Abstract
Background
Surgical removal of the lens from larval Xenopus laevis results in a rapid transdifferention of central corneal cells to form a new lens. The trigger for this process is understood to be an induction event arising from the unprecedented exposure of the cornea to the vitreous humour that occurs following lens removal. The molecular identity of this trigger is unknown.
Results
Here, we have used a functional transgenic approach to show that BMP signalling is required for lens regeneration and a microarray approach to identify genes that are upregulated specifically during this process. Analysis of the array data strongly implicates Wnt signalling and the Pitx family of transcription factors in the process of cornea to lens transdifferentiation. Our analysis also captured several genes associated with congenital cataract in humans. Pluripotency genes, in contrast, were not upregulated, supporting the idea that corneal cells transdifferentiate without returning to a stem cell state. Several genes from the array were expressed in the forming lens during embryogenesis. One of these, Nipsnap1, is a known direct target of BMP signalling.
Conclusions
Our results strongly implicate the developmental Wnt and BMP signalling pathways in the process of cornea to lens transdifferentiation (CLT) in Xenopus, and suggest direct transdifferentiation between these two anterior eye tissues.
Background
Urodele amphibians, for example the axolotl, are well known for their incredible ability to regenerate appendages, such as the limb. However, axolotls are unable to regenerate the lens of the eye following its removal (lentectomy). In contrast, the anuran amphibian Xenopus laevis, in which limb regeneration is subject to an ontogenic decline leading up to metamorphosis, is able to regenerate a new lens from the overlying central corneal cells (for review see [1,2]). This process was first described by Freeman in 1963, and involves a transdifferentiation of one cell type (corneal epithelium) to another (lens) [3]. It differs from the better-known Wolffian regeneration in adult newts, where a new lens is formed from cells of the pigmented dorsal iris epithelium and is known as cornea to lens transdifferentiation, or CLT [2].
The trigger for CLT in vivo is exposure of the outer corneal cells to an unidentified factor present in the vitreous of the eye, most likely originating from the neural retina [4,5]. In vitro, epithelial cells from any location within the lentogenic area, a region extending twice the diameter of the eye [3] can respond to the vitreous factor and initiate CLT, whereas cells outside this region are refractory to the trigger [6-8]. The limitation of lens forming ability to the lentogenic area correlates with Pax6 expression, and ectopic Pax6 in flank epidermis can confer competence to undergo CLT [9]. As with other cases of regeneration in Xenopus, there is an ontogenic decline in the ability to initiate CLT in vivo [3], however, this is thought to arise due to a mechanical barrier formed by the healing of the inner cornea rather than a loss of competence [10]. Interestingly, the close relative Xenopus tropicalis, which exhibits more rapid healing of the inner cornea following lentectomy, fails to initiate CLT in vivo although reciprocal transplants show that the central corneal cells of X. tropicalis can respond to the vitreous factor [11].
Fifty years on from the discovery of CLT, we still know little of the molecular mechanisms that drive the process. While it is generally believed that transdifferentiation occurs directly and not via proliferation of stem cells [12], a direct demonstration of this is lacking. Jon Henry and colleagues have shown that several transcription factors known to be fundamental to lens development are re-expressed during the process of CLT (Pax6, Prox1, Otx2 and Sox3), suggesting that similar regulation of gene expression drives differentiation during both development and regeneration of the lens [13,14]. Previous EST analysis of corneal cells undergoing CLT has identified several hundred transcripts from a library constructed from corneal tissue at 1-4 days after lens removal [14,15].
Despite the identification of multiple candidate pathways from these expression studies, functional analysis of potential transdifferentiation factors has so far been lacking. A single in vitro study demonstrated the ability of acidic fibroblast growth factor (aFGF) to induce lens fibre formation in cultured outer corneas, although morphological organisation of the fibres does not occur [16]. In the current study, we have used a transgenic line of Xenopus laevis to reveal a need for functional BMP signalling during the process of CLT along with a microarray strategy to identify genes and pathways that are likely to be specifically involved in the process of transdifferentiation. The microarray strategy differs from previous library based approaches in that we can specifically compare expression in wounded, non-regenerating corneas to that in corneal tissue undergoing CLT, with the aim of identifying genes associated with the regenerative process. Analysis of this microarray data indicates an important role for wingless/int1 (Wnt) pathway signalling in CLT and suggests that, as with tail and limb regeneration in Xenopus [17-19], several morphogens may be acting to trigger the regenerative process of CLT. We have identified several new candidates for CLT, some of which are also involved in lens formation during development, and many of which are associated with lens pathology, particularly cataract development.
Results
Lens regeneration is dependent on BMP signalling
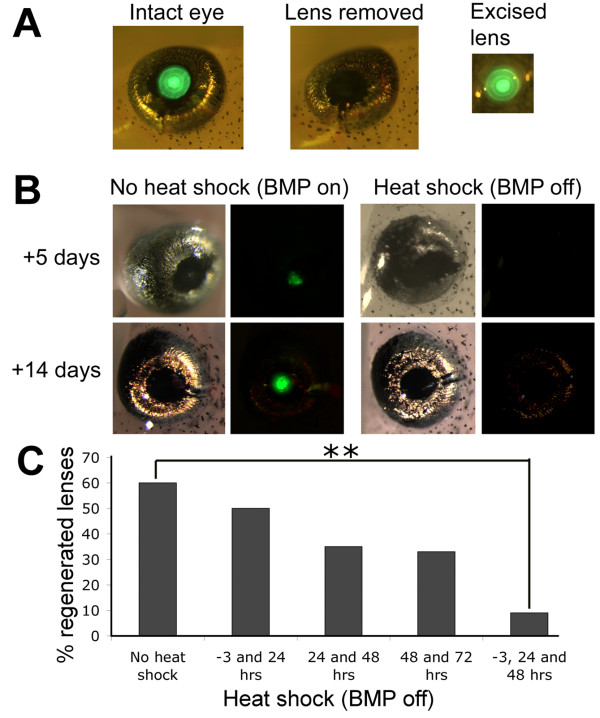
We have previously shown that overexpression of Noggin, an inhibitor of BMP signalling, can prevent regeneration of tails and limbs in Xenopus tadpoles [17,20]. The stable transgenic line, N1 contains a single insertion of the double transgene Hsp70:Noggin;γ-Crystallin:GFP. The eyes of N1 tadpoles are marked by green fluorescent protein (GFP), which accumulates in the inner lens cells regardless of the temperature at which the animals are kept (Figure 1A). When the GFP expressing lens is surgically removed and the cornea replaced, no green fluorescence can be detected. If lentectomised N1 tadpoles are raised at 24°C, GFP is first detected after 3-5 days, indicating that CLT is underway and new lens cells are expressing γ-Crystallin (Figure 1B). In our hands, CLT occurs in 60-70% of cases, and after two weeks, the new lens is morphologically indistinguishable from the original. However, if the N1 tadpoles are subjected to heat shock, by placing in water at 34°C for 30 minutes each day, CLT usually fails, and no new expression of GFP is detected, suggesting that BMP signalling must be active for transdifferentiation to occur (Figure 1B). The process of heat shocking itself did not adversely affect the regenerative potential of wild type sibling eyes or those carrying a γ-cry-RFP transgene (C.B. unpublished observations).
Figure 1.
BMP signalling is required for lens regeneration. A) Lentectomy of N1 transgenic tadpoles at stage 50. These tadpoles carry the Hsp70:Noggin; γ-Crystallin:GFP transgene and express GFP in the central lens cells, but not the outermost lens epithelial cells. Removal of the lens can be visualised by the lack of GFP in the lentectomised eye. B) Heat shock initiates expression of the BMP inhibitor noggin, attenuating BMP signalling. In around 60-70% of non heat shocked, lentectomised eyes, regeneration of the lens from the overlying cornea occurs and GFP can be seen after 3-5 days and a new lens is formed after 14 days. When the tadpoles are subjected to heat shock, no GFP is expressed and no lens regenerated. Bright field views are to the left with corresponding fluorescent views adjacent. C) Graph showing the effect of heat shock activation of Noggin on lens regeneration as determined by the presence of any detectable GFP in the eye 10 days after lentectomy. ** indicates p < 0.001. (Chi squared analysis).
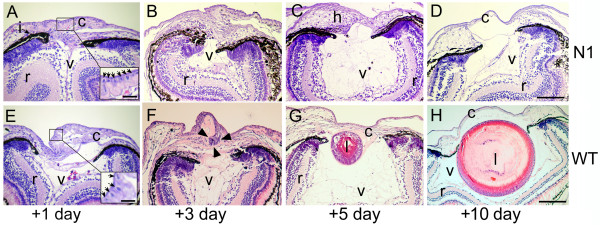
We have previously shown that each daily heat shock generates a burst of Noggin expression that is detectable for a day or two, but declines[20]. Heat shocks were applied at different times during post lentectomy recovery (Figure 1C). Heat shock at -3, +24 and +48 hours, relative to surgical removal of the lens, resulted in only 9% of eyes regenerating detectable GFP positive lens cells (n = 56), compared to non heat shocked transgenics of which 60% regenerated lens cells (n = 60). This difference was found to be highly significant by chi squared analysis (p < 0.001). Tadpoles heat shocked twice, at either 24 and 48 hours or 48 and 72 hours, regenerated lens in 35% (n = 23) and 33% (n = 15) of cases respectively, and chi squared analysis compared to unheatshocked controls showed that this was close to significant (p = 0.06 and 0.09 respectively). Heat shock of tadpoles at -3 and 24 hours resulted in 50% regeneration (n = 10), suggesting that BMP signalling needs to be sustained to inhibit lens regeneration fully. To investigate this further, we sectioned lentectomised eyes one, three, five and ten days after lentectomy and compared the histology of heat shocked N1 tadpoles to that or similarly treated WT sibling animals. Heat shocks were applied before lentectomy and for the first two days afterwards (unless fixed at day 1) (Figure 2). Freeman described five distinct phases of CLT in Xenopus laevis based on histological analysis [3]. At stage 1 (1-2 days after lentectomy), cells of the inner layer of the outer cornea have undergone a change from squamous to cuboidal epithelium. In both N1 and WT eyes one day after lentectomy, corresponding to stage 1, the vitreous appeared collapsed and the cornea very thickened (compare Figure 2A and 2E to 2H), with no obvious differences between the transgenic and WT eyes. Cuboidal cells were consistently visible in both cases (Figure 2A and 2E insets), suggesting that this initial stage of CLT does not depend on BMP signalling. Indeed, Freeman observed that this stage occurs on wounding of the cornea even if the lens is not subsequently removed [3]. We observed a slightly thicker eosin stained extracellular matrix in some, but not all, N1 samples. In later stages, though, a clear difference in the histology of the WT and N1 eyes was observed. In N1 tadpoles, in which BMP signalling is attenuated, the cells of the thickened cornea appear to become hypertrophic (Figure 2B, C). By ten days the cornea has thinned to its original state, with the cells once again appearing squamous, and the vitreous has partially reinflated despite the lack of a lens (Figure 2D). Of 8 ten day samples sectioned, one showed regeneration of a small lentoid.
Figure 2.
Histology of WT and BMP inhibited (N1) tadpole eyes following lentectomy. (A-D) Representative histological sections through the eyes of transgenic N1 tadpoles 1, 3, 5 and 7 days after lentectomy. Inset in A shows arrows pointing to columnar corneal cells (E-H) Representative sections of wild type eyes at 1, 3, 5 and 7 days after lentectomy. Inset in E indicates columnar corneal cells (arrows). Arrowheads in F indicate the forming lentoid moving into the vitreous. Both transgenic and wild type eyes were subjected to heat shock at -3, 24 and 48 hours relative to lentectomy. Abbreviations: c, cornea; h, hypertrophic cells; i, iris (pigmented epithelium); l, lens; r, retina; v, vitreous. Scale bars in D and H corresponds to 50 μm and applies to all panels. Inset scale bar in A and E corresponds to 10 μm.
In WT eyes fixed three days post lentectomy, in 5/10 samples, the transdifferenting cornea had reached early Freeman stage 3, and a cluster of aggregated cells is beginning to invade the vitreous (Figure 2F). A further 2/10 had reached mid to late Freeman stage 3 and no CLT was observed in the remaining 3 samples. At this stage the connection to the cornea is very clear. By five days, the aggregate has detached from the cornea and become a lens vesicle and primary lens fibres have begun to form (Figure 2G, Freeman stage 4). By ten days, primary and secondary lens fibres have formed and the lens appears essentially complete (Figure 2H, Freeman stage 5). The cornea has returned to its original state and is once more composed of squamous epithelial cells.
Microarray analysis of CLT I: Pattern matching based on crystallin expression (CRY list)
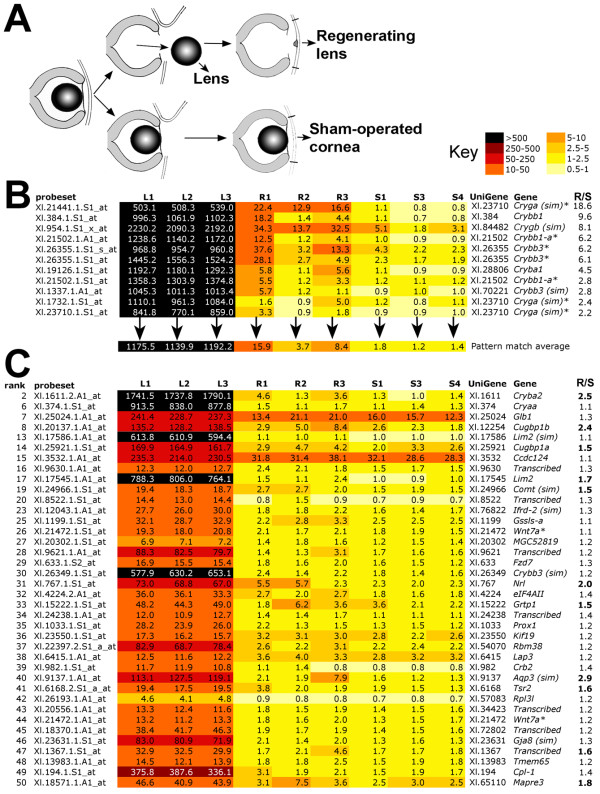
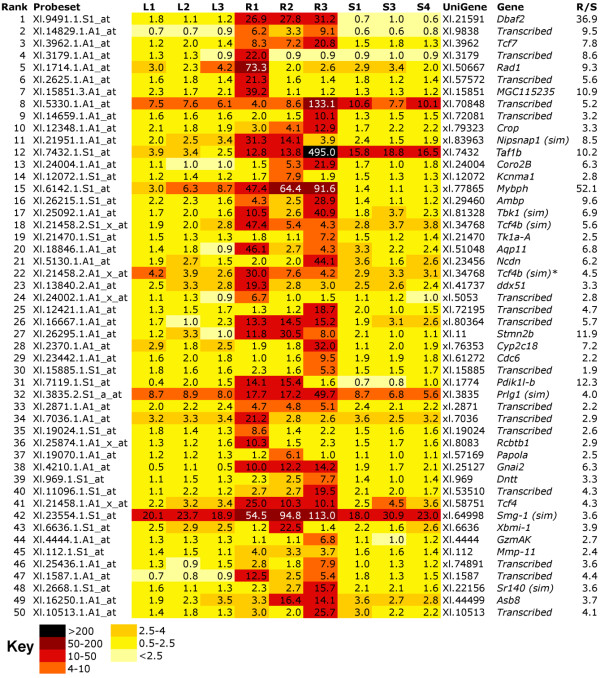
Microarray samples were prepared as shown in Figure 3A. Nine Affymetrix Xenopus laevis GeneChips were probed with triplicate biological replicates prepared either from stage 50 lens (L), corneas undergoing CLT (expected to be predominantly at early Freeman stage 3) (R) and sham operated corneas undergoing healing (S). The normalised data were first sorted by performing pattern matching using TIGR-MEV with the instruction that R (mean) is greater than any of the three S replicates. Thus, the average intensity for a particular probeset from three biological replicate samples of CLT tissue should be statistically higher than for any of the sham operated controls, in which wound healing but not CLT is occurring. At the top of this list were several probesets representing members of the crystallin (Cry) family of structural lens proteins. However, between biological replicates, there was a pattern among the crystallins in the CLT replicates whereby R1 and R3 were consistently seen to have higher expression than R2. We took this to be an indication that, in the pool of corneas that made up the R2 sample, CLT was either lagging behind or occurring in fewer of the corneas. We made an assumption that other genes being specifically switched on during CLT might follow the same pattern, and so we selected 11 probesets representing seven different UniGene sets of crystallins, and averaged the intensities for each sample (Figure 3B). These average intensities were then used with TIGR-MEV pattern matching software to search for similar patterns across the 9 samples. 1642 statistically significant (p < 0.05) matches were obtained (CRY list). The top 50 ranked matches were manually annotated and are shown in the heat map in Figure 3C. The full CRY list can be found in additional file 1 (note this is not annotated fully).
Figure 3.
Identification of genes that mimic expression of crystallins during CLT. A) Strategy for identifying CLT specific genes by microarray. Lenses were removed from stage 50 tadpoles and used for RNA extraction of samples L1-3 (differentiated lens). The tadpoles that had undergone lentectomy were allowed to recover for three days before harvesting the cornea for samples R1-3 (regenerating). Finally, a second set of tadpoles were subjected to mock surgery, the cornea was cut, lifted and replaced as for lentectomy but the inner cornea and lens were left in situ. After 3 days corneas were recovered from these animals, generating samples S1, 3 and 4 (sham operated). B) Heatmap of Affymetrix array data showing that the pattern of crystallin expression varies among replicate R samples, with R2 consistently lower. An average expression was used to search for similar patterns in the data. C) Heatmap of top 50 significant matches to the crystallin pattern, excluding the crystallin genes used to generate the pattern average. R/S is the ratio of transcript expression in R1-3 vs. S1, 3 and 4 levels. Bold type in C indicates R > 1.5 × S.
Crystallins used in the pattern match are shown in Figure 3B. Three further crystallin genes, Cryba2, Cryaa and a gene most similar to Crybb3, were detected by the match. Prox1, a known eye development gene, was also captured. The Wnt family member Wnt7a appeared twice (due to the presence of multiple probe sets captured by the filter) in the top 50, along with the Wnt receptor Frz7, which occurred a total of four times in the CRY list. Seven of the top 50 ranked genes could not be assigned to a protein family and are annotated as transcribed. Interestingly, two homologues of a CUG triplet repeat RNA binding protein, Cugbp1, were also ranked highly in the match. These proteins, also known as EDEN-BP, are members of the Bruno family and are involved in degradation of mRNA through binding to instability sites [21].
To obtain an unbiased overall view of the selected crystallin pattern matched data, 1037 genes with a high probability match (p < 0.01) were used to examine gene ontology. These formed the slightly shorter CRY* list. TC numbers, with duplicates removed, were used to match probesets to specific GO biological function categories. Table 1 shows the GO categories that were statistically over represented in the CRY* dataset. Genes associated with nuclear migration, microtubule polymerisation, development, Wnt/planar cell polarity (PCP) signalling, eye development and visual perception were among the most over represented ontologies. The two genes in "eye development (sensu vertebrata)" correspond to the lens intrinsic membrane protein Lim2 and a possible homologue, Lim2(sim). Lim2 encodes a lens specific protein probably involved in cell junctions, and mutations in humans are associated with congenital cataracts [22,23]. The inclusion of two categories involved with Wnt/PCP signalling, "R3/R4 cell fate commitment" and "establishment of wing hair orientation" ontologies seems to be entirely due to one gene, Frz7, which has two TC numbers, and a total of 4 hits in the list. However, the "frizzled signalling pathway" ontology was also significantly overrepresented (Table 1), creating a strong case for the involvement of Wnt signalling in directing the lens fate. Genes in this category included, as well as Frz7, a second receptor gene, Frz8 (2 hits), the ligand Wnt5b transcription factors Pitx3 and Tcf3, and the signalling components Axin1 and the alpha subunit of CK2.
Table 1.
Top 10 statistically over represented GO categories in the CRY* list
| GMRG_Term | TC numbers | p | Description | |
|---|---|---|---|---|
| total | subset | |||
| GO:0000065/0000743 | 16 | 6 | 0.001 | nuclear migration along microtubule/during conjugation with cellular fusion |
| GO:0007275 | 243 | 32 | 0.003 | multicellular organismal development |
| GO:0046785 | 10 | 4 | 0.006 | microtubule polymerization |
| GO:0001737 | 2 | 2 | 0.006 | establishment of wing hair orientation |
| GO:0006221 | 2 | 2 | 0.006 | pyrimidine nucleotide biosynthetic process |
| GO:0007464 | 2 | 2 | 0.006 | R3/R4 cell fate commitment |
| GO:0043010 | 2 | 2 | 0.006 | eye development (sensu Vertebrata) |
| GO:0006364 | 29 | 7 | 0.006 | rRNA processing |
| GO:0007601 | 87 | 14 | 0.008 | visual perception |
| GO:0007222 | 12 | 4 | 0.012 | frizzled signaling pathway |
TC numbers are tentative consensus numbers and each one represents a group of ESTs for one gene, as well as providing a way to link to gene ontology. Total TC therefore shows the number of genes on the Xenopus GeneChip which are linked to a particular ontology, and subset TC shows how many of these were found in the CRY* list
Several genes were validated using quantitative real time PCR (q-rtPCR), as shown in Table 2. After normalisation to the housekeeping gene ODC, 11/11 genes were confirmed as being expressed at higher levels in lens than in either corneas undergoing CLT or sham operated corneas. Of these, six were confirmed expressed at higher levels in corneas undergoing CLT vs. sham operated corneas: five crystallins and Fzd7. Two of the remaining genes (Wnt7a, Kif19) were found to be downregulated by q-rtPCR and unchanged in the array, Glb1 was unchanged in either analysis, and Cryaa was found to be upregulated by q-rtPCR but not on the array. The RNA binding protein Cugbp1b was not significantly up regulated by q-rtPCR although expression was confirmed as being higher in CLT samples.
Table 2.
Q-rtPCR validation of 11 genes from the CRY list
| Rank in array | Gene name | Lens (L/S) | CLT (R/S) |
|
|---|---|---|---|---|
| qPCR | qPCR | array | ||
| 1 | Cryba1 | 37009.6 | 38.5 | 4.5 |
| 3 | Crygb (sim) | 75864.3 | 7.3 | 8.1 |
| 4 | Crybb3 | 45918.7 | 7.8 | 6.2 |
| 6 | Cryaa | 95666.6 | 17.9 | 1.1 |
| 7 | Glb1 | 7.6 | 0.8 | 1.3 |
| 8 | Cugbp1b | 41.8 | 1.3 | 2.4 |
| 15 | Ccdc124 | 3.62 | 1.5 | 1.1 |
| 18 | Crybb1 | 22333.3 | 5.0 | 9.6 |
| 26 | Wnt7a | 39193.7 | 0.5 | 1.1 |
| 29 | Fzd7* | 5.9 | 3.9 | 1.3/2.7 |
| 36 | Kif19 | 2.1 | 0.5 | 1.2 |
Bold numbers indicate upregulation of more than 1.5 fold, numbers in italics indicate down regulation and normal font indicates no significant change. * appears twice in CRY list. L/S shows the relative (fold higher) expression in lens when compared to sham operated corneas. Likewise R/S shows relative expression in corneas undergoing CLT when compared to sham operated control corneas.
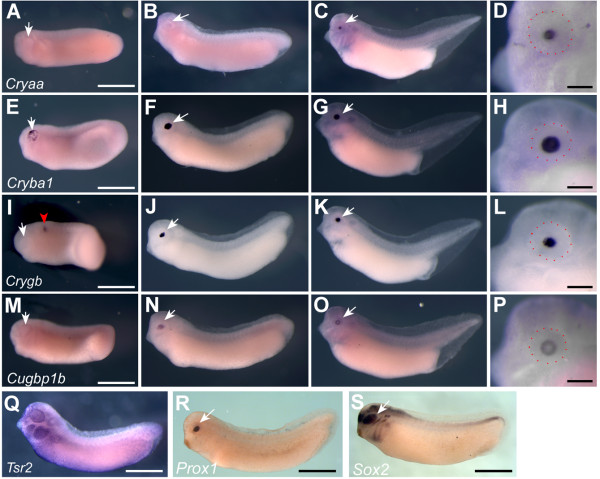
Seven genes from the CRY list were then cloned and used to determine developmental expression patterns using in situ hybridisation (Figure 4). Cryaa was expressed in central cells of the developing lens from stage 30 (Figure 4A-D) corresponding to the primary lens fibres. Cryba1 was expressed much earlier in lens development, and was first seen in punctate pattern across the whole lens placode at stage 26 (Figure 4E). Expression appeared to be in all lens cells at stage 30 (Figure 4F) and this was maintained at stage 38 (Figure 4G, H). Crygb was first detected in the hindbrain in a single stripe at stage 26 (Figure 4I). This expression was transient and was superceded by expression in the central two thirds of the lens cells by stage 30 (Figure 4J). This expression was maintained at stage 38 (Figure 4K, L). Cugbp1b expression was first seen at stage 30, specifically in lens cells (Figure 4M, N). By stage 38, expression had cleared from the central, Cryaa expressing cells, but was maintained in peripheral lens cells corresponding to the secondary lens fibres (Figure 4O, P). Tsr2 transcripts were detected in epithelial cells and were not specific to the eye (Figure 4Q). Two previously described eye genes also found in the CRY list, Prox1 and Sox2, were included here for comparison. Prox1 transcripts were seen specifically in lens cells at stage 30 as previously described [24]. Sox2 (Figure 4R) expression was seen in optic cup and lens as well as in the forming lateral line, branchial arches and neural tube (Figure 4S) [25].
Figure 4.
Expression of lens specific genes during development. In situ hybridisation of three Crystallin genes and Cugbp1b during development of the lens, indicated by dark blue staining. A-D, Cryaa expression at st. 26 (A), 30 (B) and 38 (C and D). E-H, Expression of Cryba1 throughout the lens at st. 26 (E), 30 (F) and 38 (G and H). I-L, expression of Crygb. I) at st. 26, note that red arrowhead indicates a transient stripe of expression in the hindbrain. Crygb is expressed in the central lens at st. 30 (J) and 38 (K and L). M-P) Cugbp1b transcripts are absent from the forming eye at st 26 (M), detected in the central lens cells at st.30 (N), and in the outer lens cells at st. 38 (O and P). Q) Tsr2 is expressed in epithelial cells including the cells overlying the eye, shown at stage 30. R) Prox1 expression is lens specific at stage 30, as previously described [23]. Sox2 is expressed throughout the eye as well as in the olfactory placode lateral line primordia and branchial arches as previously described [24]. White arrows indicate the position of the eye, scale bars in D, H, L, P, Q, R and S are 200 μm and red dots show the approximate margin of the eye. Scale bars in A, E, I and M apply to all other panels and correspond to 1 mm. Embryos are oriented with anterior to the left and dorsal uppermost.
Microarray analysis of CLT II: Identification of regeneration associated genes (RAG)
A second pattern match was devised to capture potential regeneration associated genes that were not highly expressed in lens (RAG list). The parameters for this were set as low expression in lens, high in samples undergoing CLT (R) and low in sham operated controls (S). 1516 probesets passed this filter with a cut-off p value of < 0.05 and the complete RAG list can be found in additional file 2. The top 50 ranked genes were re-annotated manually and their expression profile is shown in Figure 5. Sixteen of the 50 genes could not be annotated and are listed as transcribed. The top ranking gene DBAf2 encodes the Xenopus MHC class II α chain, which could suggest a sustained immune response in CLT. Of particular interest is the third ranking gene, Tcf7, also known as Tcf1 in Xenopus. Tcf7 is a HMG-box transcription factor that represses target gene expression in the absence of Wnt signalling [26].
Figure 5.
Heat map of top 50 genes, where expression is higher in samples undergoing CLT (R) than in stage matched sham operated corneas (S) or lenses (L). Darker colour indicates higher expression. R/S indicates the ratio of transcript expression in R1-3 vs. S1, 3 and 4 levels. Where possible, the genes have been manually annotated with reference to the probe source files. "Transcribed" is used to denote genes where no annotation was possible. "(sim)" indicates high similarity to the named gene.
Also included in the RAG list were the matrix metalloproteinases Mmp11 and Mmp14, transcription factors Pitx2, Pitx2a, Six1, Pax6, and the signalling proteins Fgf8b and Wnt6, which both appear twice. Tcf4, which encodes a bHLH transcription factor unrelated to Wnt signalling, appears three times on the list and a closely related gene that we have annotated as Tcf4b (similar) appears twice.
Gene ontology of 794 unique TC numbers corresponding to high probability hits (p < 0.01) from the RAG list was analysed (RAG*). Statistically over-represented gene ontologies are shown in Table 3. Strikingly, the most significantly over-represented ontology group was the Wnt receptor signalling pathway. Eight from a possible 41 genes assigned to this group were present in the RAG* list. These genes included three members of the Pitx family of transcription factors, Pitx1, Pitx2 and Pitx2a as well as Wnt pathway components Dvl2 and Lrp6, each of which is present twice in the RAG list. Also included in this group were the genes encoding the ligand Wnt2, the HMG group transcription factor Tcf7l2 (appears twice, formerly known as Tcf4 but not to be confused with the bHLH transcription factor described above) and Btrc, an F-box/WD-40 protein associated with ubiquitination. The ontology analysis is not exhaustive: Wnt3 and Wnt6 were also present in the RAG list but had not been assigned a TC number, and were therefore not picked up by the ontology screen. As well as Wnt signalling, two other gene ontology terms, "patterning of blood vessels" and "chromatin assembly or disassembly" were also highly significantly over-represented (Table 3). The gene ontology term "patterning of blood vessels" scored highly because of inclusion of the Pitx genes 1, 2 and 2a, which are also involved in this process. The genes in this category "chromatin assembly or disassembly" that were present in the analysis were Unkempt1, Histone 2 ab and 2a, Smarcc2, Mta2, Hsp90ab1 and Cbx1.
Table 3.
Top 10 statistically over represented GO categories in the RAG* list.
| GMRG_Term | TC numbers | p | Description | |
|---|---|---|---|---|
| total | subset | |||
| GO:0016055 | 41 | 8 | 0.003 | Wnt receptor signaling pathway |
| GO:0001569 | 6 | 3 | 0.004 | patterning of blood vessels |
| GO:0006333 | 40 | 7 | 0.009 | chromatin assembly or disassembly |
| GO:0030001 | 3 | 2 | 0.011 | metal ion transport |
| GO:0045010 | 3 | 2 | 0.011 | actin nucleation |
| GO:0006729 | 3 | 2 | 0.011 | tetrahydrobiopterin biosynthetic process |
| GO:0009591 | 15 | 4 | 0.011 | perception of mechanical stimulus |
| GO:0007368 | 10 | 3 | 0.019 | determination of left/right symmetry |
| GO:0006826 | 10 | 3 | 0.019 | iron ion transport |
| GO:0050930 | 4 | 2 | 0.020 | induction of positive chemotaxis |
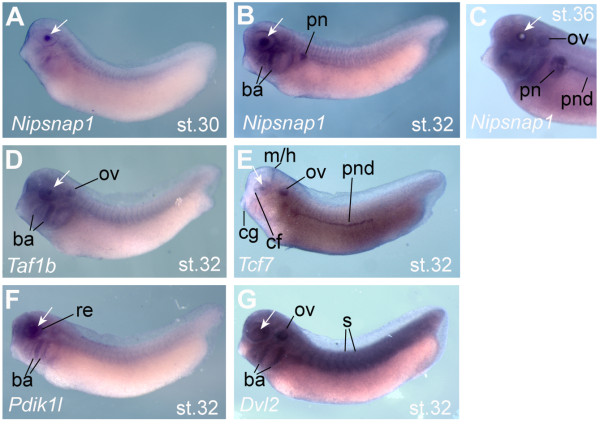
Validation was performed for 13 genes with 9 confirmed as statistically increased in CLT and two more showing the same trend as the array (Table 4). Five genes were examined for expression in developing lens. Although none of these were found to be entirely lens specific at tailbud stages, the BMP target gene Nipsnap1 was expressed only in the lens at stage 30, with expression in branchial arches, otic vesicle and pronephros appearing around stage 32 (Figure 6A-C). Taf1b was expressed in the lens, branchial arches and otic vesicle from stage 32 (Figure 6D). The Wnt target gene and pathway-specific transcription factor Tcf7 was also expressed in developing lens at stage 32 but was also seen in the choroid fissure of the eye cup, midbrain hindbrain boundary, otic vesicle, cement gland and pronephritic duct at stage 32 (Figure 6E). Pdik1l expression was restricted to the developing eye (lens and retina) and branchial arches, beginning at stage 30, (Figure 6F) and the Wnt pathway gene Dvl2 was expressed almost ubiquitously (Figure 6G).
Table 4.
Q-rtPCR validation of 13 genes from the RAG list.
| Rank in array | Gene name | Lens (L/S) | CLT (R/S) | |
|---|---|---|---|---|
| qPCR | qPCR | array | ||
| 3 | Tcf7 | 0.6 | 5.7 | 7.8 |
| 10 | Crop | 0.7 | 1.4 | 3.3 |
| 11 | Nipsnap1 (sim) | 0.1 | 176.3 | 8.5 |
| 12 | Taf1b | 1.0 | 8.0 | 10.2 |
| 15 | Mybph | 0.0 | 0.2 | 52.1 |
| 31 | Pdik1l-b | 1.1 | 10.3 | 12.3 |
| 38 | Gnai2 | 5.5 | 56.1 | 6.3 |
| 41 | Tcf4 | 0.0 | 4.4 | 4.3 |
| 45 | Mmp-11 | 1.8 | 100.0 | 2.4 |
| 56 | Creld1 | 0.0 | 1.6 | 5.2 |
| 71 | Amph | 24.2 | 74.6 | 2.9 |
| 479 | Mmp14 | 0.1 | 3.4 | 2.3 |
| 1391 | Lmo4 | 4.11 | 2.2 | 1.4 |
Bold numbers indicate upregulation of more than 1.5 fold, numbers in italics indicate down regulation and normal font indicates no significant change. L/S shows the relative (fold higher) expression in lens when compared to sham operated corneas. Likewise R/S shows relative expression in corneas undergoing CLT when compared to sham operated control corneas.
Figure 6.
Expression of selected transdifferentiation genes during development. In situ hybridisation of five regeneration associated genes (RAG). Dark blue staining marks expression, and a white arrow indicates the position of the lens. A) Nipsnap1 expression appears in lens cells at stage 30 (A) and is also found in the pronephros (pn) and branchial arches (ba) from stage 32 (B). At stage 36, Nipsnap1 expression is seen in the pronephros and pronephritic duct (pnd) and otic vesicle (ov) as well as in the periphery of the lens (C). D) Tbp1b is expressed predominantly in lens, otic vesicle and branchial arches at stage 32. E) Tcf7 is expressed in lens and choroid fissure of the retina (cf), the midbrain-hindbrain junction (m/h), cement gland (cg), otic vesicle and pronephritic duct at at stage 32. F) Pdik1l is expressed in the developing lens and retina (re) and branchial arches at stage 32. G) Dvl2 is expressed in eye tissues but is not specific, with expression extending throughout the embryo including the somites (s), otic vesicle and branchial arches. Embryos are oriented with anterior to the left and dorsal uppermost.
Pluripotency genes are not upregulated in 3 day corneas undergoing CLT
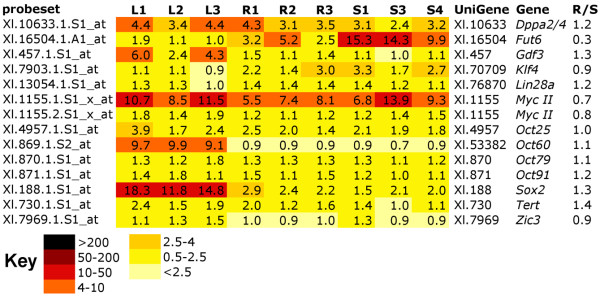
Thirteen genes associated with pluripotency were present on the GeneChip array. These included Klf4 (biklf-A), c-Myc, Sox2, and four POU genes, Oct25, Oct60, Oct79 and Oct91, which seem, in Xenopus, to substitute for the pluripotency gene Oct3/4 of mammals [27], Dppa2/4, Gdf3, Fut6 (Ssea1 homologue), Lin28, Tert and Zic3. However, with the exception of Sox2, none of these were captured in our screen (Figure 7). Sox2 was expressed at highest levels in the lens samples (8 fold higher levels than in sham operated cornea controls) but was not significantly elevated in corneas undergoing CLT when compared to the same controls. Sox2 then may be involved in lens differentiation but does not seem to be indicative of dedifferentiation in this case. Another pluripotency associated gene, Fut6, was significantly upregulated in sham operated corneas when compared to corneas undergoing CLT (3.7 fold increase), with no expression in lens. Since we only observed one time point, it is possible that we have missed a change in pluripotency gene expression. Further analysis of earlier and later stages of CLT will be needed to exclude this possibility.
Figure 7.
Heat Map showing that expression of pluripotency associated genes does not change during CLT. Darker colour indicates higher levels of expression. R/S indicates the ratio of transcript expression in R1-3 vs. S1, 3 and 4 levels. Where possible, the genes have been manually annotated with reference to the probe source files.
Discussion
BMP signalling is essential for CLT
BMP signalling has not been previously linked to transdifferentiation of the cornea into lens. However, Faber et al demonstrated that BMP signalling is required for mammalian primary lens cell differentiation both in vivo and in vitro [28]. Furthermore, BMP4 is essential for lens development in mice and acts upstream of the transcription factor and early lens placode marker Sox2 [29,30], which in turn directly regulates crystallins [31]. Finally, BMP7 is also expressed in the developing lens and regulates expression of Pax6, which also regulates crystallins [29,30].
We have previously used the transgenic line N1, in which noggin overexpression is controlled by the inducible Hsp70 heat shock promoter, to demonstrate a requirement for functional BMP signalling in limb and tail regeneration in Xenopus laevis [17,20]. Both tail and limb regeneration involve epimorphic type regeneration, the regrowth of a patterned organ comprised of multiple tissues. Here, we use the same line to show that a distinct type of regenerative process, that of transdifferentiation of cornea to lens, also depends on active BMP signalling. Noggin overexpression, which blocks BMP signalling, appeared to have no effect on the early corneal thickening and cell shape changes associated with wound healing. However, prolonged inhibition of BMP signalling prevented the subsequent progression of cells to transdifferentiating aggregates that eventually form the new lens. Instead, the extra cells seem to become hypertrophic and die. Cellular hypertrophy was also observed in the AEC of poorly regenerating Xenopus limb buds [32], suggesting that BMP may be a survival signal for cells during regeneration.
In support of this, we have shown that increased expression of Nipsnap1, a known direct BMP target gene [33], is associated with CLT. The expression of Nipsnap1 in developing lens suggests that it may act downstream of BMPs in specifying the lens fate. In another study of Xenopus CLT, Malloch et al detected transcripts from a gene annotated as similar to BMP5 [15]. CLT in Xenopus differs from the better studied Wolffian regeneration of the lens in newts, which takes place via dedifferentiation of the pigmented epithelial cells (PECs) of the dorsal iris rather than transdifferentiation from the more closely related cornea [2]. Despite this difference, similar signals may be involved in the process. In a recent study, a large number of ESTs were generated from Cynops pyrrhogaster dorsal iris PECs undergoing dedifferentiation after lentectomy [34]. In this dataset, crystallins were not identified, suggesting that the dedifferentiated PECs had not yet begun to transdifferentiate into lens cells. Maki and colleagues detected multiple members of the BMP and TGFβ growth factor pathways but report that components of the Wnt, Fgf and hedgehog signalling pathways were not detected. In newts, however, BMP inhibition leads to enhanced regeneration, in conflict with our observations in Xenopus. Functional evidence showed that inhibition of BMP induces formation of a lens from the ventral iris, which does not normally regenerate [35]. This difference in the role of BMP may reflect inherent differences between Wolffian regeneration in newts and CLT in Xenopus, possibly due to the requirement for dedifferentiation in the newt.
Wnt signalling pathway components are upregulated during lens regeneration
Wnt/β-catenin signalling (via the canonical pathway) is known to be important in driving lens cell differentiation in mammals, with several Wnt ligands, along with their receptors (Frizzled family) and the Lrp5/6 co-receptors expressed during development of the lens [36-38]. Reporter strains in mice have shown a period of Tcf/Lef activity in the lens epithelium as it develops [39,40], and it is thought that canonical Wnt signalling is required for initial formation of the lens epithelium. In contrast, the Wnt/PCP pathway is thought to play a role in the later lens fibre differentiation from the lens epithelium [41]. Regulation of Wnt signalling via inhibition by secreted frizzled related proteins (Sfrp) also occurs in mammalian lens development [38]. Sfrp2 expression under a lens specific crystallin promotor in mice led to cataract formation [42].
Here, we present evidence that the developmental role of canonical wnt signalling in mammalian anterior eye formation is recapitulated in lens regeneration from the cornea in Xenopus. Gene ontology analysis of our RAG list demonstrated that regeneration of the lens in Xenopus is accompanied by significantly increased expression of several components of the Wnt signalling pathway. Although the gene ontology for the CRY list suggested involvement of Wnt/PCP pathway, this result is entirely due to the expression of a single Wnt receptor, Frz7, and there is therefore insufficient evidence to implicate Wnt/PCP rather than canonical Wnt signalling, in CLT. The antagonist Sfrp2 was also recovered from the CRY list and was expressed at 1.5 fold higher levels in regenerating CLT corneas than in sham operated controls.
Wnt signalling components were also recovered from an EST collection generated from tissue undergoing the early stages of CLT. Wnt7b and two genes related to the Wnt modulator Sfrp5 were identified by Malloch and colleagues [15]. Furthermore, Wolffian regeneration of the newt lens, while occurring from the neural crest derived pigmented epithelial cells of the dorsal iris rather than the cornea, may well employ some of the same mechanisms (Reviewed in [43]). Experiments have demonstrated that if Wnt is made available, the ventral iris can also regenerate a lens [44].
Pitx genes, which may be Wnt targets, are upregulated during lens regeneration
Three members of the bicoid related homeobox transcription factor Pitx were captured in the RAG list: Pitx2, Pitx2a and Pitx1. A fourth, Pitx3, was captured in the CRY list and was expressed in lens tissue as well as 1.4 × higher in corneas undergoing CLT than in control corneas. Pitx factors have not previously been implicated in vertebrate regeneration although a recent report identified a role in asexual reproduction/regeneration of the ascidian Botryllus schlosseri [45]. Mutations in Pitx genes in humans are known to cause eye developmental defects, particularly affecting the anterior eye structures: cornea, iris and lens. Pitx2 mutations cause Axenfeld-Rieger syndrome type 1, a congenital malformation syndrome affecting the anterior eye [46,47] iridogoniodysgenesis type 2 (iris hypoplasia) [48], Peters' anomaly (defective cornea) [49] and ring dermoid of the cornea [50]. Mutations in the related gene Pitx3 are known to cause congenital cataracts [51]. Pitx2 is induced by the canonical Wnt signalling pathway directly via Lef1 [52]. Pitx genes may therefore act downstream of Wnt signalling in lens regeneration in Xenopus.
Lens crystallins are markers of differentiation and are expressed during CLT
Eleven probesets representing seven UniGenes belonging to the lens crystallin family were used to search for genes with similar patterns of expression across the nine datasets. Searching our microarray data for similarly expressed genes uncovered three more crystallins: Cryaa, cryba2 and crybb3 (similar). Of these, only cryba2 showed a significantly higher expression in corneas undergoing CLT than in sham-operated corneas, i.e. a regenerative response. The expression of three selected crystallin genes was observed at different times during the process of embryonic lens formation (Figure 4). The α-crystallin cryaa was expressed late, between lens stage 4 and 5 according to McDevitt and Brahma [53]. The β-crystallin Cryba1 was the earliest of the three to be expressed in the lens placode, beginning at lens stage 1-2 and the γ-crystallin crygb was expressed slightly later at lens stage 3-4. This pattern of developmental expression is somewhat reflected in the corneas undergoing CLT, with α-crystallins unchanged between regenerating and sham operated corneas and all identified β and γ-crystallins on the array being upregulated during CLT. Therefore we believe we have captured corneas in the act of transdifferentiating, just as the first crystallins become expressed. Furthermore, we observe a correlation in timing of α, β and γ-crystallin expression that reflects that seen during lens development. However, others have reported that the timing of crystallin expression differs between regeneration and development [54].
Genes associated with congenital cataract formation have similar expression profiles to crystallins in our microarray data
Formation of congenital cataract often results from mutation of genes involved in the formation of the anterior eye, which includes the lens. Many cases of congenital cataract result from mutations in the crystallin genes discussed above. Our search for regeneration specific gene upregulation revealed changes in several other genes known to be involved the formation of congenital cataracts. One such gene, Pitx3, is discussed earlier. The b-Zip transcription factor L-maf ranks 31st in the CRY list, and is 2-fold upregulated in corneas undergoing CLT relative to sham operated controls (Figure 3). L-maf is expressed in the lens placode and later in fibre cells in Xenopus, and directly activates the expression of several lens crystallins [55]. Mutations in a human homologue of this gene cause a type of congenital cataract called CCA4 (OMIM#610202) [56].
Lens intrinsic membrane protein 2 (Lim2) is ranked 17th on the CRY list and a potential homologue, Lim2 similar, is ranked 13th. Lim2, but not its homolgue, was upregulated in CLT. Lim2 protein is very abundant in the human lens, and mutations in Lim2, also known as MP19, are associated with congenital or early onset cataract [23]. The forkhead transcription factor Lens1 (FoxE3) was ranked 218th in the CRY list and expression was 1.6 fold higher in CLT corneas than in sham operated controls. FoxE3 is associated with ocular dysgenesis and cataracts in humans (OMIM#601094)[57].
Cugbp1 is an RNA binding protein targeting CUG repeats. Xenopus laevis appears to have two homolgues of this gene, Cugbp1b was ranked 8th in the CRY list and expression was 2.4 fold higher in CLT corneas relative to controls, and Cugbp1a was ranked 14th with a 1.5 fold higher relative expression. While Cugbp1 is not directly linked to the formation of cataracts, expansion of CTG repeats in the 3'UTR of the human DMPK gene cause myotonic dystrophy, a form of adult muscular dystrophy that is accompanied by cataract formation [58]. Here, we have shown that Cugbp1b is expressed specifically in the forming lens and is upregulated during CLT, suggesting it could be involved in the pathology of myotonic dystrophy. However, the Cugbp1 duplication appears to be unique to Xenopus laevis and an eye developmental role has not yet been described for these CUG binding proteins in other vertebrates.
Genes associated with pluripotency are not upregulated in three day CLT tissue but chromatin modification may occur
Recent transcriptome analysis of Wolffian regeneration of the newt lens identified several genes associated with reprogramming, such as histone deacetylases and the oncogene c-myc, highly suggestive of dedifferentiation [34,59]. Furthermore, expression of a subset of pluripotency associated genes (Klf4, Sox2 and c-myc) was found to be increased during newt lens and limb regeneration [60]. In contrast, recent evidence has shown that pluripotency genes were not up regulated during zebrafish fin regeneration, although reduction of either Oct4 or Sox2 activity prevented fins from regenerating [61]. Our microarray data, while limited to only one stage of the process, suggests that CLT occurs without dedifferentiation in Xenopus. Thirteen genes associated with pluripotency were present on the GeneChip array. However, with the exception of Sox2, none of these were up regulated during CLT (Figure 7). Sox2 was expressed at highest levels in the lens samples but was not significantly elevated in corneas undergoing CLT compared to sham operated corneas. Sox2 then may be involved in lens differentiation but does not seem to be indicative of dedifferentiation in this case. Similarly, Sox2 expression was found in limbs and tails before amputation [61]. Another pluripotency associated gene, Fut6, was significantly upregulated in sham operated corneas when compared to corneas undergoing CLT (3.7x), with no expression in lens. Christen et al recently showed the same result in Xenopus limb blastema vs. pseudoblastema, with what they call Fut1 (the same gene) being upregulated regardless of regenerative success [61]. While limited to a single timepoint in the regenerative process, our results show no evidence for pluripotency gene up regulation during CLT and we therefore suggest that returning to a pluripotent state is not part of the CLT process, unlike the lens regeneration from newt PECs. We do note, however, that genes associated with chromatin assembly and disassembly were statistically overrepresented in the RAG list, suggesting that epigenetic changes may be taking place during CLT.
Conclusions
We have shown a functional role for BMP signalling in the process of lens regeneration from the cornea (CLT) in Xenopus laevis tapoles, and identified a new role for Nipsnap1, a of BMP signalling, in the process of lens development. Furthermore, we present strong evidence for the involvement of Wnt signalling and Pitx transcription factors in CLT. Our microarray analysis has identified many genes that are involved in lens pathology, in particular the development of congenital cataract. Finally, we have shown that although there may be alterations to chromatin, there is no evidence for a return to pluripotency, or dedifferentiation, seen in 3 day corneas undergoing CLT.
Methods
Transgenic frogs
The N1 stable line of transgenic Xenopus has been previously described [17]. Briefly, the animals contain a transgene (Hsp70:Noggin; γ-Crystallin:GFP) comprised of two linked parts, the first containing X. laevis Noggin coding sequence under the control of the Hsp70 promoter, and the second the green fluorescent protein (GFP) coding sequence under the control of the lens specific promoter γ-crystallin. The line is derived from a single insertion founder made by sperm nuclear injection using the method of [62] Kroll and Amaya 1996 modified as in Beck et al (2003) [17]. All animal experiments were subject to New Zealand's animal welfare standards for vertebrates and were reviewed by the University of Otago Animal Ethics Committee (AEC). The AEC approved all experiments under protocols AEC78/06 and AEC78/09.
Lentectomy
Stage 50-51 tadpoles were anaesthetised in MS222 (1/4000 w/v) in 0.1 × MMR, then placed on their left sides on damp paper towels for surgery. The outer cornea was first snipped with Vannas iridectomy scissors from the posterior side and cut dorsally and ventrally before lifting up as a flap. The inner cornea was ruptured and the lens gently removed using forceps, and the flap of cornea and epidermis was replaced gently over the eye. Animals were allowed to recover in 0.1 × MMR overnight before returning to a marine biotech aquarium and fed as normal. For sham-operated animals, the flap of outer cornea/epidermis was raised as for lentectomy but then replaced without further intervention.
Histology
Tadpoles were fixed overnight in cold ethanol/glycine fixative (70% ethanol, 15 mM glycine pH 2.0) at -20°C, dehydrated in methanol and embedded in paraffin wax. 5 μm sections were cut using a Leica microtome and stained with haematoxylin and eosin. At least five animals were sectioned for each timepoint and condition reported.
Microarray analysis
St. 50-51 tadpoles were either subjected to lentectomy or sham operated as described above. After three days, corneas (containing some epidermis) from lentectomised eyes (R) or sham operated eyes (S) were dissected from tadpoles under anaesthesia in MS222 using Vannas iridectomy scissors and fine forceps and transferred to small pieces of dry Whatman 3 M filter paper, cut with a hole punch, in groups of 8-10. The paper discs were immediately transferred to RNA later in a 1.5 ml centrifuge tube and stored on ice until RNA isolation. Groups of 8-10 dissected lenses (L) were placed directly in RNAlater (Qiagen). Three biological replicate samples were prepared in each case and RNA was isolated using an RNaqueous micro kit (Ambion) with brief manual homogenisation in buffer before removing the paper discs manually. An amplification step (in vitro transcription) was performed using 50 ng of starting material and the samples were labelled with Biotin using a Nugen Ovation kit according to the manufacturers instructions. Three biological replicate pools for each treatment (L, R and S) were hybridised to Xenopus laevis GeneChips (Affymetrix, version 1) and washed using protocol WS2v4_450 before scanning on a 7G Plus GeneChip Scanner 3000 (Affymetrix). Data was normalised for the nine samples using an RMA algorithm with the software GenePattern. Heatmaps were prepared in Microsoft Excel using a custom macro to colour cells according to their values.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) [63] and are accessible through GEO Series accession number GSE28014.
Annotation and gene naming
Annotation was done manually by searching NCBI UniGene with the accession number of the source sequence used to design the Affymetrix probesets. Gene nomenclature follows that used by UniGene: where this differs from common usage, it has been highlighted in the text where practical. Genes that could not be annotated confidently are marked as transcribed. Genes with high similarity to a known gene are marked (similar) or (sim).
Gene ontology analysis
Xenopus TC (Tentative Consensus) numbers and Gene Ontology (GO) assignments for biological function were obtained for the Affymetrix Xenopus laevis GeneChip using Resourcerer v13.0 [64], December 2006 release. Lists of TC numbers were generated for both CRY and RAG lists and duplicate TC numbers (arising when the GeneChip contained multiple probe sets for one gene) removed using the online BAR duplicate remover tool [65]. Genemerge v1.2 [66] was used to determine GO groups that were statistically over-represented in either list compared to the genes represented on the array.
Q-rtPCR
Primers for quantitative real time PCR were designed, where possible, to different exons, to avoid amplification of genomic DNA. The NCBI program Spidey [67] was used to predict intron-exon boundaries by comparing X. laevis cDNA sequence to X. tropicalis genomic and transcript sequence from the Joint Genome Institute [68]. Primer sequences, annealing temperatures and product sizes can be found in additional file 3.
Cloning and ISH
Primer pairs for amplification of cDNA can be found in additional file 3.
Crystallins were amplified from reverse transcribed st 50 lens RNA and all other cDNAs from reverse transcribed RNA isolated from mixed embryo stages (13, 19, 33 and 37), using Mango TAQ (Bioline). PCR products were cloned directly into T-tailed PCRIIscript vector using a TOPO kit (Invitrogen) and transformed into chemically competent TOP10 E.coli (Invitrogen). Insertions were verified by DNA sequencing, performed by Otago University's Gene Analysis Service. Digoxygenin labelled ribonucleotide probes were made by linearising plasmids with XhoI and transcribing using SP6 polymerase labelled with digoxigenin-UTP labelling mix (Roche). DNase I (Invitrogen) was used to remove templates following transcription and the probes were precipitated with 2.5 M LiCl. Whole-mount in situ hybridisation of albino embryos and tadpoles was performed as described previously [69]. Proteinase K treatment was 10 μg/ml for 10 minutes for embryos.
Authors' contributions
RCD performed microarray hybridisation, analysis and validation. CWB designed the study, performed embryo experiments and in situ hybridisation, and drafted the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
Excel spreadsheet of CRY list.
Excel spreadsheet of RAG list.
Excel spreadsheet of primers used.
Contributor Information
Robert C Day, Email: robert.day@otago.ac.nz.
Caroline W Beck, Email: caroline.beck@otago.ac.nz.
Acknowledgements
The authors acknowledge the assistance of Ellen Hansen with preparation of histological sections, James McEwan for Prox1 and Sox2 in situ hybridisation, and Amy Armstrong for care of the frog colony at Otago.
References
- Beck CW, Izpisua Belmonte JC, Christen B. Beyond early development: Xenopus as an emerging model for the study of regenerative mechanisms. Dev Dyn. 2009;238:1226–48. doi: 10.1002/dvdy.21890. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Tsonis PA. Molecular and cellular aspects of amphibian lens regeneration. Prog Retin Eye Res. 2010;29:543–55. doi: 10.1016/j.preteyeres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G. Lens regeneration from the cornea in Xenopus laevis. J Exp Zool. 1963;154:39–65. doi: 10.1002/jez.1401540105. [DOI] [PubMed] [Google Scholar]
- Reeve JC, Wild AE. secondary lens formation from the cornea following implantation of larval tissues betwen the inner and outer cornea of Xenopus laevis tadpoles. J Embrol Exp Morph. 1981;63:121–132. [PubMed] [Google Scholar]
- Filoni S, Bosco L, Cioni C. The role of neural retina in lens regeneration from cornea in larval Xenopus laevis. Acta Embryol Morphol Exp. 1982;3:15–28. [PubMed] [Google Scholar]
- Reeve JG, Wild AE. Lens regeneration from cornea of larval Xenopus laevis in the presence of the lens. J Embryol Exp Morphol. 1978;48:205–14. [PubMed] [Google Scholar]
- Bosco L, Filoni S, Cioni C. Lens formation from cornea in the presence of the old lens in larval Xenopus laevis. J Exp Zool. 1980;213:9–14. doi: 10.1002/jez.1402130103. [DOI] [PubMed] [Google Scholar]
- Cannata SM, Arresta E, Bernardini S, Gargioli C, Filoni S. Tissue interactions and lens-forming competence in the outer cornea of larval Xenopus laevis. J Exp Zoolog Part A Comp Exp Biol. 2003;299:161–71. doi: 10.1002/jez.a.10275. [DOI] [PubMed] [Google Scholar]
- Gargioli C, Giambra V, Santoni S, Bernardini S, Frezza D, Filoni S, Cannata SM. The lens-regenerating competence in the outer cornea and epidermis of larval Xenopus laevis is related to pax6 expression. J Anat. 2008;212:612–20. doi: 10.1111/j.1469-7580.2008.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filoni S, Bernardini S, Cannata SM, D'Alessio A. Lens regeneration in larval Xenopus laevis: experimental analysis of the decline in the regenerative capacity during development. Dev Biol. 1997;187:13–24. doi: 10.1006/dbio.1997.8598. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Elkins MB. Cornea-lens transdifferentiation in the anuran, Xenopus tropicalis. Dev Genes Evol. 2001;211:377–87. doi: 10.1007/s004270100163. [DOI] [PubMed] [Google Scholar]
- Filoni S. Retina and lens regeneration in anuran amphibians. Semin Cell Dev Biol. 2009;20:528–34. doi: 10.1016/j.semcdb.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Schaefer JJ, Oliver G, Henry JJ. Conservation of gene expression during embryonic lens formation and cornea-lens transdifferentiation in Xenopus laevis. Dev Dyn. 1999;215:308–18. doi: 10.1002/(SICI)1097-0177(199908)215:4<308::AID-AJA3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Henry JJ, Carinato ME, Schaefer JJ, Wolfe AD, Walter BE, Perry KJ, Elbl TN. Characterizing gene expression during lens formation in Xenopus laevis: evaluating the model for embryonic lens induction. Dev Dyn. 2002;224:168–85. doi: 10.1002/dvdy.10097. [DOI] [PubMed] [Google Scholar]
- Malloch EL, Perry KJ, Fukui L, Johnson VR, Wever J, Beck CW, King MW, Henry JJ. Gene expression profiles of lens regeneration and development in Xenopus laevis. Dev Dyn. 2009;238:2340–56. doi: 10.1002/dvdy.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco L, Venturini G, Willems D. In vitro lens transdifferentiation of Xenopus laevis outer cornea induced by Fibroblast Growth Factor (FGF) Development. 1997;124:421–8. doi: 10.1242/dev.124.2.421. [DOI] [PubMed] [Google Scholar]
- Beck CW, Christen B, Slack JM. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell. 2003;5:429–39. doi: 10.1016/S1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- Lin G, Slack JM. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316:323–35. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Pearl EJ, Barker D, Day RC, Beck CW. Identification of genes associated with regenerative success of Xenopus laevis hindlimbs. BMC Dev Biol. 2008;8:66. doi: 10.1186/1471-213X-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CW, Christen B, Barker D, Slack JM. Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech Dev. 2006;123:674–88. doi: 10.1016/j.mod.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–87. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Li X, Yang Y, Church RL. Localization of lens intrinsic membrane protein MP19 and mutant protein MP19(To3) using fluorescent expression vectors. Mol Vis. 2002;8:372–88. [PubMed] [Google Scholar]
- Ponnam SP, Ramesha K, Tejwani S, Matalia J, Kannabiran C. A missense mutation in LIM2 causes autosomal recessive congenital cataract. Mol Vis. 2008;14:1204–8. [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Mochii M, Yamamoto TS, Takahashi TC, Eguchi G, Okada TS. Pax-6 and Prox 1 expression during lens regeneration from Cynops iris and Xenopus cornea: evidence for a genetic program common to embryonic lens development. Differentiation. 1999;65:141–9. doi: 10.1046/j.1432-0436.1999.6530141.x. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev Biol. 2004;271:439–66. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Nusse R. A versatile transcriptional effector of Wingless signaling. Cell. 1997;89:321–3. doi: 10.1016/S0092-8674(00)80210-X. [DOI] [PubMed] [Google Scholar]
- Morrison GM, Brickman JM. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development. 2006;133:2011–22. doi: 10.1242/dev.02362. [DOI] [PubMed] [Google Scholar]
- Faber SC, Robinson ML, Makarenkova HP, Lang RA. Bmp signaling is required for development of primary lens fiber cells. Development. 2002;129:3727–37. doi: 10.1242/dev.129.15.3727. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–88. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Hogan BL. BMP4 is essential for lens induction in the mouse embryo. Genes Dev. 1998;12:3764–75. doi: 10.1101/gad.12.23.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Uchikawa M, Kamachi Y. Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int J Dev Biol. 2004;48:819–27. doi: 10.1387/ijdb.041868hk. [DOI] [PubMed] [Google Scholar]
- Barker DM, Beck CW. Overexpression of the transcription factor Msx1 is insufficient to drive complete regeneration of refractory stage Xenopus laevis hindlimbs. Dev Dyn. 2009;238:1366–78. doi: 10.1002/dvdy.21923. [DOI] [PubMed] [Google Scholar]
- Peiffer DA, Von Bubnoff A, Shin Y, Kitayama A, Mochii M, Ueno N, Cho KW. A Xenopus DNA microarray approach to identify novel direct BMP target genes involved in early embryonic development. Dev Dyn. 2005;232:445–56. doi: 10.1002/dvdy.20230. [DOI] [PubMed] [Google Scholar]
- Maki N, Martinson J, Nishimura O, Tarui H, Meller J, Tsonis PA, Agata K. Expression profiles during dedifferentiation in newt lens regeneration revealed by expressed sequence tags. Mol Vis. 2010;16:72–8. [PMC free article] [PubMed] [Google Scholar]
- Grogg MW, Call MK, Okamoto M, Vergara MN, Del Rio-Tsonis K, Tsonis PA. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature. 2005;438:858–62. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump RJ, Ang S, Chen Y, von Bahr T, Lovicu FJ, Pinson K, de Iongh RU, Yamaguchi TP, Sassoon DA, McAvoy JW. A role for Wnt/beta-catenin signaling in lens epithelial differentiation. Dev Biol. 2003;259:48–61. doi: 10.1016/S0012-1606(03)00179-9. [DOI] [PubMed] [Google Scholar]
- Ang SJ, Stump RJ, Lovicu FJ, McAvoy JW. Spatial and temporal expression of Wnt and Dickkopf genes during murine lens development. Gene Expr Patterns. 2004;4:289–95. doi: 10.1016/j.modgep.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Chen Y, Stump RJ, Lovicu FJ, McAvoy JW. Expression of Frizzleds and secreted frizzled-related proteins (Sfrps) during mammalian lens development. Int J Dev Biol. 2004;48:867–77. doi: 10.1387/ijdb.041882yc. [DOI] [PubMed] [Google Scholar]
- Liu H, Mohamed O, Dufort D, Wallace VA. Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev Dyn. 2003;227:323–34. doi: 10.1002/dvdy.10315. [DOI] [PubMed] [Google Scholar]
- Liu H, Thurig S, Mohamed O, Dufort D, Wallace VA. Mapping canonical Wnt signaling in the developing and adult retina. Invest Ophthalmol Vis Sci. 2006;47:5088–97. doi: 10.1167/iovs.06-0403. [DOI] [PubMed] [Google Scholar]
- Chen Y, Stump RJ, Lovicu FJ, McAvoy JW. A role for Wnt/planar cell polarity signaling during lens fiber cell differentiation? Semin Cell Dev Biol. 2006;17:712–25. doi: 10.1016/j.semcdb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stump RJ, Lovicu FJ, Shimono A, McAvoy JW. Wnt signaling is required for organization of the lens fiber cell cytoskeleton and development of lens three-dimensional architecture. Dev Biol. 2008;324:161–76. doi: 10.1016/j.ydbio.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Mizuno N, Kondoh H. Determinative roles of FGF and Wnt signals in iris-derived lens regeneration in newt eye. Dev Growth Differ. 2008;50:279–87. doi: 10.1111/j.1440-169X.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Mizuno N, Takada R, Takada S, Kondoh H. Determinative role of Wnt signals in dorsal iris-derived lens regeneration in newt eye. Mech Dev. 2006;123:793–800. doi: 10.1016/j.mod.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Tiozzo S, De Tomaso AW. Functional analysis of Pitx during asexual regeneration in a basal chordate. Evol Dev. 2009;11:152–62. doi: 10.1111/j.1525-142X.2009.00316.x. [DOI] [PubMed] [Google Scholar]
- Fitch N, Kaback M. The Axenfeld syndrome and the Rieger syndrome. J Med Genet. 1978;15:30–4. doi: 10.1136/jmg.15.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU. et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–9. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Alward WL, Semina EV, Kalenak JW, Heon E, Sheth BP, Stone EM, Murray JC. Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol. 1998;125:98–100. doi: 10.1016/S0002-9394(99)80242-6. [DOI] [PubMed] [Google Scholar]
- Doward W, Perveen R, Lloyd IC, Ridgway AE, Wilson L, Black GC. A mutation in the RIEG1 gene associated with Peters' anomaly. J Med Genet. 1999;36:152–5. [PMC free article] [PubMed] [Google Scholar]
- Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, Cai F, Pan Q, Long Z, Dai H. et al. Mutation in PITX2 is associated with ring dermoid of the cornea. J Med Genet. 2004;41:e129. doi: 10.1136/jmg.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry V, Yang Z, Addison PK, Francis PJ, Ionides A, Karan G, Jiang L, Lin W, Hu J, Yang R. et al. Recurrent 17 bp duplication in PITX3 is primarily associated with posterior polar cataract (CPP4) J Med Genet. 2004;41:e109. doi: 10.1136/jmg.2004.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Wynshaw-Boris A, Rose DW, Rosenfeld MG. Pitx genes during cardiovascular development. Cold Spring Harb Symp Quant Biol. 2002;67:81–7. doi: 10.1101/sqb.2002.67.81. [DOI] [PubMed] [Google Scholar]
- McDevitt DS, Brahma SK. Ontogeny and localization of the crystallins during embryonic lens development in Xenopus laevis. J Exp Zool. 1973;186:127–40. doi: 10.1002/jez.1401860204. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Mochii M, Takahashi TC, Eguchi G, Okada TS. Lens regeneration in Xenopus is not a mere repeat of lens development, with respect to crystallin gene expression. Differentiation. 1999;64:143–9. doi: 10.1046/j.1432-0436.1999.6430143.x. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Yasuda K. Distinct roles of maf genes during Xenopus lens development. Mech Dev. 2001;101:155–66. doi: 10.1016/S0925-4773(00)00585-2. [DOI] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F. et al. Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet. 2002;11:33–42. doi: 10.1093/hmg/11.1.33. [DOI] [PubMed] [Google Scholar]
- Semina EV, Brownell I, Mintz-Hittner HA, Murray JC, Jamrich M. Mutations in the human forkhead transcription factor FOXE3 associated with anterior segment ocular dysgenesis and cataracts. Hum Mol Genet. 2001;10:231–6. doi: 10.1093/hmg/10.3.231. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. Deconstructing myotonic dystrophy. Science. 2000;289:1701–2. doi: 10.1126/science.289.5485.1701. [DOI] [PubMed] [Google Scholar]
- Maki N, Tsonis PA, Agata K. Changes in global histone modifications during dedifferentiation in newt lens regeneration. Mol Vis. 2010;16:1893–7. [PMC free article] [PubMed] [Google Scholar]
- Maki N, Suetsugu-Maki R, Tarui H, Agata K, Del Rio-Tsonis K, Tsonis PA. Expression of stem cell pluripotency factors during regeneration in newts. Dev Dyn. 2009;238:1613–6. doi: 10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Robles V, Raya M, Paramonov I, Belmonte JC. Regeneration and reprogramming compared. BMC Biol. 2010;8:5. doi: 10.1186/1741-7007-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–83. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- NCBI Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo/
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897–9. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- BAR duplicate remover tool. http://www.bar.utoronto.ca/ntools/cgi-bin/ntools_duplicate_remover.cgi
- Castillo-Davis CI, Hartl DL. GeneMerge--post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–2. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- NCBI Spidey mRNA to genomic alignment tool. http://www.ncbi.nlm.nih.gov/spidey/index.html
- JGI Xenopus tropicalis genome. http://genome.jgi-psf.org/Xentr4/Xentr4.home.html
- Beck CW, Slack JM. Analysis of the developing Xenopus tail bud reveals separate phases of gene expression during determination and outgrowth. Mech Dev. 1998;72:41–52. doi: 10.1016/S0925-4773(98)00015-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel spreadsheet of CRY list.
Excel spreadsheet of RAG list.
Excel spreadsheet of primers used.