the untreated cells did not (compare cell pellets 2 and 3 to 1 in Figure 2a). Subsequently, cells were lysed and HPG–Ub proteins in the cell lysates were captured with Ni-NTA-agarose beads. The beads were then heated to 95 °C for 5 min and the supernatants were analyzed by SDS-PAGE. In the in-gel fluorescence analysis, clear florescent bands were observed for samples derived from cell pellets 2 and 3, but not for the sample derived from pellet 1 (left panel, Figure 2b), indicating selective cross-coupling of 1a with HPG–Ub inside E. coli cells. The identities of the fluorescent HPG–Ub bands were confirmed by Coomassie blue staining of the same gel (right panel, Figure 2b). The lower abundance of HPG-Ub protein in seen sample 2 was probably due to the inhibition of protein expression by the palladium complex and 1a when they were added separately.20
Figure 2.
Fluorescent labeling of HPG–Ub by fluorescein iodide 1a in E. coli cells via copper-free Sonogashira cross-coupling. (a) Fluorescence image of the cell pellets upon excitation at 365 nm. Pellet 1, cells collected after treatment with 100 µM of fluorescein iodide 1a but not palladium complex; Pellet 2, cells collected after treatment with 100 µM 1a, 1 mM palladium complex, 5 mM sodium ascorbate in Na2HPO4 buffer at 37 °C for 4 h; Pellet 3, cells collected after treatment of the pre-activated mixture of palladium complex (1 mM) and 1a (100 µM). See Supporting Information for the treatment details. (b) In-gel fluorescence (left panel) and Coomassie blue staining (right panel) of SDS-PAGE analysis of the proteins captured by Ni-NTA-agarose beads from the lysates of the three cell pellets.
In conclusion, we have discovered a new palladium complex for the aqueous copper-free Sonogashira cross-coupling reactions. This palladium complex promoted the efficient cross-coupling of a wide range of aryl iodides with the terminal alkyne-containing peptide and protein substrate in good to excellent yields. The ability of this cross-coupling reaction to functionalize a metabolically encoded alkyne-containing protein in E. coli was also demonstrated, indicating a potential utility of this reaction in monitoring and manipulating proteins in cellular systems. Efforts to further optimize this palladium-catalyzed cross-coupling reaction for cellular applications, e.g., by immobilizing the complex onto nanoparticles to improve cell penetration and reduce complex loading,21 are currently underway.
Supplementary Material
ACKNOWLEDGMENT
We gratefully acknowledge the National Institutes of Health (R01 GM 085092) for financial support.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthetic schemes for new ligands, experimental procedures, and characterization of all compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. Nat. Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lim RK, Lin Q. Chem. Commun. 2010;46:1589–1600. doi: 10.1039/b925931g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chalker JM, Bernardes GJL, Davis BG. Acc. Chem. Res. ASAP; 2011. DOI: 10.1021/ar200056q. [DOI] [PubMed] [Google Scholar]
- 3.(a) Carrico IS, Carlson BL, Bertozzi CR. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]; (b) Zeng Y, Ramya TN, Dirksen A, Dawson PE, Paulson JC. Nat. Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]; (c) Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 5.(a) Agard NJ, Prescher JA, Bertozzi CR. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]; (b) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc. Natl. Acad. Sci. USA. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ning X, Guo J, Wolfert MA, Boons G-J. Angew. Chem. Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Proc. Natl. Acad. Sci. USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Blackman ML, Royzen M, Fox JM. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]; (b) Nilsson BL, Kiessling LL, Raines RT. Org. Lett. 2000;2:1939–1941. doi: 10.1021/ol0060174. [DOI] [PubMed] [Google Scholar]; (c) Kohn M, Breinbauer R. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 7.Song W, Wang Y, Qu J, Madden MM, Lin Q. Angew. Chem. Int. Ed. 2008;47:2832–2835. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]; (b) Song W, Wang Y, Qu J, Lin Q. J. Am. Chem. Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]; (c) Wang Y, Song W, Hu WJ, Lin Q. Angew. Chem. Int. Ed. 2009;48:5330–5333. doi: 10.1002/anie.200901220. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Song W, Wang Y, Yu Z, Vera CI, Qu J, Lin Q. ACS Chem. Biol. 2010;5:875–885. doi: 10.1021/cb100193h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang J, Zhang W, Song W, Wang Y, Yu Z, Li J, Wu M, Wang L, Zang J, Lin Q. J. Am. Chem. Soc. 2010;132:14812–14818. doi: 10.1021/ja104350y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lim RK, Lin Q. Acc. Chem. Res. ASAP; 2011. DOI: 10.1021/ar200021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Lin YA, Chalker JM, Floyd N, Bernardes GJL, Davis BG. J. Am. Chem. Soc. 2008;130:9642–9643. doi: 10.1021/ja8026168. [DOI] [PubMed] [Google Scholar]; (b) Ai H-w, Shen W, Brustad E, Schultz PG. Angew. Chem. Int. Ed. 2010;49:935–937. doi: 10.1002/anie.200905590. [DOI] [PubMed] [Google Scholar]; (c) Lin YA, Chalker JM, Davis BG. J. Am. Chem. Soc. 2010;132:16805–16811. doi: 10.1021/ja104994d. [DOI] [PubMed] [Google Scholar]
- 9.(a) Herrerías CI, Yao X, Li Z, Li CJ. Chem. Rev. 2007;107:2546–2562. doi: 10.1021/cr050980b. [DOI] [PubMed] [Google Scholar]; (b) Doucet H, Hierso J–C. Angew. Chem. Int. Ed. 2007;46:834–871. doi: 10.1002/anie.200602761. [DOI] [PubMed] [Google Scholar]; (c) Firouzabadi H, Iranpoor N, Gholinejad M. J. Mol. Catal. A: Chem. 2010;321:110–116. [Google Scholar]
- 10.For a recent example, see: Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem. Int. Ed. 2011;50:8051–8056. doi: 10.1002/anie.201101817.
- 11.Kodama K, Fukuzawa S, Nakayama H, Sakamoto K, Kigawa T, Yabuki T, Matsuda N, Shirouzu M, Takio K, Yokoyama S, Tachibana K. ChemBioChem. 2007;8:232–238. doi: 10.1002/cbic.200600432. [DOI] [PubMed] [Google Scholar]
- 12.(a) Speers AE, Adam GC, Cravatt BF. J. Am. Chem. Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]; (b) Kennedy DC, Lyn RK, Pezacki JP. J. Am. Chem. Soc. 2009;131:2444–2445. doi: 10.1021/ja809451w. [DOI] [PubMed] [Google Scholar]
- 13.(a) Chalker JM, Wood CSC, Davis BG. J. Am. Chem. Soc. 2009;131:16346–16347. doi: 10.1021/ja907150m. [DOI] [PubMed] [Google Scholar]; (b) Spicer CD, Davis BG. Chem. Commun. 2011;47:1698–1700. doi: 10.1039/c0cc04970k. [DOI] [PubMed] [Google Scholar]
- 14.Li JH, Zhang XD, Xie YX. Eur. J. Org. Chem. 2005:4256–4259. [Google Scholar]
- 15.Vijay-Kumar S, Bugg CE, Cook WJ. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 16.van Hest JCM, Kiick KL, Tirrell DA. J. Am. Chem. Soc. 2000;122:1282–1288. [Google Scholar]
- 17.(a)Our preliminary studies indicated that excess amount of the palladium complex was necessary to achieve high conversions due to nonspecific sequestration of the palladium complex by the proteins; see Figure S1 in Supporting Information for details. Similar phenomena were also observed in the palladium-catalyzed Suzuki-Miyaura cross-coupling reactions with protein substrates; see reference 13. To further alleviate this problem, we also pre-incubated the palladium complex with the aryl iodide (pre-activation) to minimize the effect of non-specific binding. (b) 2.4% DMSO was used in the reaction to help dissolve fluorescein iodide. (c) We found by adding the palladium complex/1a mixture pre-incubated for 1 h (“pre-activated”) into the HPG-Ub solution in two portions instead of one improved the reaction yield.
- 18.The greater than 5 kDa increase in apparent MW is typically observed in electrophoresis of PEGylated proteins: see ref. 7c as well as Lim RK, Lin Q. Chem. Commun. 2010;46:7993–7995. doi: 10.1039/c0cc02863k. Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG. Bioorg. Med. Chem. Lett. 2004;14:5743–5745. doi: 10.1016/j.bmcl.2004.09.059.
- 19.(a) Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; 1994. [Google Scholar]; (b) Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]; (c) Antos JM, Francis MB. Curr. Opin. Chem. Biol. 2006;10:253–262. doi: 10.1016/j.cbpa.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 20.In our cytotoxicity studies based on colony formation assay and growth curve measurement, the pre-activated palladium complex/1a mixture and 1a itself didn’t show toxicity to E. coli cells while palladium complex and palladium complex/1a mixture added separately showed some level of cytotoxicity. See Figure S4 in Supporting Information for details.
- 21.Yusop RM, Unciti-Broceta A, Johansson EMV, Sánchez-Martín RM, Bradley M. Nature Chem. 2011;3:239–243. doi: 10.1038/nchem.981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.