Abstract
Background
Paphiopedilum is a horticulturally and ecologically important genus of ca. 80 species of lady's slipper orchids native to Southeast Asia. These plants have long been of interest regarding their chromosomal evolution, which involves a progressive aneuploid series based on either fission or fusion of centromeres. Chromosome number is positively correlated with genome size, so rearrangement processes must include either insertion or deletion of DNA segments. We have conducted Fluorescence In Situ Hybridization (FISH) studies using 5S and 25S ribosomal DNA (rDNA) probes to survey for rearrangements, duplications, and phylogenetically-correlated variation within Paphiopedilum. We further studied sequence variation of the non-transcribed spacers of 5S rDNA (5S-NTS) to examine their complex duplication history, including the possibility that concerted evolutionary forces may homogenize diversity.
Results
5S and 25S rDNA loci among Paphiopedilum species, representing all key phylogenetic lineages, exhibit a considerable diversity that correlates well with recognized evolutionary groups. 25S rDNA signals range from 2 (representing 1 locus) to 9, the latter representing hemizygosity. 5S loci display extensive structural variation, and show from 2 specific signals to many, both major and minor and highly dispersed. The dispersed signals mainly occur at centromeric and subtelomeric positions, which are hotspots for chromosomal breakpoints. Phylogenetic analysis of cloned 5S rDNA non-transcribed spacer (5S-NTS) sequences showed evidence for both ancient and recent post-speciation duplication events, as well as interlocus and intralocus diversity.
Conclusions
Paphiopedilum species display many chromosomal rearrangements - for example, duplications, translocations, and inversions - but only weak concerted evolutionary forces among highly duplicated 5S arrays, which suggests that double-strand break repair processes are dynamic and ongoing. These results make the genus a model system for the study of complex chromosomal evolution in plants.
Background
Paphiopedilum, a genus of approximately 80 species indigenous to tropical and subtropical Southeast Asia, is among the most widely grown and hybridized of all orchids. Species of Paphiopedilum are also ecologically important narrow endemics in various mainland and island habitats, which range from montane rainforest to seaside cliffs [1]. Karyological studies of Paphiopedilum have revealed considerable chromosomal variation, which ranges from 2n = 26 to 2n = 42, in aneuploid increments suggestive of centric fission [2]. Basic molecular phylogenetic information on the genus is available [3]. Subgenus Parvisepalum, which is sister to the rest of the genus, has 2n = 26 metacentric chromosomes, whereas the type subgenus Paphiopedilum includes both clades of 2n = 26 species and two distinct lineages of species that bear greater than 26 chromosomes, with the number of telocentrics equal to twice the number of metacentrics that ostensibly split [3]. Haploid genome size is extremely large in these orchids, ranging from 16.1 to 35.1 megabases (Mb)[4]. Chromosome number has been shown to be positively correlated with genome size [4], so rearrangement processes must include either insertion or deletion of DNA segments.
General issues in plant chromosomal evolution include the contribution of rearrangements to genome structure and size. Rearrangement processes involve double-strand break repair, which occurs frequently at hotspots in pericentromeric and telomeric regions [5,6]. Gene duplications may be caused by unequal crossing over, retrotransposition, or genome duplication [7]. Tandem repeats duplication or segmental duplication is one of the possible outcomes of unequal crossing over [7,8]. These phenomena may be investigated empirically through use of Fluorescence In Situ Hybridization (FISH) on highly repetitive DNA loci subject to concerted evolution, such as the 18S-5.8S-25S (45S) and 5S ribosomal DNA (rDNA) arrays, which may show duplication or evidence for rearrangement-producing heterologous recombination [9]. Infrageneric comparative rDNA FISH analyses, in which mobility and patterning have been systematically investigated as species-specific karyotype markers, are common in the literature [10-14]. We use such analyses here to document chromosomal dynamics in Paphiopedilum. FISH has been applied previously to Paphiopedilum, but in a limited manner only, and especially in hybrids [15,16].
Both 45S and 5S rDNAs in plants are characterized by intergenic spacers. 5S rDNA non-transcribed spacer (5S-NTS) sequences have seen some use as phylogenetic markers [17-21]. However, most studies of 5S-NTS to-date have employed direct sequencing of PCR products, and there is evidence that the NTS both within and among arrays can show polymorphism. We have cloned 5S-NTS segments in Paphiopedilum in order to study past and ongoing gene duplication events and the possibility of gene conversion both within arrays and among duplicated loci.
We briefly report distribution patterns of rDNA signals from a phylogenetic systematic perspective [22] according to accepted section-level classification. We do not aim to provide complete karyotypic comparisons, nor a full cytotaxonomic treatment; rather, we concern ourselves with demonstrable evidence for dynamic rearrangements during the evolution of Paphiopedilum. 5S-NTS sequence data are also compared with a phylogenetic hypothesis in order to ascertain duplication history of paralogs.
Results
Distribution patterns of ribosomal DNA by Fluorescence In Situ Hybridization, according to phylogeny and section-level classification
Section Parvisepalum
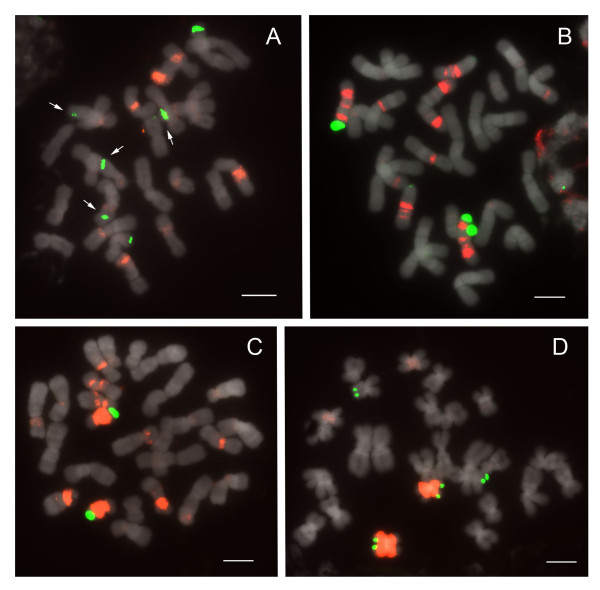
Section Parvisepalum is the sister group of all other Paphiopedilum species (Figure 1). Two to four 25S rDNA signals are apparent (Figure 2) among 2n = 26 chromosomes, with two signals most parsimoniously interpretable as the basal condition since this state is shared by the outgroup genera Mexipedium and Phragmipedium (unpublished data;[23]). With 2 signals being the inferred primitive condition, rearrangement by duplication is observed in Paphiopedilum armeniacum, P. emersonii and P. hangianum, which have more loci. 5S rDNA patterns are stable, showing 2 subtelomeric signals that are usually closely linked with one pair of 25S signals (Table 1). In P. delenatii, translocation of either the 5S or 25S rDNA locus has occurred. This phenomenon is also seen in P. malipoense, with its two chromosomes that show hemizygous 25S and 5S rDNA signals, respectively.
Figure 1.
Section-level phylogenetic tree of genus Paphiopedilum. Section-level phylogenetic tree based on rDNA ITS sequences published by Cox [3].
Figure 2.
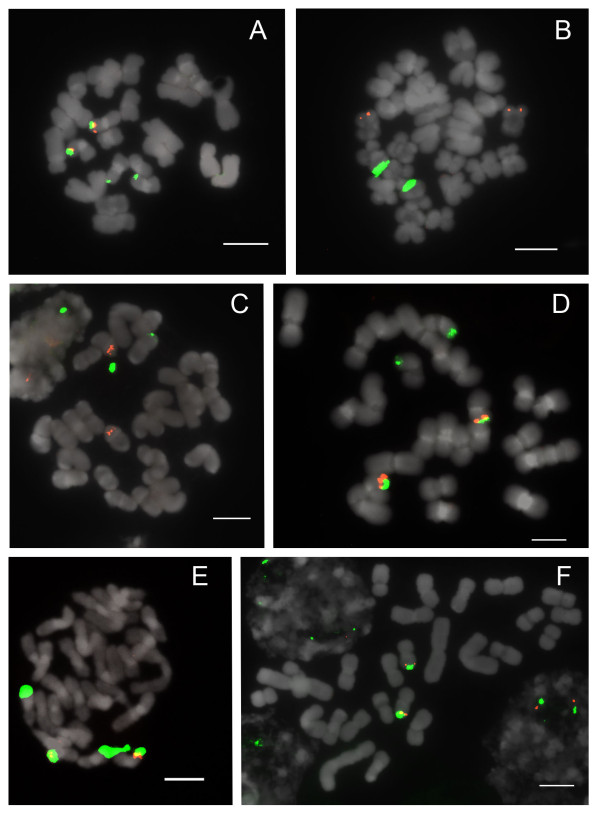
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Parvisepalum. (A) Paphiopedilum emersonii, (B) P. delenatii, (C) P. malipoense, (D) P. hangianum, (E) P. armeniacum, (F) P. micranthum. 25S rDNA (green) and 5S rDNA (red) probes were simultaneously detected in all Paphiopedilum species. Chromosomes were counterstained with DAPI. All scale bars = 10 μm.
Table 1.
Paphiopedilum species studied, diploid chromosome numbers, rDNA FISH patterns, and 5S-NTS sequence polymorphic sites
| Number of rDNA sites | Positions of rDNA sitesb | |||||||
|---|---|---|---|---|---|---|---|---|
| 5S | 25S+5S | |||||||
| Taxon | 2n | 25S | major | visible sitesa | Co-localization | 5S | 25S | 5S-NTS Polymorphic sites |
| Paphiopedilum | ||||||||
| Subg. Parvisepalum | ||||||||
| Sect. Parvisepalum | ||||||||
| armeniacum | 26 | 4 | 2 | 2 | 2 | st | t | 104 |
| delenatii | 26 | 2 | 2 | 2 | 0 | st | t | 178 |
| emersonii | 26 | 4 | 2 | 2 | 2 | st | t | 124 |
| hangianum | 26 | 4 | 2 | 2 | 2 | st | t | 120 |
| malipoense | 26 | 2 | 2 | 2 | 1 | st | t | 94 |
| micranthum | 26 | 4 | 2 | 2 | 2 | st | t | 59 |
| Subg. Paphiopedilum | ||||||||
| Sect. Concoloria | ||||||||
| bellatulum | 26 | 2 | 2 | 2 | 0 | i | t | 118 |
| niveum | 26 | 2 | 2 | 2 | 0 | i | t | 198 |
| Sect. Cochlopetalum | ||||||||
| liemianum | 32 | 2 | 4 | 22 | 2 | st, i, p, c | t | 162 |
| moquettianum | 34 | 2 | 4 | 20 | 2 | st, i, p, c | t | 225 |
| primulinum | 32 | 2 | 4 | 25 | 2 | st, i, p, c | t | 71 |
| victoria-regina | 34 | 2 | 4 | 24 | 2 | st, i, p, c | t | 137 |
| Sect. Paphiopedilum | ||||||||
| druryi | 30 | 2 | 4 | 16 | 0 | st, i, p, c | t | 184 |
| fairrieanum | 26 | 2 | 2 | 14 | 2 | st, i, p, c | t | 146 |
| henryanum | 26 | 2 | 2 | 17 | 2 | st, i, p, c | t | 180 |
| hirsutissimum | 26 | 2 | 6 | 21 | 2 | st, i, p, c | t | 182 |
| tigrinum | 26 | 2 | 6 | 17 | 2 | st, i, p, c | t | 141 |
| Sect. Coryopedilum | ||||||||
| adductum | 26 | 9 | 4 | 28 | 6 | st, i, p, c | t, st | 180 |
| gigantifolium | 26 | 6 | 6 | 32 | 6 | st, i, p, c | t | 210 |
| glanduliferum | 26 | 4 | 4 | 26 | 4 | st, i, p, c | t | 202 |
| randsii | 26 | 4 | 4 | 30 | 4 | st, i, p, c | t, st | 187 |
| sanderianum | 26 | 2 | 4 | 16 | 0 | st, i, p, c | t | 143 |
| stonei | 26 | 2 | 4 | 25 | 2 | st, i, p, c | t | 114 |
| supardii | 26 | 9 | 4 | 26 | 7 | st, i, p, c | t | 226 |
| Sect. Pardalopetalum | ||||||||
| dianthum | 26 | 2 | 4 | 28 | 2 | st, i, p, c | t | 251 |
| haynaldianum | 26 | 4 | 4 | 8 | 2 | st, i, p, c | t | 110 |
| lowii | 26 | 6 | 4 | 28 | 4 | st, i, p, c | t, st | 161 |
| parishii | 26 | 4 | 4 | 34 | 4 | st, i, p, c | t | 189 |
| Sect. Barbata | ||||||||
| acmodontum | 38 | 2 | 4 | 4 | 0 | i | t | 169 |
| curtisii | 36 | 2 | 2 | 2 | 0 | i | t | 164 |
| dayanum | 36 | 2 | 4 | 6 | 0 | i, p | t | 169 |
| hennisianum | 34 | 2 | 2 | 6 | 0 | i | t | 153 |
| purpuratum | 40 | 2 | 4 | 8 | 0 | st, i | t | 138 |
| sangii | 38 | 2 | 4 | 18 | 0 | st, i | t | 118 |
| sukhakulii | 40 | 2 | 2 | 13 | 0 | st, i | t | 151 |
| venustum | 40 | 2 | 4 | 8 | 2 | i | t | 109 |
| wardii | 42 | 2 | 4 | 4 | 0 | i | t | 159 |
a Minimum numbers of visible 5S rDNA FISH signals, including numbers of both major and visible dispersed sites.
b st, subtelomeric; t, telomeric; i, interstitial; p, pericentromeric; c, centromeric
Section Concoloria
Species of section Concoloria show two 25S and 5S signals (Table 1), each on separate chromosomes (2n = 26 total), similarly to Paphiopedilum delenatii of section Parvisepalum, except in that the 5S signals are interstitially instead of subtelomerically placed (Figure 3).
Figure 3.
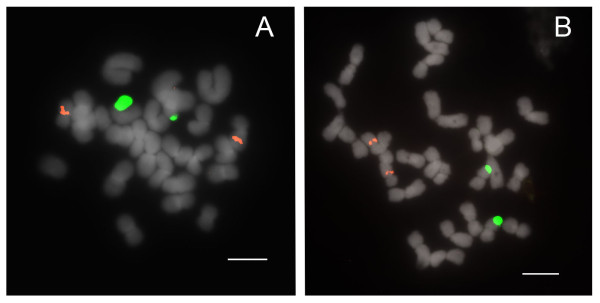
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Concoloria. (A) Paphiopedilum bellatulum, (B) P. niveum.
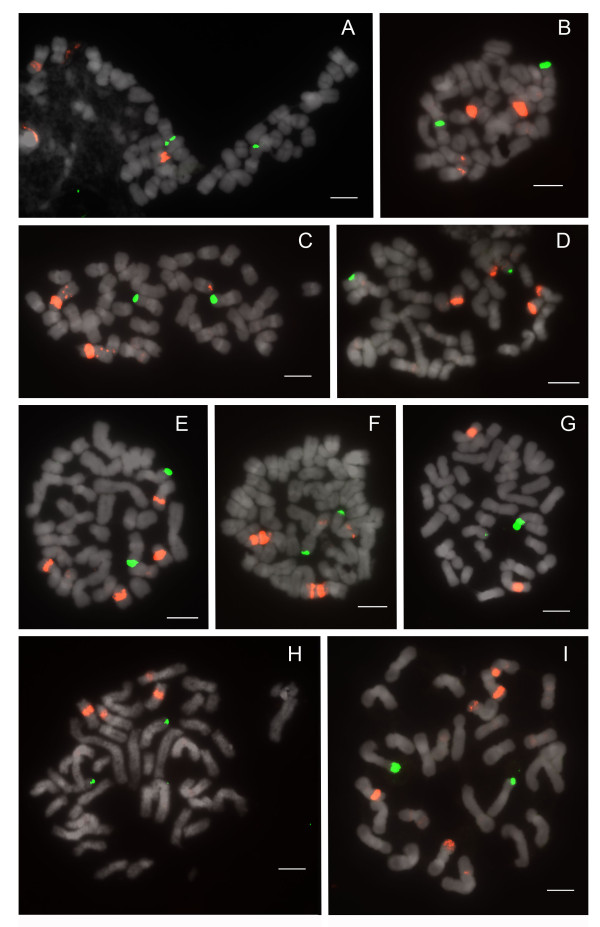
Section Cochlopetalum
Section Cochlopetalum displays an aneuploid number of chromosomes, the telocentrics of which have been suggested to descend via centric fission from 25 diploid metacentrics [2]. According to phylogenetic relationships known at present (Figure 1), and the centric fission hypothesis, sections Cochlopetalum and Barbata (with telocentrics descended from 26 diploid metacentrics) have evolved aneuploid increase independently. All four species studied here have two telomeric 25S rDNA signals, and 4 major 5S rDNA signals (Figure 4; Table 1). All 4 species have multiple dispersed 5S signals, rather unlike species of sections Parviflora and Concoloria, and these, like the major loci, are mostly subtelomeric, pericentromeric and centromeric in position. The 2 species with 2n = 32 chromosomes, Paphiopedilum liemianum (Figure 4C) and P. primulinum (Figure 4A), both have two 5S bands localized on the same chromosomes as the 25S signals, whereas only a single 5S band is seen on the same chromosome in the 2n = 34 species P. moquettianum (Figure 4B) and P. victoria-regina (Figure 4D).
Figure 4.
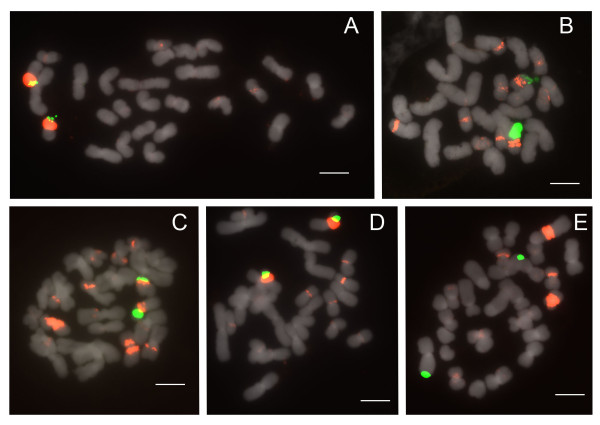
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Cochlopetalum. (A) Paphiopedilum primulinum, (B) P. moquettianum, (C) P. liemianum, (D) P. victoria-regina.
Section Paphiopedilum
All 5 species of section Paphiopedilum studied show two 25S signals in the telomeric region (Figure 5; Table 1). All species, which are 2n = 26 except for P. druryi (Figure 5E) at 2n = 30, show at least 2 specific 5S rDNA bands, as many as 6, and numerous dispersed signals in the pericentromeric and centromeric regions. In all but P. druryi the major signals are closely linked with the 25S arrays. In P. druryi, 4 of the major signals appear to be located on different arms and on morphologically different chromosomes that may only be partly homologous (this condition was observed in at least 4 cells).
Figure 5.
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Paphiopedilum. (A) Paphiopedilum fairrieanum, (B) P. hirsutissimum, (C) P. tigrinum, (D) P. henryanum, (E) P. druryi.
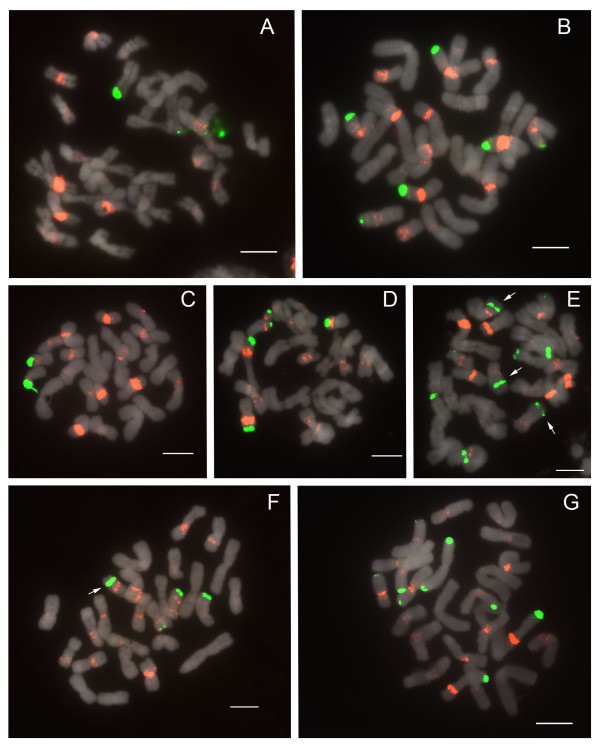
Sections Coryopedilum and Pardalopetalum
In current phylogenetic results, section Pardalopetalum is derived within section Coryopedilum (Figure 1); as such, they will be discussed together here. Together, the Coryopedilum/Pardalopetalum clade, all species having 2n = 26, is the most dynamic in Paphiopedilum regarding chromosomal rearrangements (Figure 6, 7; Table 1). 25S signals vary from 2 to 9, the latter showing hemizygosity. Signals in all species except Paphiopedilum lowii (Figure 7A), P. adductum (Figure 6E) and P. randsii (Figure 6F) are telomeric. 1-4 subtelomeric 25S signals were observed in P. lowii, P. adductum and P. randsii. In P. supardii (Figure 6G), one hemizygous chromosome has telomeric 25S signals on each arm. P. adductum also shows 25S hemizygosity, and both this species and P. supardii show the maximum number of signals. Species of the Coryopedilum/Pardalopetalum group show at least 4 major 5S rDNA signals (up to 8 in P. parishii (Figure 7B)) and multiple dispersed repeats in pericentromeric and centromeric regions. In the Pardalopetalum group, all species show at least 2 strong (up to 5) 5S bands located on one chromosome. Close linkage with 25S occurs throughout the group, other than in P. sanderianum (Figure 6A), either with major or minor 5S bands, and appearing in different placements along chromosome arms.
Figure 6.
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Coryopedilum. (A) Paphiopedilum sanderianum, (B) P. gigantifolium, (C) P. stonei, (D) P. glanduliferum, (E) P. adductum, (F) P. randsii, (G) P. supardii. Arrows indicate subtelomeric 25S rDNA signals.
Figure 7.
FISH of 25S and 5S rDNA to metaphase chromosomes of Paphiopedilum section Pardalopetalum. (A) Paphiopedilum lowii, (B) P. parishii, (C) P. dianthum, (D) P. haynaldianum. Arrows indicate subtelomeric 25S rDNA signals.
Section Barbata
Species of section Barbata, which have 2n = 28-42 and the largest genome sizes, show constancy in 25S rDNA distribution, with 2 telomeric signals (Figure 8; Table 1). Major 5S signals number 2-4, and extremely few dispersed repeats were observed. Most 5S loci are not centromeric, whereas telomeric, subtelomeric, pericentromeric, and interstitial placements are observed. Only Paphiopedilum curtisii (Figure 8G) and P. hennisianum(Figure 8B) have two major 5S signals, and the first species shows no dispersed repeats. P. sukhakulii (Figure 8C), P. venustum (Figure 8F) and P. wardii (Figure 8A) show linked 5S signals. Only in P. venustum is close linkage of 25S and 5S observed, and then only involving a minor 5S band. Because Barbata is the most derived section in the genus (Figure 1), either its species have lost 25S and 5S rDNA loci, since Cochlopetalum, Paphiopedilum, Coryopedilum, and Pardalopetalum usually have more, or the species of the latter sections have increased the number of rDNA loci independently given the low number in sections Parvisepalum and Concoloria.
Figure 8.
FISH distribution pattern of 25S and 5S rDNA on metaphase chromosomes of Paphiopedilum section Barbata. (A) Paphiopedilum wardii, (B) P. hennisianum, (C) P. sukhakulii, (D) P. purpuratum, (E) P. dayanum, (F) P. venustum, (G) P. curtisii, (H) P. acmodontum, (I) P. sangii.
Diversity of 5S ribosomal DNA non-transcribed spacer sequences
We investigated duplication history correlated with the dynamic rearrangements observed in 5S rDNA loci. In order to survey sequence variation in 5S-NTS, random clones, 7 (Paphiopedilum niveum) or 8 (all others) per species, were sequenced (Additional file 1). Only a few clones were identical to each other (2 sequences from P. acmodontum, 2 from P. henryanum, 2 from P. hirsutissimum, 2 from P. stonei, 4 from P. dayanum, 4 from P. malipoense, and one sequence each of P. stonei and P. supardii). Sequences of 5S-NTS ranged from 283 bp (P. micranthum 1) to 455 bp (P. bellatulum 5). Given extensive sequence divergence of 5S-NTS and our desire not to manually adjust alignment [24], an objective alignment was accomplished using MAFFT and default settings. Numbers of polymorphic loci within species, and phylogenetic relationships, were assessed in order to estimate the strength of gene conversion and the extent of paralogy, respectively. Numbers of polymorphic sites within species positively correlated with minimum numbers of visible 5S signals (P < 0.01, R^2 = 0.21; Figure 9), suggesting that interlocus gene conversion is relatively weak. A phylogenetic tree outgroup-rooted using Phragmipedium besseae showed 2 major groups of sequences: section Parvisepalum versus the remainder of the genus. The single tree of maximum likelihood is shown (as a phylogram, Additional file 2), as is the majority-rule consensus tree based on 100 bootstrap replicates (Additional file 3). Some large species-specific clades were observed, as well as some section-specific clades. Overall, however, the phylogenetic tree was poorly representative of phylogenetic relationships due to extensive duplication of 5S loci.
Figure 9.
The relationship between polymorphism of 5S-NTS sequences and numbers of observed 5S rDNA signals. Line indicates trend derived from linear regression analysis based on 5S-NTS within-species polymorphic sites and minimum numbers of visible 5S signals (data from Table 1.). P < 0.01, R^2 = 0.21.
Discussion
Variation in numbers and chromosomal locations of rDNA
Variation in numbers and distribution patterns of rDNA loci among related species is commonly observed in many different plant genera, including Brassicaceae [10], Cyperaceae [11], Asteraceae [25,26], Leguminosae [27], Pinus[28], and Rosaceae [14]. Plants typically show some degree of conservatism of rDNA repeat duplication, such that when multiple loci do appear, species are commonly polyploid relatives of diploids. There is no evidence at all, however, for polyploidy in Paphiopedilum, where the only chromosome number differences are aneuploid, in a series reflective of centric fission or fusion.
In general, FISH patterns of 25S rDNA loci are reported to be more polymorphic than those of the 5S rDNA [12-14,26,28-32]. Conversely, in all sections of Paphiopedilum, except for Parvisepalum and Concoloria, 5S rDNA sites showed much more variability both in number and physical location than did 25S rDNA sites.
The most parsimonious ancestral number of 25S rDNA sites in Paphiopedilum is two, based on outgroup comparison to the genera Mexipedium and Phragmipedium (unpublished results;[3,22]). Duplication of 25S rDNA sites was observed only in three of the seven sections of Paphiopedilum: Parvisepalum (2n = 26), Coryopedilum (2n = 26) and Pardalopetalum (2n = 26) (Table 1). The physical positions of 25S rDNA loci are relatively conservative. In most Paphiopedilum species we analyzed, 25S rDNA signals are located in terminal chromosome positions. Variation was only observed in three species, Paphiopedilum adductum, P. randsii and P. lowii, which showed 1-4 subtelomeric 25S rDNA signals (Figures 6E, F and 7A, respectively). The ancestral number of 5S rDNA sites, again by outgroup comparison, is 2 (unpublished results from Mexipedium and Phragmipedium), and is only observed in sections Parvisepalum and Concoloria. Massive duplication and amplification of 5S rDNA loci, leading to large-scale polymorphism of numbers, sizes and physical positions of signals, was found prevalent in the remaining five sections. The numbers and distribution of rDNA loci vary widely among plants; however, usually less than one-third of chromosomes display either 45S rDNA or 5S rDNA [13]. It is therefore noteworthy that in some lineages of Paphiopedilum, up to 24 of the 26 chromosomes bear at least one rDNA locus, and a single chromosome can bear up to five major 5S rDNA loci.
Apparently, there is no strong correlation between the increase in the number of rDNA sites and the increase in the number of chromosomes or genome size. A similar situation has also been described in many other diploid species, e.g. the diploid lineage of Brassicaceae [10], Cyperaceae [11,12], Iris[13], and Rosaceae [14]. The massive duplication of rDNA loci in Paphiopedilum sections Cochlopetalum, Paphiopedilum, Coryopedilum and Pardalopetalum could partly contribute to the increase of genome size. Perhaps paradoxically, species with the smallest (P. exul; section Paphiopedilum) and largest (P. dianthum; section Pardalopetalum) haploid genome sizes are both members of groups that show considerable 25S and 5S locus duplication in our FISH experiments. These two species differ more than two-fold in genome size, 16.1 to 35.1 Mb, respectively [33]. If we assume that the number of distinct genes among Paphiopedilum species is roughly constant, this would suggest that genome size increase is primarily due to repetitive element amplification, but that since rDNA duplication is associated with both smaller and larger genomes in the genus, size differences may be more logically traceable to other repetitive DNAs, such as mobile elements. However, a possible tendency for elimination of rDNA loci was found in section Barbata, which has the greatest average genome sizes and chromosome numbers [4]. The number of 25S rDNA loci in Barbata remains two through all the species we studied, while the distribution pattern of 5S rDNA is less dispersed than its sister group, Coryopedilum plus Pardalopetalum. Due to the derived phylogenetic position of section Barbata (Figure 1), it is most parsimonious to conclude that unique chromosomal conditions seen in the group would be similarly derived (autapomorphic). As such, centric fission in Barbata appears to be associated with loss of rDNA loci, while in other systems, centric fission has led to rDNA gains [34]. Elimination of rDNA loci during chromosomal evolution has been documented in, e.g., Brassicaceae and Rosaceae [10,14]. The mechanism that accounts for such loss of rDNA loci, however, remains unclear. A presumed evolutionary loss of abundant terminal nucleolar organizing regions (NOR) in Arabidopsis has been hypothesized to be the consequence of an ancient fusion event [35]. In the case of section Barbata, additional traceable chromosome markers are needed to provide further evidence that chromosomal rearrangements are related to rDNA loss.
A combination of different mechanisms causes high mobility of rDNA
Different mechanisms have been postulated to account for the mobility and polymorphism of numbers, sizes and positions of rDNA sites, such as transposon-mediated transpositional events [36-38], and chromosome rearrangements (translocation, inversion, duplication, deletion) caused by homologous or non-homologous unequal crossing-over and gene conversion [9,28,30,36]. These processes could act alone or in combination, and they do not necessarily imply changes in overall chromosome morphology [31,34].
The great degree of 5S repeat dispersion seen in sections Cochlopetalum, Paphiopedilum, Coryopedilum and Pardalopetalum has, to our knowledge, only been observed in the monocots Alstroemeria, Tulipa, and Iris[13,39,40]. The original seeding of rDNA repeats to ectopic locations in the genome could be the result of transposable element activity or perhaps incorporation of array segments into breakpoints as part of non-homologous end joining during DNA repair. Indeed, some of the signals we observed may be pseudogenes transported within the genomes by retroelements, therefore leading to the false interpretation that we are visualizing entire and active rDNA arrays. Both subtelomeric and pericentromeric regions are well known as hot spots of breakpoints and are also enriched for TEs [5,6]. Considering the abundant minor loci we observed in these regions, a contribution of transpositions to the dispersed distribution pattern is tenable, and TEs containing 5S rDNA-derived sequences have in fact been observed in many plants [41] and animals [42]. It is nonetheless possible that due to the similarity of rDNA arrays, chromosomal rearrangement could be induced via heterologous recombination, and in turn, rearrangement could generate repeated sequences through unequal crossovers. After generation of a novel locus, in situ amplification cycles via rearrangement could lead to the origin of FISH-detectable loci. Furthermore, hemizygous 5S rDNA sites have been widely observed in many Paphiopedilum species. A double-strand break occurring in a hemizygous region would increase the probability of causing other rearrangements, owing to the absence of a homologous template for its repair [5]. The lack of dispersed repeats in the basalmost section Parvisepalum may reflect either a lack of seeding events or slow amplification processes that do not yield hybridization-visible arrays. However, in the case of 5S rDNA, there is in fact strong evidence for NTS sequence diversity, which could either be accounted for by the presence or small loci below the FISH detection limit or perhaps by considerable within-array diversity. One future experimental approach to determine whether considerable intra-array diversity indeed exists would be to perform FISH using 5S-NTS-specific probes.
Diversification of 25S rDNA distribution patterns is also observed in Paphiopedilum, but the numbers of loci and degree of dispersion is much lower than for 5S rDNA. Therefore, 5S rDNA might be more frequently seeded by TEs via transpositional events, or, amplification or maintenance of 5S rDNA loci via rearrangement could be more effective and tolerated during the chromosome evolution process. The different evolutionary tendencies between 25S and 5S rDNA might be caused by their function and sequence divergence or localization in distinct nuclear compartments [43].
5S-NTS sequences highlight interlocus and intralocus diversity and weak concerted evolutionary forces
Previous studies of other angiosperm species have suggested that intralocus 5S rDNA diversity occurs. Within-array 5S rDNA diversity appears very likely in Paphiopedilum as well, since many species (e.g., all Parvisepalum and Concoloria) have only one observable 5S locus. For example, 6 species of section Parvisepalum are represented in our phylogenetic analysis by 6-8 distinct sequence variants. These 5S-NTS variants can be concluded to occur within at least partial arrays, pseudogenized or not, since the amplified pieces include sections of 5S rDNA at their 5' and 3' ends. Recent within-species duplication events may be indicated by single-species clades of 5S-NTS sequences, such as P. dayanum, P. lowii, P. sangii, but these could just as well indicate within-array variation, as single-species clades of Parvisepalum (e.g., P. malipoense) and Concoloria (P. bellatulum) most likely do. In many cases, it can be readily seen that duplication of 5S loci has occurred prior to speciation, for example, within Coryopedilum (a large group of sequences representing P. sanderianum, P. stonei, and P. supardii; similarly also within a group of P. adductum and P. randsii sequences). In some cases, ancient duplications must be much older than the major phylogenetic groups of Paphiopedilum, since, for example, P. delenatii shares sequence variants similar to other Parvisepalum species yet has at least one other variant that is more similar to sequences from all other sections. We investigated the possibility of contamination regarding this finding, but discovered similar repeats across 8 distinct P. delenatii accessions (results not shown). Another explanation for multispecies clades, e.g., within well-defined groups such as Parvisepalum could be ancient hybridization.
We observed that increasing within-species 5S NTS sequence diversity correlates with increasing minimum numbers of visible 5S rDNA loci in Paphiopedilum (Figure 9); therefore we infer that interlocus concerted evolution is weak within the genus. Our conclusion concurs with previous findings in many plant genera, such as Gossypium[17], Triticum[18], Chenopodium[19], Nicotiana [20] and Pinus[27]. So far, to our knowledge, noticeable interlocus concerted evolution of 5S rDNA arrays has not been demonstrated in plants.
The best supported hypothesis to explain weak homogenization forces on 5S rDNA arrays is that the chromosomal location of rDNA arrays has a substantial impact on interlocus concerted evolution [17,20,44-47]. Arrays located in subtelomeric regions are thought to undergo stronger interlocus homogenization forces than ones located in proximal regions. Potential evidence was observed in section Barbata, in which all of six species studied possess two 5S loci. These six species can be categorized into two groups according to the locations of 5S loci. One group harboring proximal 5S loci includes P. wardii, P. dayanum, P. venustum and P. acmodontum, while the other group harboring subtelomeric 5S loci includes P. purpuratum and P. sangii (Figure 8; Table 1). Considering that all six species are closely related and possess the same number of loci, it can be logically assumed that the difference in sequence polymorphism between the two groups is caused by the different locations of the 5S loci. The fact that the average number of polymorphic sites in the proximal-loci group (151.5) is 18% more than that in the subtelomeric-loci group (128), indicates that proximally located loci seem less homogenized than the subtelomerically located loci.
Additionally, we found that not only interlocus but also intralocus concerted evolution is also influenced by chromosomal localization. In section Concoloria, two closely related species, P. bellatulum and P. niveum, both have one 5S locus, but with different localizations. The difference in sequence polymorphism between the two species may be caused by the different locations of the 5S loci. P. niveum, which has a pericentromeric locus, showed 1.68 - fold more polymorphic sites than P. bellatulum, which has a subtelomeric locus (Table 1).
It is well-known that meiotic homologous recombination has been largely suppressed in pericentromeric and centromeric regions. Unequal crossovers between sister chromatids and gene conversion documented in the centromeres of many organisms have been postulated as the major homogenization force for tandem repeats located in these areas [48-52]. A plausible explanation for this has been proposed previously: if unequal crossover events between rDNAs of two chromosomes occurred in the proximal region to centromeres, this may result in the exchange of not only a fraction of the rDNA but also the centromeres themselves. Such an event is more likely to have significantly greater negative consequences to the organism than if the event occurred in the subtelomeric region, which then might result in exchange of telomeres [17,47]; loss of centromeres would prohibit cell division, whereas loss of telomeres might not restrict mitosis or meiosis. As such, centromerically-located rDNA arrays are expected to show weaker homogenization forces, since fewer individuals with unequal crossovers in this region are expected to survive. In contrast, the subtelomeric region is characterized by a higher rate of interchromosomal exchange [5], thus stronger concerted evolutionary forces could be expected in this region.
All species of section Parvisepalum, as with P. bellatulum, have subtelomeric 5S loci, some of which are closely linked with 25S loci. If 5S localization correlates significantly with homogenization, as with 25S, which is always telomeric-subtelomerically located, we should expect subtelomeric 5S repeats to show decreased sequence diversity due to stronger concerted evolutionary forces. However, this is not the case, since variation in the number of polymorphic sites is not significantly different by section (with or without Pardalopetalum included in Coryopedilum; single factor ANOVA P = 0.06 and 0.1, respectively). We therefore infer that localization of the 5S rDNA arrays only partially contributes to the weak concerted evolution observed in Paphiopedilum.
There are several other hypothesized mechanisms that could lead to the weak concerted evolutionary force on 5S rDNA arrays. For example, ongoing chromosomal rearrangement such as insertion, deletion, or transposition could occur within arrays too frequently for interlocus concerted evolution to be effective. Another possibility is that concerted evolutionary processes homogenize 5S rDNA arrays at rates lower than the rate of speciation, thus novel mutations cannot be fixed or removed and high levels of intralocus polymorphism are expected within arrays [17]. Additionally, the base composition and secondary structure of rDNA sequences may also affect the rate of concerted evolution [53]. It is unknown whether weak concerted evolutionary forces are shared by other Paphiopedilum tandem repeats, or if this is characteristic of 5S rDNA arrays only. This issue can be elucidated by further studies on other tandem repeats, such as 25S rDNA arrays.
Conclusions
Paphiopedilum species display many chromosomal rearrangements - for example, duplications, translocations, and inversions - but only weak concerted evolutionary forces among highly duplicated 5S arrays, which suggests that double-strand break repair processes are dynamic and ongoing. These results make the genus a model system for the study of complex chromosomal evolution in plants.
Methods
Plant materials
Thirty-seven species of the Paphiopedilum genus covering all seven sections were analyzed in this study. Information on the species and sections is provided in Table 1. Actively growing roots were used for chromosome preparation, while leaves were used for genomic DNA extraction.
Chromosome preparation
Root tips were pre-treated with 0.004 M 8-hydroxyquinoline for 4-6 h at 10°C, and fixed in freshly prepared fixative (3:1 ethanol: acetic acid) for 48 h at 10°C. The fixed root tips were then rinsed thoroughly with tap water and macerated in an enzyme mixture containing 2% cellulase (Onozuka R-10, Rpi) and 1% pectolyase (Aspergillus japonicus Y-23, MP) at 37°C for 30 min. After re-fixation in fixative for 15 min, the meristematic cells were squashed in a drop of 45% acetic acid under a coverslip (22 × 22 mm) on a microscope slide. Slides were then dipped into liquid nitrogen and air dried after the coverslips were carefully removed by a blade.
Probe labelling and Fluorescence in situ hybridization (FISH)
25S rDNA, a 2.3-kb ClaI subclones of the 25S rDNA coding region of Arabidopsis thaliana [54] and 5S rDNA (pTa794) [55]were used as probes. 25S rDNA was labelled with biotin-16-dUTP (Roche) and 5S rDNA was labelled with digoxigenin-11-dUTP (Roche), all by nick translation method using the kit from Roche. The hybridization buffer consisted of 50% deionized formamide, 2 × SSC, 50 mM sodium phosphate (pH 7.0), 10% dextran sulfate and sheared salmon sperm DNA (Invitrogen) in 100 × excess of labeled probes. The 25S and 5S rDNA probes were mixed to a final concentration of about 2 ng/μl and then denatured at 94°C for 10 min before being used. Slides with metaphase spreads were treated with 70% deionized formamide in 2 × SSC at 70°C for 2 min. Denatured probes in hybridization buffer were then applied to the slides, which were incubated at 37°C for 10 h in a humid chamber. Post-hybridization washes and immunodetection were carried out in an automated in situ hybridization instrument, the InsituPro VSi (Intavis Bionanalytical Instruments). The slides were washed in 2 × SSC at room temperature for 5 min and twice in 2 ×SSC at 50°C for 10 min. Fluorescence signal was detected using anti-Digoxigenin-Rhodamine conjugate (Roche) and streptavidin-fluorescein conjugate (Invitrogen). The preparations were mounted and counterstained in Vectashield containing 1.5 μg/ml DAPI (4', 6-diamidino-2-phenylindole) (Vector Laboratories). Images were taken by a Zeiss AxioCam MRm black-and-white CCD camera on a Zeiss Imager. Z1 fluorescence microscope and then processed uniformly using Zeiss AxioVision software. FISH signals were false-colored, and DAPI fluorescence was left in gray-tone.
PCR amplification, cloning and sequencing
Total genomic DNA was extracted from fresh leaves using Qiagen DNeasy Plant Mini kit. The 5S-NTS region was amplified by PCR using the universal degenerate primers: 5'-TGGGAAGTCCTYGTGTTGCA-3' and 5'-KTMGYGCTGGTATGATCGCA-3'[56]. Touchdown amplification was performed as follows: an initial step at 94°C for 5 min, followed by 10 cycles of 94°C for 1 min, annealing for 1 min (start at 60°C, and decreased by 1°C per cycle), and 72°C for 1 min, then 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, the final step at 72°C was extended to 10 min. After gel purification using QIAquick Gel Extraction Kit (Qiagen), PCR products were ligated into pDrive Cloning vector and transformed into QIAGEN EZ competent cells (Qiagen PCR Cloning kit). Recombinant clones were screened by colony direct PCR method and were sequenced 7-8 clones per each species using T7 (5'-TAATACGACTCACTATAGGG-3') primer.
Data analysis
Sequences were aligned using the MAFFT (Multiple Alignment using Fast Fourier Transform) web server at the European Bioinformatics Institute [57]. Default parameters were used: gap opening penalty = 1.53, gap extension penalty = 0.123, tree rebuilding number = 1, maxiterate = 0, and perform FFTS = localpair. The sequence alignment is available as a supplementary FASTA file (Additional file 4).
Within-species 5S-NTS polymorphism was estimated, based on the aforementioned multiple alignment, using DnaSP version 5.10.01 [58]. The relationship between numbers of polymorphic sites and minimum numbers of visible 5S rDNA signals was investigated using linear regression analysis (in Microsoft Excel).
Phylogenetic reconstruction was performed using maximum likelihood optimization available through the RaxML BlackBox web server [59] running RaxML version 7.2.8 [60]. Default settings were used. The 8 Phragmipedium besseae sequences were indicated as the outgroup. RaxML was called using the following commands: raxml -# 100 -n pasted -o bess1, bess2, bess3, bess4, bess5, bess6, bess7, bess8 -f a -m GTRGAMMA -x 564547904 -p 564547904 -s 0VaDTW. All search information, as was output on the web site, is included in Additional file 5.
Authors' contributions
TL and VAA conceived of the study, TL performed all experiments, TL and VAA analyzed data, and both TL and VAA prepared the manuscript. All auhors read and approved the final manuscript.
Supplementary Material
GenBank data deposition information of 5S-NTS sequences
5S-NTS sequences: the single tree of maximum likelihood
5S-NTS sequences: the majority-rule consensus tree based on 100 bootstrap replications
The 5S-NTS sequence alignment using MAFFT, provided in FASTA format
Report from RAxML phylogenetic analysis of 5S-NTS sequences
Contributor Information
Tianying Lan, Email: tianying@buffalo.edu.
Victor A Albert, Email: vaalbert@buffalo.edu.
Acknowledgements
We thank R. Hasterok and B. Liu for providing rDNA clones. This study was supported by funds from the University at Buffalo.
References
- Cribb P. Natural History Publications. 2. Borneo; 1998. The genus Paphiopedilum. [Google Scholar]
- Karasawa K. Karyomorphological studies in Paphiopedilum. Orchidaceae. Bulletin of the Hiroshima Botanic Garden. 1979;2:1–149. [Google Scholar]
- Cox A, Pridgeon AM, Albert VA, Chase MW. Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae) Nuclear rDNA ITS sequences Plant Syst Evol. 1997;208(3-4):197–223. [Google Scholar]
- Cox A, Abedelnour G, Bennett MD, Leitch IJ. Genome size and karyotype evolution in the slipper orchids (Cypripedioideae: Orchidaceae) American Journal of Botany. 1998;85(5):681–687. doi: 10.2307/2446538. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437(7055):94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplcation: an update. Trends in Ecology and Evolution. 2003;18(6):292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- Hurles M. Gene duplication: The genomic trade in spare parts. PLoS Biol. 2004;2(7):0900–0904. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120(3-4):351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen WK, Maluszynska J. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany. 2006;97(2):205–216. doi: 10.1093/aob/mcj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva C, Quintas CC, Vanzela AL. Distribution of 45S and 5S rDNA sites in 23 species of Eleocharis (Cyperaceae) Genetica. 2010;138(9-10):951–957. doi: 10.1007/s10709-010-9477-5. [DOI] [PubMed] [Google Scholar]
- Sousa A, Barros e Siva AE, Cuadrado A, Loarce Y, Alves MV, Guerra M. Distribution of 5S and 45S rDNA sites in plants with holokinetic chromosomes and the "chromosome field" hypothesis. Micron. 2011;42(6):625–631. doi: 10.1016/j.micron.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Martinez J, Vargas P, Luceno M, Cuadrado A. Evolution of Iris subgenus Xiphium based on chromosome numbers, FISH of nrDNA (5S, 45S) and trnL-trnF sequence analysis. Plant Syst Evol. 2010;289:223–235. doi: 10.1007/s00606-010-0345-7. [DOI] [Google Scholar]
- Mishima M, Ohmido N, Fukui K, Yahara T. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae) Chromosoma. 2002;110(8):550–558. doi: 10.1007/s00412-001-0175-z. [DOI] [PubMed] [Google Scholar]
- Lee Y, Chung MC. Identification of genome relationships among Paphiopedilum species by genomic and fluorescent in situ hybridization. Acta Horiticluturae. 2008;766:331–334. [Google Scholar]
- Lee Y, Chang FC, Chung MC. Chromosome pairing affinities in interspecific hybrids reflect phylogenetic distances among lady's slipper orchids (Paphiopedilum) Annals of Botany. 2011;108(1):113–121. doi: 10.1093/aob/mcr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R, Zhao X, Paterson AH, Wendel JF. Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol. 1996;42(6):685–705. doi: 10.1007/BF02338802. [DOI] [PubMed] [Google Scholar]
- Kellogg E, Appels R. Intraspecific and interspecific variation in 5S RNA genes are decoupled in diploid wheat relatives. Genetics. 1995;140(1):325–343. doi: 10.1093/genetics/140.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan P, Kolano BA, Maluszynska J, Coles ND, Bonifacio A, Rojas J, Coleman CE, Stevens MR, Fairbanks DJ, Parkinson SE, Jellen EN. Molecular and cytological characterization of ribosomal RNA genes in Chenopodium quinoa and Chenopodium berlandieri. Genome. 2006;49(7):825–839. doi: 10.1139/G06-033. [DOI] [PubMed] [Google Scholar]
- Fulnecek J, Lim KY, Leitch AR, Kovarík A, Matyásek R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88(1):19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhang DM, Wang XQ, Ma XF, Wang XR. Intragenomic and interspecific 5S rDNA sequence variation in five Asian pines. American Journal of Botany. 2003;90:17–24. doi: 10.3732/ajb.90.1.17. [DOI] [PubMed] [Google Scholar]
- Albert V. Cladistic relationships of the slipper orchids (Cypripedioideae: Orchidaceae) from congruent morphological and molecular data. Lindleyana. 1994;9:115–132. [Google Scholar]
- Anisimova M, Cannarozzi GM, Liberles DA. Finding the balance between the mathematical and biological optima in multiple sequence alignment. Trends in Evolutionary Biology. 2010;2(1):e7. [Google Scholar]
- Garcia S, Panero JL, Siroky J, Kovarik A. Repeated reunions and splits freature the highly dynamic evolution of 5S and 35S ribosomal RNA genes (rDNA) in the Asteraceae family. BMC Plant Biology. 2010;10:176. doi: 10.1186/1471-2229-10-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska H, Tate JA, Matyasek R, Leitch AR, Soltis DE, Soltis PS, Kovarik A. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evolutionary Biology. 2010;10:291. doi: 10.1186/1471-2148-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscone E, Klein F, Lambrow M, Fuchs J, Schweizer D. Quantitative karyotyping and dual-color FISH mapping of 5S and 18S-25S rDNA probes in the cultivated Phaseolus species (Leguminosae) Genome. 1999;42(6):1224–1233. doi: 10.1139/g99-070. [DOI] [PubMed] [Google Scholar]
- Cai Q, Zhang D, Liu Z-L, Wang X-R. Chromosomal localization of 5S and 18S rDNA in five species of subgenus Strobus and their implications for genome evolution of Pinus. Annals of Botany. 2006;97(5):715–722. doi: 10.1093/aob/mcl030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson R, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L) and its putative diploid ancestors. Chromosoma. 1996;105(1):55–61. doi: 10.1007/BF02510039. [DOI] [PubMed] [Google Scholar]
- Linares C, Gonzalez J, Ferrer E, Fominaya A. The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5. 8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome. 1996;39(3):535–542. doi: 10.1139/g96-068. [DOI] [PubMed] [Google Scholar]
- Thomas H, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E, King IP. Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Research. 1996;4(7):486–490. doi: 10.1007/BF02261775. [DOI] [PubMed] [Google Scholar]
- Datson P, Murray BG. Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosome Research. 2006;14(8):845–857. doi: 10.1007/s10577-006-1092-z. [DOI] [PubMed] [Google Scholar]
- Ksiazczyk T, Taciak M, Zwierzykowski Z. Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. J Appl Genet. 2010;51(4):449–460. doi: 10.1007/BF03208874. [DOI] [PubMed] [Google Scholar]
- Plant DNA C-values database (release 5.0, Dec. 2010) http://www.kew.org/cvalues/
- Hall K, Parker JS. Stable chromosome fission associated with rDNA mobility. Chromosome Research. 1995;3(7):417–422. doi: 10.1007/BF00713891. [DOI] [PubMed] [Google Scholar]
- Lysak M, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA. 2006;103(13):5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altinkut A, Raskina O, Nevo E, Belyayev A. En/Spm-like transposons in Poaceae species: transposase sequence variability and chromosomal distribution. Cellular & Molecular Biology Letters. 2006;11(2):214–230. doi: 10.2478/s11658-006-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Wobus U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma. 1985;92:143–148. doi: 10.1007/BF00328466. [DOI] [Google Scholar]
- Raskina O, Belyayev A, Nevo E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Research. 2004;12(2):153. doi: 10.1023/b:chro.0000013168.61359.43. [DOI] [PubMed] [Google Scholar]
- Baeza C, Schrader O, Budahn H. Characterization of geographically isolated accessions in five Alstroemeria L. species (Chile) using FISH of tandemly repeated DNA sequences and RAPD analysis. Plant Syst Evol. 2007;269(1-2):1–14. doi: 10.1007/s00606-007-0591-5. [DOI] [Google Scholar]
- Mizuochi H, Marasek A, Okazaki K. Molecular cloning of Tulipa fosteriana rDNA and subsequent FISH analysis yields cytogenetic organization of 5S rDNA and 45S rDNA in T. gesneriana and T. fosteriana. Euphytica. 2007;155(1-2):235–248. doi: 10.1007/s10681-006-9325-y. [DOI] [Google Scholar]
- Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA. 2008;105(15):5833–5838. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20(5):694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- Mantovani M, Abel LD, Moreira-Filho O. Conserved 5S and variable 45S rDNA chromosomal localisation revealed by FISH in Astyanax scabripinnis (Pisces, Characidae) Genetica. 2005;123(3):211–216. doi: 10.1007/s10709-004-2281-3. [DOI] [PubMed] [Google Scholar]
- Wendel J, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proc Natl Acad Sci USA. 1995;92(1):280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Sang T. Physical mapping of ribosomal RNA genes in peonies (Paeonia, Paeoniaceae) by fluorescent in situ hybridization: implications for phylogeny and concerted evolution. Am J Bot. 1999;86(5):735–740. doi: 10.2307/2656583. [DOI] [PubMed] [Google Scholar]
- Fukushima K, Imamura K, Nagano K, Hoshi Y. Contrasting patterns of the 5S and 45S rDNA evolutions in the Byblis liniflora complex (Byblidaceae) J Plant Res. 2011;124(2):231–244. doi: 10.1007/s10265-010-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush T, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175(2):477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P, Henikoff S. Centromeres convert but don't cross. PLoS Biol. 2010;8(3):e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Wing RA, Bennetzen JL, Jackson SA. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 2007;23(3):134–139. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8(3):e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelhauer D, Schwarz T. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous alpha-satellite DNA array. Genome Res. 2002;12(12):1815–1826. doi: 10.1101/gr.451502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizès G. Human centromeric alphoid domains are periodically homogenized so that they vary substantially between homologues: mechanism and implications for centromere functioning. Nucleic Acids Res. 2006;34(6):1912–1924. doi: 10.1093/nar/gkl137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar J, Glémin S, Galtier N. GC-biased gene conversion impacts ribosomal DNA evolution in vertebrates, angiosperms and other eukaryotes. Mol Biol Evol. 2011. [DOI] [PubMed]
- Unfried I, Gruendler P. Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res. 1990;18(13):4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980;8(21):4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Bennett MD, Dyer TA. Use of the polymerase chain reaction to detect spacer size heterogeneity in plant 5S-rRNA gene clusters and to locate such clusters in wheat (Triticum aestivum L) Theor Appl Genet. 1992;83(6-7):684–690. doi: 10.1007/BF00226685. [DOI] [PubMed] [Google Scholar]
- MAFFT web server at the European Bioinformatics Institute. http://www.ebi.ac.uk/Tools/msa/mafft/
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- RaxML BlackBox web server. http://phylobench.vital-it.ch/raxml-bb/index.php
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank data deposition information of 5S-NTS sequences
5S-NTS sequences: the single tree of maximum likelihood
5S-NTS sequences: the majority-rule consensus tree based on 100 bootstrap replications
The 5S-NTS sequence alignment using MAFFT, provided in FASTA format
Report from RAxML phylogenetic analysis of 5S-NTS sequences