Abstract
Objective
Metastasis is the most common cause of death of prostate cancer patients. Identification of specific metastasis biomarkers and novel therapeutic targets is considered essential for improved prognosis and management of the disease. MicroRNAs (miRNAs) form a class of non-coding small RNA molecules considered to be key regulators of gene expression. Their dysregulation has been shown to play a role in cancer onset, progression and metastasis, and miRNAs represent a promising new class of cancer biomarkers. The objective of this study was to identify down- and up-regulated miRNAs in prostate cancer that could provide potential biomarkers and/or therapeutic targets for prostate cancer metastasis.
Methods
Next generation sequencing technology was applied to identify differentially expressed miRNAs in a transplantable metastatic versus a non-metastatic prostate cancer xenograft line, both derived from one patient's primary cancer. The xenografts were developed via subrenal capsule grafting of cancer tissue into NOD/SCID mice, a methodology that tends to preserve properties of the original cancers (e.g., tumor heterogeneity, genetic profiles).
Results
Differentially expressed known miRNAs, isomiRs and 36 novel miRNAs were identified. A number of these miRNAs (21/104) have previously been reported to show similar down- or up-regulation in prostate cancers relative to normal prostate tissue, and some of them (e.g., miR-16, miR-34a, miR-126*, miR-145, miR-205) have been linked to prostate cancer metastasis, supporting the validity of the analytical approach.
Conclusions
The use of metastatic and non-metastatic prostate cancer subrenal capsule xenografts derived from one patient's cancer makes it likely that the differentially expressed miRNAs identified in this study include potential biomarkers and/or therapeutic targets for human prostate cancer metastasis.
Introduction
Prostate cancer is the most common cancer in men and the second leading cause of cancer deaths in the United States [1]. While considerable advances have been made in the treatment of localized, organ-confined tumors, prostate cancer is currently incurable once it has progressed to metastasis, and most deaths from this disease are due to metastases that are highly resistant to conventional therapies. Currently, prostate-specific antigen (PSA) is a major serum biomarker used for the detection and monitoring of prostate cancer progression. However, the prognostic value of increased PSA levels is limited, since advanced prostate cancer can be associated with very low or normal PSA values. There is therefore an urgent need for new, more specific biomarkers which can be used to predict cancer progression on their own or in cooperation with a current biomarker such as PSA [2]. Furthermore, novel therapeutic targets associated with prostate cancer metastasis are urgently needed.
MicroRNAs (miRNAs) are small non-coding RNAs (17 to 27 nucleotides) that negatively regulate the expression of target genes by binding to 3′ untranslated regions (UTRs) of mRNAs and inhibiting translation or promoting mRNA degradation [3]. Recent studies have shown dysregulation of miRNAs in human tumors indicating a role for such molecules in cancer pathogenesis, including cancer onset, progression and metastasis [4], [5]. Thus far, only a small number of studies have investigated miRNA expression in prostate cancer, and only a few have dealt with metastasis of this disease. Differences in the expression profiles of miRNAs so far identified may have prognostic value for the various aspects of the disease and a better understanding of the role of miRNAs in the development and progression of prostate cancer is needed [6]. Further research may also lead to identification of new miRNAs that are specifically related to prostate cancer progression and metastasis. Such metastasis-associated miRNAs may serve as metastatic biomarkers and/or new targets for therapy of metastatic disease.
Studies aimed at identifying genetic factors with key roles in prostate cancer metastasis have been impeded by a lack of optimal experimental models. While xenograft models based on established cancer cell lines representing different stages of cancer progression can be useful for identifying mechanisms underlying metastasis, they do not adequately mimic clinical disease [7]. Efforts have therefore focused on use of patients' prostate cancer tissues. However, the typical heterogeneity of such tissues, consisting of both non-metastatic and potentially metastatic subpopulations, makes it difficult to identify factors such as genes that underlie the development of metastasis [8]. Moreover, it is difficult to obtain metastatic prostate cancer tissues from patients for experimental purposes, since they are not routinely or feasibly biopsied or resected from patients, and rapid autopsy programs are extremely expensive and difficult to manage. To overcome the above hurdles, we developed next generation patient-derived prostate cancer xenograft models, that more closely resemble the clinical situation, by using subrenal capsule grafting of patients' cancer tissue into immuno-deficient mice. This methodology favors retention of the properties of the original cancers [9]–[11]. Furthermore, it has been possible to establish transplantable, metastatic and non-metastatic prostate cancer sublines from heterogeneous xenografts [12], [13]. Use of metastatic and non-metastatic xenografts has already been effective in the identification of prostate cancer metastasis-associated genes [13].
Illumina's massively parallel DNA sequencing by synthesis technology is a widely-adopted next-generation sequencing platform. It supports parallel sequencing using a proprietary reversible terminator-based method that enables detection of single bases as they are incorporated into growing DNA strands. A fluorescently-labeled terminator is imaged as each dNTP is added and then cleaved to allow incorporation of the next base. Since all four reversible terminator-bound dNTPs are present during each sequencing cycle, natural competition minimizes incorporation bias, leading to true base-by-base sequencing [14].
In the present study, Illumina next generation sequencing technology was utilized to compare the miRNA profiles of a transplantable metastatic versus a non-metastatic prostate cancer xenograft line, both derived via subrenal capsule grafting [10]–[12] from one patient's primary cancer tissue. Differentially expressed known and novel miRNAs were found that may have specific roles in the metastasis of prostate cancer.
Materials and Methods
Patient-derived prostate cancer xenograft models
NOD/SCID mice used for xenografting were bred and maintained at the British Columbia Cancer Research Centre Animal Facility (Vancouver, Canada). All experimental protocols were approved by the University of British Columbia Animal Care Committee (A10-0100). A prostate cancer biopsy specimen was obtained at the BC Cancer Agency with the patient's written informed consent. Ethical approval was provided by the University of British Columbia - British Columbia Cancer Agency Research Ethics Board (UBC BCCA REB #H04-60131).
The establishment of transplantable prostate cancer tissue xenograft lines via subrenal capsule grafting has been described previously [9]. In the present study, a recently prepared metastatic prostate cancer xenograft line, LTL-313H [15], and a non-metastatic counterpart, LTL-313B (unpublished), were used that had been derived from different loci of one patient's prostate cancer biopsy sample (www.livingtumorlab.com). Both lines were PSA- and AR-positive as shown via immunohistochemistry ([15]; unpublished data). They were routinely maintained under renal capsules of male NOD/SCID mice supplemented with testosterone, as previously described [9]. The LTL-313H xenografts showed invasion of the mouse host kidney and cancer cells were detected in the lungs of the hosts after 3 months of grafting. In contrast, the LTL-313B xenografts showed no obvious invasion of the mouse kidney and did not show any distant metastases (data not shown).
Small RNA library construction and cDNA sequencing
LTL-313H and LTL-313B xenograft tissues were collected and RNA was extracted using TRIzol (Invitrogen, Mississauga, ON, Canada) according to the manufacturer's instructions. The RNA was submitted to the Genome Sciences Centre at the British Columbia Cancer Agency (www.bcgsc.bc.ca) for small-RNA cDNA library construction and sequencing as previously described [16] with minor modifications. Each library had a specific index sequence in its 5′ adaptor, i.e. “ACATCGA” for the LTL-313H library and “CGTGATA” for the LTL-313B library; both libraries were mixed and the sequencing was run in one flow cell in the Illumina's platform.
Small RNA mapping and differential expression detection
The 5′ indexed cDNA sequences were used to distinguish the origin of the RNAs. 3′ Adaptor sequences were removed from all reads and those remaining tags that were 16 to 27 nucleotides in length and expressed at a tag count of 2 or more in each library were used for further analysis. The trimmed sequences were mapped to miRBase 15 human stem-loop sequences (http://www.mirbase.org/) using the Novoalign (www.novocraft.com) program allowing up to 3 mismatches. Those that matched an miRBase sequence were then grouped as: 1) known mature miRNA and miRNA*, 2) putative miRNA*, not previously reported in the miRBase, 3) loop sequences and 4) sequences which matched the loop-sequence but did not have known mature sequences. The sequences matching known miRNAs were further clustered, based on their starting positions, and counted. The most abundant variation/starting position tags were used for comparison between libraries. Tag counts were normalized to the total counts of those sequences which matched the miRBase 15 stem-loop sequences and the two libraries were compared for differential expression of the sequences using the Fisher's exact test with Bonferroni's correction. Sequences were deemed significantly differentially expressed by the two libraries if the p-value was <0.001 and there was at least a 2–fold change in the sequences' normalized counts.
Novel miRNA identification
The sequences that did not match known miRNA stem-loop sequences were filtered out with known transcripts sequences downloaded from UCSC [17]. To identify novel miRNA candidates amongst the remaining unmatched sequences, the miRanalyzer program [18] was used (http://web.bioinformatics.cicbiogune.es/microRNA/miRanalyser.php). The output candidates were checked one by one for homologies to known non-coding RNA, and non-homologous sequences were taken as novel miRNA candidates.
miRNA target prediction and pathway analysis
The target genes for each differentially expressed miRNA were predicted using MicroCosm version 5 [19], [20] with a threshold of p = 0.001. As some of the genes were potentially regulated by both up-regulated and down-regulated miRNAs, we focused for further analysis on the genes that were potentially regulated only by up-regulated or down-regulated miRNAs. Identification of KEGG pathways associated with potential target genes was carried out using DAVID 6.7 (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/). In addition, we compared the target gene lists with gene expression data by microarray assay using the same xenograft tissues to identify those putative targets that might be regulated at the mRNA level.
Microarray gene expression analysis
The RNA that was used for the miRNA sequencing library was also used for mRNA-based gene expression analysis using the Agilent's Human GE 44K platform at the Vancouver Prostate Centre Microarray Facility (www.mafpc.ca). All data are MIAME compliant and the raw data have been deposited in GEO (accession number GSE28029). The expression signal was transformed to z-score and calculated z-ratio and the mRNAs with more than 1.96 of z-ratio were dealt up-regulated and less than −1.96 as down-regulated [21]. The data were also filtered by Flag.
Cell culturing
The 22Rv1 prostate cancer cell line was cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere containing 5% CO2.
miRNA precursor transfection
The precursor sequence of mir-486, shown in the miRBase, and a non-silencing negative control were subcloned into the pcDNA6.2-GW/EmGFP miR plasmid (Invitrogen). 22Rv1 cells were seeded in 12 well-plates at a density of 1.0×105 cells per well, 24 hours in advance of transfection. Transfection was carried out using Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. The transfection efficiency was validated by GFP signal monitoring using an inverted fluorescence microscope system (Zeiss). To confirm increased levels of mature target miRNA, a portion of the transfected cells was used for validation by quantitative (qPCR). To this end, total RNA was extracted using a miRNeasy mini kit (Qiagen) and the quantity of RNA determined by nanodrop spectrophotometry (Thermo Scientific). Portions (25 ng) of each of the total RNA preparations were reverse-transcribed to cDNA using a Universal cDNA Synthesis Kit (Exiqon) following the manufacturer's instructions. The cDNA was diluted and mixed with microRNA LNA PCR primers and SYBR Green master mix (Exiqon). qPCR was carried out using an ABIPrism 7900HT (Applied Biosystems) following the manufacturer's instructions. The ΔΔCT was used for calculating the fold changes relative to the control and U6 was used as an endogenous control.
MTT assay
The transfected cells were seeded in 96-well plates at a density of 1×104 cells/well. MTT solution (20 µl of 5 mg/ml) was added to the cultures (200 µl volumes) for a 4 hr incubation at 37°C. Following removal of the culture medium, the remaining crystals were dissolved in DMSO and absorbance at 570 nm was measured.
Migration/invasion assay
BioCoat Matrigel invasion chambers (BD Biosciences) were used to measure tissue invasiveness of cells. In the upper chambers, 1×105 cells/well were plated in 0.50 ml of serum-free medium. In the lower chambers, 0.75 ml of medium/10% FBS was delivered. The chambers were incubated for 30 hr at 37°C in a humidified atmosphere with 5% CO2. The cells that remained in the upper chamber were removed and the transmigrated cells fixed in methanol and stained with crystal violet and stained cells were counted by microscopic analysis. Tumor cell invasion was expressed as the percentage of cells that had passed through the Matrigel-coated membranes relative to the number of cells that had passed through the uncoated membranes (invasion index). All assays were performed in triplicate.
Results
miRNA sequencing and annotation
Small RNAs were isolated from metastatic LTL-313H and non-metastatic LTL-313B prostate cancer tissue xenografts and processed to allow deep sequencing using the Illumina's platform. The reads with adaptor index sequences “ACATCGA” and “CGTGATA” were given LTL-313H origin and LTL-313B origin, respectively. More than 10 million total reads were obtained for each of the libraries. These reads were compared with the sequence data in the miRBase 15 microRNA Sequence Database. For the metastatic and non-metastatic prostate cancer tissue libraries, 3,445,642 and 2,272,677 tags, respectively, were fully mapped to human miRNA stem-loop sequences present in the miRBase database (Table 1). The completely matched reads were annotated, according to their position in the stem-loop structure. A shift of up to 2 bases in the starting and ending positions was allowed for sequences to be annotated as isomers of known mature miRNAs (isomiRs). The total numbers of known miRNAs plus miRNA*s in the metastatic and non-metastatic libraries were 447 and 509, respectively (Table 1). The most highly expressed miRNA (and isomiR) was the miR-148a with total counts of 270,801 and 763,877 reads per metastatic and non-metastatic libraries, respectively. When the isomiRs were grouped using the same starting position, the miR-148a remained the most abundant miRNA in the non-metastatic library with a total count of 846,468, whereas in the metastatic library miR-21 was most abundant with a total count of 310,102.
Table 1. Small-RNA library sequencing summary.
| Metastatic library | Non-metastatic library | |
| index tag sequence | ACATCGA | CGTGATA |
| total cDNA reads | 10,525,988 | 11,644,175 |
| reads mapped to miRNA miRBase stem-loop sequences | 2,272,677 | 3,445,642 |
| unique sequences | 1,875,353 | 2,941,722 |
| most abundant miRNA | miR-148a (270,801) | miR-148a (763,877) |
| most abundant miRNA grouped at starting position, total | miR-21 (310,102) | miR-148a (846,468) |
| total known miRNA plus miRNA* | 447 | 509 |
miRNA and miRNA* expressions
In the miRBase, miRNAs derived from a precursor are designated miRNA, the predominantly expressed arm, and miRNA* the less-expressed, opposite arm. If both arms are similarly expressed, they are referred to as 5p and 3p arms. The expression of known miRNAs in the prostate cancer xenografts was in general higher than that of miRNA*s. In a number of cases, miRNA*s (as identified by miRBase) were more expressed than the corresponding miRNAs (Table 2). Thus miR-144* was substantially more expressed than miR-144 in both metastatic and non-metastatic libraries; similarly miR-126* was more expressed than miR-126, but only in the non-metastatic library. Differences were also found in the expression patterns of 3p and 5p arm miRNAs (Table 2). The 3p arms of miR-28 and miR-339 showed higher expressions than the corresponding 5p arms in the metastatic line, whereas they showed lower expressions than the corresponding 5p arms in the non-metastatic line. In the metastatic line, the miR-542 showed up-regulation of the 3p arm but down-regulation of the 5p arm. Fragments that were counterparts of known mature miRNAs, but that had not previously been reported to the miRBase database, were designated “putative novel miRNA*” species (Table 3). A total of 32 of such putative miRNA*s was observed showing at least two reads in one of the two libraries. Some of these miRNA*s also showed higher expression than their corresponding miRNAs, e.g., miR-1277* and miR-1307*.
Table 2. Levelsa of miRNA and miRNA* showing dysregulation in (i) miRNA∶miRNA* ratios in metastatic and non-metastatic xenografts and (ii) the levels of miRNA or miRNA* in the two xenograft lines.
| Expression level | |||
| miRNA | metastatic | non-metastatic | fold change |
| miR-7 | 72.35 | 22.5 | 3.22 |
| miR-7-1* | 2.76 | 24.54 | −8.88 |
| miR-126 | 26005.24 | 1799.7 | 14.45 |
| miR-126* | 1099.03 | 3798.06 | −3.46 |
| miR-144 | 915.16 | 134.98 | 6.78 |
| miR-144* | 5429.24 | 418.37 | 12.98 |
| miR-335 | 22.58 | 54.63 | −2.42 |
| miR-335* | 65.9 | 91.74 | 1.39 |
| miR-374a | 1638.17 | 674.3 | 2.43 |
| miR-374a* | 954.79 | 825.93 | 1.16 |
| miR-28-3p | 317.5 | 188.15 | 1.69 |
| miR-28-5p | 184.78 | 1264.17 | −6.84 |
| miR-339-3p | 65.43 | 102.55 | −1.57 |
| miR-339-5p | 35.94 | 189.03 | −5.26 |
| miR-542-3p | 100 | 26.88 | 3.72 |
| miR-542-5p | 0.92 | 12.27 | −13.31 |
Tag counts were normalized to the total counts of sequences that matched the miRBase 15 stem-loop sequences and are expressed as number per 1,000,000 tags.
Table 3. Putative miRNA*s, i.e. fragments complementary to known miRNAs but not reported in the miRBase.
| Expression levels | ||||||
| putative miRNA* sequence | matched to miRNA stem-loop | starting position | putative miRNA* | miRNA | ||
| metastatic | non-metastatic | metastatic | non-metastatic | |||
| CTGTACAACCTTCTAGCTTTCC | hsa-let-7c† | 56 | 4.15 | 1.17 | 2039.08 | 1584.38 |
| TCAATAAATGTCTGTTGAAT | hsa-mir-95 | 15 | 0 | 0.88 | 44.70 | 973.18 |
| TATACAACTTACTACTTTCC | hsa-mir-98 | 81 | 0 | 0.58 | 105.99 | 79.47 |
| CGGGGCCGTAGCACTGTCTGA | hsa-mir-128-1 | 15 | 0.92 | 0 | 42.39 | 5.26 |
| ATGTAGGGATGGAAGCCATGA | hsa-mir-135a-2 | 61 | 1.38 | 1.75 | 379.25 | 588.99 |
| TGGAAACATTTCTGCACAAACT | hsa-mir-147b | 12 | 0 | 1.17 | 1.38 | 0.29 |
| GTCATTTTTGTGATGTTGCAG | hsa-mir-153-2 | 14 | 12.44 | 1.46 | 219.34 | 554.81 |
| AGTGGTTCTTAACAGTTCAACA | hsa-mir-203 | 27 | 0 | 6.14 | 116.12 | 296.54 |
| AGCCCCTGCCCACCGCACACTG | hsa-mir-210 | 28 | 1.38 | 1.17 | 13967.09 | 302.97 |
| GCTCTGACGAGGTTGCACTACT | hsa-mir-301b | 10 | 1.38 | 0.88 | 4.15 | 3.21 |
| GTTCCTGCTGAACTGAGCCAGT | hsa-mir-3074 | 12 | 0 | 0.58 | 0.46 | 0.00 |
| AGGGACTTTTGGGGGCAGATGTG | hsa-mir-365-1 | 16 | 1.38 | 1.17 | 16.59 | 22.50 |
| GCGACGAGCCCCTCGCACAAACC | hsa-mir-375 | 5 | 0 | 0.88 | 6414.45 | 11574.16 |
| TGTGTTGCATGTGTGTATATGT | hsa-mir-466 | 14 | 0.92 | 0 | 0 | 0 |
| CAAAAGCAATCGCGGTTTTTGC | hsa-mir-548e | 15 | 0 | 3.80 | 0.46 | 0 |
| CAAAAACTGCAATTACTTTTGC | hsa-mir-548h-3 | 65 | 0 | 0.58 | 0 | 0 |
| TTTGGTGCATATTTACTTTAGG | hsa-mir-559 | 57 | 2.76 | 0.58 | 0 | 0 |
| ATCAAGGATCTTAAACTTTGCC | hsa-mir-561 | 26 | 3.23 | 1.46 | 0 | 0.88 |
| TCGCGGTTTGTGCCAGATGAC | hsa-mir-579 | 25 | 0.92 | 0 | 0.46 | 2.34 |
| ACAACCCTAGGAGAGGGTGCCATT | hsa-mir-652 | 20 | 0 | 1.17 | 11.06 | 16.65 |
| GGACCTTCCCTGAACCAAGGA | hsa-mir-659 | 23 | 2.76 | 1.17 | 0 | 0 |
| ACCTCCTGTGTGCATGGATT | hsa-mir-660 | 52 | 0 | 1.17 | 32.26 | 68.37 |
| CGGCCCCACGCACCAGGGTAAG | hsa-mir-874 | 10 | 0.92 | 0 | 3.23 | 2.05 |
| CGGGAACGTCGAGACTGGAGC | hsa-mir-1247 | 77 | 0.92 | 0.58 | 0 | 0.29 |
| CTATCTTCTTTGCTCATCCTTG | hsa-mir-1255a | 69 | 0 | 1.17 | 2.76 | 1.75 |
| AGTTGGCATGGCTCAGTCCAAGT | hsa-mir-1269 | 25 | 0 | 5.55 | 9.22 | 7.89 |
| TATATATATATATGTACGTATG | hsa-mir-1277 | 11 | 16.59 | 46.45 | 0 | 2.05 |
| ATCTCACTTTGTTGCCCAGG | hsa-mir-1285-1 | 13 | 0.92 | 0 | 0 | 2.05 |
| CTCTAGCCACAGATGCAGTGAT | hsa-mir-1287 | 55 | 0 | 2.05 | 0.92 | 7.89 |
| GAGTGGGGCTTCGACCCTAACC | hsa-mir-1296 | 59 | 0 | 0.58 | 10.60 | 12.27 |
| CCACCTCCCCTGCAAACGTC | hsa-mir-1306 | 15 | 1.38 | 1.75 | 0 | 0.58 |
| TCGACCGGACCTCGACCGGCTCG | hsa-mir-1307 | 41 | 82.48 | 126.50 | 11.98 | 26.59 |
putative let-7c* sequence is not identical to the let-7c* in the miRBase but shows complementarity to let-7c.
Novel miRNA candidates
One advantage of utilizing a sequencing approach for miRNA profiling is the opportunity to identify novel miRNAs or miRNA*s. To this end, we used an miRanalyzer, a microRNA detection and analysis tool [18], in combination with homology searches to identify known transcripts, including non-coding RNAs (e.g., rRNA, tRNA, etc.). Using miPred software to distinguish real pre-miRNAs from other hairpin sequences with similar stem-loops [22], we identified 36 novel miRNA candidates. Their sequences, chromosome locations and number of reads in the metastatic and non-metastatic libraries are presented in Table 4 and Table S1. Comparative analysis of the two libraries showed significant differential expression of some of these novel miRNAs, including down-regulated miR-5680-3p and miR-5681a-3p.
Table 4. Novel miRNA candidates.
| reads in | ||||
| ID | Sequence | chromosomal location | metastatic | non-metastatic |
| hsa-miR-5680-3p | GAGAAATGCTGGACTAATCTGC | 8q22.3 | 28 | 318 |
| hsa-miR-5681a-3p | AGAAAGGGTGGCAATACCTCTT | 8q21.11 | 4 | 66 |
| hsa-miR-5682-3p | GTAGCACCTTGCAGGATAAGGT | 3q13.33 | 2 | 29 |
| hsa-miR-548aw-5p | GTGCAAAAGTCATCACGGTT | 9q34.13 | 26 | |
| hsa-miR-5683-5p | TACAGATGCAGATTCTCTGACTTC | 6p25.1 | 25 | 22 |
| hsa-miR-5684-5p | AACTCTAGCCTGAGCAACAG | 19p13.13 | 2 | 2 |
| hsa-miR-548ax-5p | AGAAGTAATTGCGGTTTTGCCA | Xp22.2 | 12 | |
| hsa-mir-5685-5p | ACAGCCCAGCAGTTATCACGGG | 6p12.1 | 9 | |
| hsa-miR-5692c-3p | AATAATATCACAGTAGGTGTAC | 5q31.1 | 8 | |
| 7q21.3 | ||||
| hsa-miR-5686-5p | TATCGTATCGTATTGTATTGT | 10q24.1 | 8 | |
| hsa-miR-5687-3p | TTAGAACGTTTTAGGGTCAAAT | 5q11.2 | 6 | |
| hsa-miR-5688-3p | TAACAAACACCTGTAAAACAGC | 3p12.1 | 5 | |
| hsa-miR-5681b-5p | AGGTATTGCCACCCTTTCTAGT | 8q21.11 | 4 | |
| hsa-miR-548at-5p | AAAAGTTATTGCGGTTTTGGC | 17q21.31 | 4 | |
| hsa-miR-5689-5p | AGCATACACCTGTAGTCCTAGA | 6p24.3 | 4 | |
| hsa-miR-5690-5p | TCAGCTACTACCTCTATTAGG | 6p21.31 | 3 | |
| hsa-miR-5691-5p | TTGCTCTGAGCTCCGAGAAAGC | 11p15.4 | 3 | |
| hsa-miR-5692a-5p | CAAATAATACCACAGTGGGTGT | 7q21.3 | 3 | |
| 8p23.1 | ||||
| hsa-miR-4666b-5p | TTGCATGTCAGATTGTAATTCCC | Xp21.2 | 3 | |
| hsa-miR-5693-3p | GCAGTGGCTCTGAAATGAACTC | 13q14.3 | 2 | |
| hsa-miR-5694-5p | CAGATCATGGGACTGTCTCAG | 14q23.3 | 2 | |
| hsa-miR-5695-3p | ACTCCAAGAAGAATCTAGACAG | 19p13.13 | 2 | |
| hsa-miR-5696-5p | CTCATTTAAGTAGTCTGATGCC | 2q11.2 | 2 | |
| hsa-miR-5697-5p | TCAAGTAGTTTCATGATAAAGG | 1p36.22 | 2 | |
| hsa-miR-5698-5p | TGGGGGAGTGCAGTGATTGTGG | 1q21.3 | 2 | |
| hsa-miR-5699-3p | TCCTGTCTTTCCTTGTTGGAGC | 10p15.3 | 2 | |
| hsa-miR-5700-5p | TAATGCATTAAATTATTGAAGG | 12q22 | 2 | |
| hsa-miR-5701-5p | TTATTGTCACGTTCTGATT | 15q11.2 | 2 | |
| hsa-miR-5702-3p | TGAGTCAGCAACATATCCCATG | 2q36.3 | 2 | |
| hsa-miR-5703-3p | AGGAGAAGTCGGGAAGGT | 2q36.3 | 2 | |
| hsa-miR-5692b-5p | AATAATATCACAGTAGGTGT | 21q22.3 | 2 | |
| hsa-miR-5704-5p | TTAGGCCATCATCCCATTATGC | 3q22.1 | 2 | |
| hsa-miR-5705-3p | TGTTTCGGGGCTCATGGCCTGTG | 4q22.1 | 2 | |
| hsa-miR-5706-5p | TTCTGGATAACATGCTGAAGCT | 5q23.1 | 2 | |
| hsa-miR-5707-5p | ACGTTTGAATGCTGTACAAGGC | 7q36.3 | 2 | |
| hsa-miR-5708-5p | ATGAGCGACTGTGCCTGACC | 8q21.13 | 2 | |
Potential metastasis-associated miRNAs
Comparative analysis of the metastatic and non-metastatic xenograft miRNA libraries revealed a total of 104 differentially expressed miRNAs or miRNA*s with 55 down-regulated and 49 up-regulated in the metastatic line (Table 5,6,7). Of the down-regulated miRNAs, 24 miRNAs showed a >5-fold decrease, including four miRNAs, i.e. miR-205, miR-503, miR-708 and miR-2115*, which were undetectable in the metastatic line. Two miRNAs, i.e. miR-24-2* and miR-101*, showed increased expression in a one-base-shift form. A one-base-shift form of miR-203 showed some increased expression in the metastatic line relative to reference miR-203, whereas in the non-metastatic line it showed a lower expression. Of the up-regulated miRNAs, 23 miRNAs showed a >5-fold change in normalized counts. One-base-shift forms of miR-9*, miR-148b* and miR-1246 showed higher expression than the reference forms in both metastatic and non-metastatic lines.
Table 5. miRNAs down-regulated in the metastatic library§.
| references | ||||||
| prostate cancer | metastasis in | |||||
| miRNA | metastatic | non-metastatic | fold change | prostate cancer | other types of cancer | |
| miR-7-1* | 2.76 | 24.54 | −8.88 | |||
| miR-15b | 489.84 | 2660.69 | −5.43 | [28] # | ||
| miR-16 | 999.03 | 17741.34 | −17.76 | [23]–[25], [38] # | [25] | |
| miR-24 | 1046.96 | 8903.54 | −8.50 | [26]–[28], [76] # | [61] | |
| miR-24-2*† | 5.07 | 22.50 | −4.44 | |||
| miR-26b | 1964.89 | 8991.18 | −4.58 | [28] # | ||
| miR-28-5p | 184.78 | 1264.17 | −6.84 | |||
| miR-29a | 446.06 | 13028.23 | −29.21 | [26], [28], [29] | ||
| miR-29c | 299.99 | 1643.68 | −5.48 | |||
| miR-33b | 22.12 | 62.81 | −2.84 | |||
| miR-34a | 76.03 | 194.87 | −2.56 | [77], [26] # | [33] | [78] |
| miR-95 | 57.60 | 1128.90 | −19.60 | [26] # | ||
| miR-101*† | 4.61 | 105.76 | −22.95 | |||
| miR-106b | 906.41 | 6545.23 | −7.22 | [30] # | ||
| miR-126* | 1099.03 | 3798.06 | −3.46 | [28] # | [34] | |
| miR-145 | 103.68 | 297.71 | −2.87 | [23], [24], [27], [29], [30] | [35] | [61] |
| miR-146b-5p | 1335.88 | 2765.57 | −2.07 | [26] # | [79], [61] # | |
| miR-185 | 84.79 | 448.76 | −5.29 | [60] | ||
| miR-186 | 1666.28 | 5838.21 | −3.50 | [61] # | ||
| miR-188-5p | 0.92 | 9.64 | −10.46 | |||
| miR-191 | 1822.50 | 12911.66 | −7.08 | [26] # | ||
| miR-193a-3p | 35.94 | 196.91 | −5.48 | |||
| miR-195 | 116.58 | 560.65 | −4.81 | [23], [26] # | ||
| miR-196a | 33.64 | 70.99 | −2.11 | [26] # | ||
| miR-200b* | 146.54 | 499.30 | −3.41 | |||
| miR-200c* | 12.44 | 54.34 | −4.37 | |||
| miR-203 | 147.92 | 434.73 | −2.94 | [26] # | [61] | |
| miR-203‡ | 224.41 | 334.81 | −1.49 | [26] # | ||
| miR-205 | 0.00 | 31.26 | N/A | [24], [31], [32] | [36] | |
most abundant miRNA started one base upstream from the mature form of miRBase.
most abundant miRNA started one base downstream from the mature form of miRBase.
Differential expressions all had a >2-fold change and a p<0.001 and are considered statistically significant.
Results from the literature that do not match the down- or up-regulation found in the present study.
Table 6. miRNAs down-regulated in the metastatic library§(Continued from Table 5).
| references | ||||||
| prostate cancer | metastasis in | |||||
| miRNA | metastatic | non-metastatic | fold change | prostate cancer | other types of cancer | |
| miR-324-5p | 27.19 | 61.94 | −2.28 | |||
| miR-331-3p | 2.76 | 14.32 | −5.18 | [80] | ||
| miR-335 | 22.58 | 54.63 | −2.42 | [62] | ||
| miR-339-5p | 35.94 | 189.03 | −5.26 | |||
| miR-342-3p | 61.75 | 247.46 | −4.01 | |||
| miR-361-5p | 103.68 | 259.73 | −2.51 | |||
| miR-363 | 1298.10 | 8476.40 | −6.53 | |||
| miR-424 | 22.12 | 76.25 | −3.45 | [28] | ||
| miR-425 | 1869.04 | 10985.75 | −5.88 | |||
| miR-454 | 10.60 | 32.72 | −3.09 | |||
| miR-497 | 164.05 | 373.67 | −2.28 | [23] | ||
| miR-503 | 0.00 | 7.30 | N/A | [23] # | ||
| miR-542-5p | 0.92 | 12.27 | −13.31 | |||
| miR-556-5p | 1.84 | 13.15 | −7.13 | [26] # | ||
| miR-582-5p | 49.77 | 203.93 | −4.10 | |||
| miR-590-5p | 12.44 | 213.86 | −17.19 | |||
| miR-627 | 2.30 | 93.20 | −40.45 | |||
| miR-651 | 11.06 | 54.63 | −4.94 | |||
| miR-652 | 12.90 | 33.89 | −2.63 | |||
| miR-660 | 51.15 | 110.14 | −2.15 | |||
| miR-664 | 8.76 | 30.97 | −3.54 | |||
| miR-708 | 0.00 | 42.36 | N/A | |||
| miR-1180 | 3.69 | 15.48 | −4.20 | |||
| miR-1269 | 11.06 | 42.66 | −3.86 | |||
| miR-1287 | 1.84 | 12.85 | −6.97 | |||
| miR-2115* | 0.00 | 7.89 | N/A | |||
| miR-3065-5p | 14.75 | 117.74 | −7.98 | |||
Differential expressions all had a >2-fold change and a p<0.001 and are considered statistically significant.
Results from the literature that do not match the down- or up-regulation found in the present study.
Table 7. miRNAs up-regulated in the metastatic library§.
| references | ||||||
| prostate cancer | metastasis in | |||||
| miRNA | metastatic | non-metastatic | fold change | prostate cancer | other types of cancer | |
| let-7d | 671.86 | 322.84 | 2.08 | [26] | ||
| let-7g | 8690.38 | 3448.64 | 2.52 | [23] # | ||
| let-7g* | 42.39 | 3.21 | 13.19 | |||
| let-7i | 851.11 | 400.55 | 2.12 | [26] | [61] | |
| miR-7 | 72.35 | 22.50 | 3.22 | |||
| miR-9 | 20121.64 | 1386.00 | 14.52 | [65],[61] # | ||
| miR-9*‡ | 352.98 | 35.64 | 9.90 | |||
| miR-17 | 12228.92 | 5600.39 | 2.18 | |||
| miR-18a | 1023.45 | 142.57 | 7.18 | |||
| miR-18b | 90.78 | 26.59 | 3.41 | |||
| miR-20b* | 89.40 | 7.60 | 11.77 | |||
| miR-27a | 1568.59 | 721.34 | 2.17 | [26], [28] # | ||
| miR-27b | 1406.39 | 425.38 | 3.31 | [28] # | ||
| miR-30a | 30144.22 | 10956.83 | 2.75 | [61] # | ||
| miR-30a* | 1300.40 | 532.90 | 2.44 | |||
| miR-31 | 80.64 | 3.80 | 21.23 | [30], [24] # | [67], [81] | |
| miR-34c-5p | 2190.22 | 51.13 | 42.84 | |||
| miR-99a | 1017.92 | 330.43 | 3.08 | [23] # | ||
| miR-106a | 2790.65 | 845.51 | 3.30 | [26],[28] [38] # | ||
| miR-125b | 1699.46 | 287.19 | 5.92 | [38]–[40], [23], [24], [27], [29] # | [61] | |
| miR-125b-2* | 15.67 | 1.46 | 10.73 | [57] | ||
| miR-126 | 26005.24 | 1799.70 | 14.45 | [54] # | ||
| miR-128 | 81.10 | 15.48 | 5.24 | [26] # | ||
| miR-136 | 40.55 | 7.89 | 5.14 | |||
| miR-138 | 38.25 | 1.46 | 26.18 | |||
| miR-140-5p | 2458.87 | 723.97 | 3.40 | |||
| miR-142-5p | 104.60 | 26.00 | 4.02 | [61] | ||
| miR-144 | 915.16 | 134.98 | 6.78 | |||
| miR-144* | 5429.24 | 418.37 | 12.98 | |||
| miR-148b*† | 258.97 | 111.31 | 2.33 | |||
| miR-151-3p | 4377.22 | 1866.89 | 2.34 | [66] | ||
| miR-152 | 2662.55 | 582.56 | 4.57 | [28] # | ||
| miR-181a-2* | 19.81 | 0.88 | 22.61 | |||
| miR-200a | 7217.64 | 1066.09 | 6.77 | [58] # | ||
| miR-210 | 16686.78 | 347.38 | 48.04 | [23] | ||
| miR-218 | 140.55 | 70.12 | 2.00 | [25],[26] # | ||
| miR-223* | 16.13 | 0.29 | 55.20 | |||
| miR-301a | 60.83 | 21.62 | 2.81 | [37] | ||
| miR-340* | 46.08 | 16.36 | 2.82 | |||
| miR-374a | 1638.17 | 674.30 | 2.43 | |||
| miR-379 | 104.14 | 35.94 | 2.90 | |||
| miR-449a | 18.43 | 1.17 | 15.77 | [59] # | ||
| miR-450a | 191.24 | 24.83 | 7.70 | [61] | ||
| miR-451 | 4504.40 | 2197.62 | 2.05 | |||
| miR-486-3p | 9.22 | 0.88 | 10.52 | |||
| miR-486-5p | 430.86 | 54.93 | 7.84 | |||
| miR-542-3p | 100.00 | 26.88 | 3.72 | |||
| miR-744* | 17.05 | 3.80 | 4.49 | |||
| miR-1246‡ | 95.39 | 22.50 | 4.24 | |||
most abundant miRNA started one base upstream from the mature form of miRBase.
most abundant miRNA started one base downstream from the mature form of miRBase.
Differential expressions all had a >2-fold change and a p<0.001 and are considered statistically significant.
Results from the literature that do not match the down- or up-regulation found in the present study.
Some of the differentially expressed miRNAs have previously been associated with prostate cancer, prostate cancer metastasis or metastasis of other types of cancer (Table 5,6,7). Of the down-regulated miRNAs a number have been reported to be down-regulated in prostate cancer relative to benign prostate tissues, i.e. miR-16 [23]–[25], miR-24 [26]–[28], miR-29a [26], miR-145 [23], [24], [27], [29], [30], and miR-205 [24], [31], [32]. The down-regulation of miR-16 [25], miR-34a [33], miR-126* [34], miR-145 [35] and miR-205 [36] correlated with the development of prostate cancer metastasis. Of the up-regulated miRNAs (in the metastatic library), miR-210 has been reported to be up-regulated in prostate carcinomas relative to BPH samples [23] and miR-301 has been linked to prostate cancer metastasis [37]. In some cases, miRNAs that were found to be up-regulated in the present study have been reported to be either up-regulated in prostate carcinomas compared to normal prostate tissue [38]–[40], or down-regulated [23], [24], [27]–[29]. Furthermore, some of the differentially expressed miRNAs have been reported to play a role in the metastasis of other types of cancer, for example, the up-regulated miRNAs, let-7i, miR-9, miR-30a, miR-125b, miR-142-5p, miR-151-3p, miR-450a and the down-regulated miRNAs, miR-24, mir-145, miR-146b-5p, miR-185, miR-186, miR-203 and miR-335.
Putative target genes for differentially expressed miRNAs
As a first step in the identification of miRNAs with potential significance in the metastatic process, we identified putative target genes for each of the differentially expressed miRNAs using Microcosm analysis, a target prediction program with a specific algorithm and coverage of miRNA, including varieties in star arms; a threshold p-value = 0.001 was maintained to get more reliable target identification (Microcosm) [41]. Putative target genes were identified for 49 out of 55 down-regulated miRNAs and for 47 out of 49 up-regulated miRNAs (Table S2); they were annotated using the DAVID program. The putative target genes of the down-regulated miRNAs were associated with a variety of KEGG pathways including “Fc gamma R-mediated phagocytosis” and “ECM-receptor interaction (Table S3). For the putative target genes of the up-regulated miRNAs, pathways such as “Pathways in cancer”, “Focal adhesion” and “Purine metabolism” were noted.
Comparison of putative miRNA target genes with genes differentially expressed in the metastatic and non-metastatic xenograft lines
Although miRNAs are thought to alter protein levels, they have in some cases been shown to also affect mRNA levels [3]. In view of this, the two xenograft lines were examined for differential mRNA expression. As shown in the Table S4, 622 mRNAs were down-regulated and 348 mRNAs were up-regulated in the metastatic line. Some of the genes identified by differential mRNA expression were potentially targeted by both up- and down-regulated miRNAs. In view of this, further analysis was restricted to genes that were potentially targeted by either up-regulated or down-regulated miRNAs. The group that showed up-regulated mRNAs associated with only down-regulated miRNAs consisted of 85 mRNAs; the group that showed down-regulated mRNAs associated with only up-regulated miRNAs consisted of 58 mRNAs. Among the mRNAs up-regulated in the metastatic line, some have been reported to have a role in tissue invasion and/or metastasis of a variety of cancer cells, including mRNA expressed by FSCN1 [42], VEGFA [43] FGFR1 [44], ADAMTS1 [45], CCL2 [46] and VIM [47] genes. Similarly, mRNAs down-regulated in the metastatic line, including mRNAs expressed by the CTGF [48] and SERPINB5 [49] genes, have been found to be down-regulated in various metastatic cancers, attesting to the reliability of our analyses (Table S5).
Increased levels of mature miR-486 in transfected cells
At 24 hours following transfection of 22Rv1 cells with pcDNA6.2-GW/EmGFP-mir486 or pcDNA6.2-GW/EmGFP-control sequence, more than 90% of the cells were found to be GFP positive. The mir-486 precursor had been properly processed to the mature miR-486 form as indicated by qPCR. Relative to the control, the expression levels of both miR-486-5p and -3p arms were 11.6 fold higher, indicating that the majority of the cells expressed elevated miR-486 levels.
Invasiveness of miR-486-transfected 22Rv1 cells
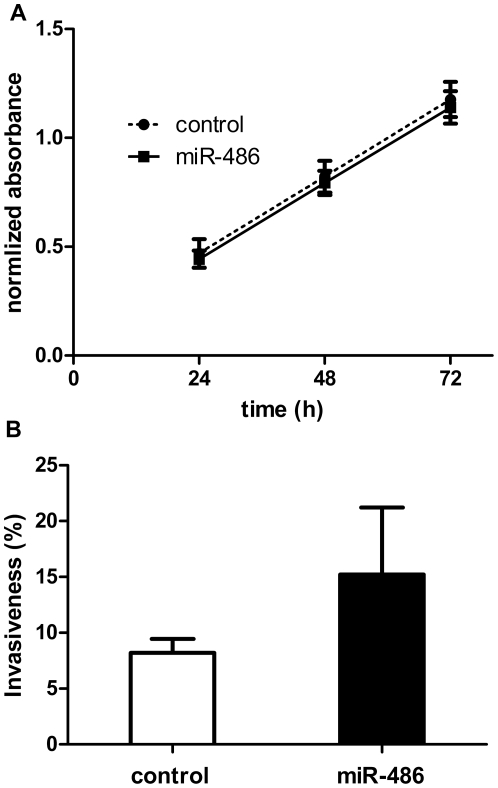
The proliferation rate of miR-486-transfected and control sequence-transfected cells was similar as indicated by the MTT assay (Fig. 1A). However, the miR-486-transfected cells showed an increase of about 85% in tissue invasiveness relative to the control cells (Fig. 1B). Although this result has borderline significance (p = 0.08), it indicates that increased expression of miR-486 enhances tissue invasiveness.
Figure 1. Effects of overexpression of miR-486 on proliferation and tissue invasiveness of 22Rv1 human prostate cancer cells.
A) As indicated by MTT assay, there was no significant difference between the growth of miR-486-transfected cells and control cells over a 72-hr period. B) Tissue invasiveness, as measured by Matrigel invasion, of miR-486 transfected cells and control cells. Data are expressed as percent invasiveness ± S.D. and show increased invasiveness of miR-486-transfected cells (85% ; p = 0.08).
Discussion
MicroRNAs have been implicated in the regulation of gene expression at the post-transcriptional level in almost every biological event, and there is an increasing body of evidence that altered expressions of specific miRNAs are involved in the development and progression of cancers [4], [5]. Using next generation sequencing for small RNA identification, the present study was aimed at identifying differentially expressed known and novel miRNAs in metastatic versus non-metastatic prostate cancer xenografts that could play a role in the progression of prostate cancer to the metastatic form. The transplantable cancer lines that were used appear to be highly suitable for the purpose since they had been derived from one patient's cancer and thus possessed a common genetic background. Furthermore, they had been developed via subrenal capsule grafting of cancer tissue into NOD/SCID mice, a methodology that tends to preserve important properties of the original cancers (e.g., tumor heterogeneity, genetic profiles) [9]–[11], [50]. As well, the maintenance of the tumor lines in the same type of graft site (under the kidney capsule) ensured that their growth was not markedly influenced by micro-environmental differences that can have an important impact on cancer development [51]. Similarly, the same type of graft site would minimize differences in miRNA production by host cells present in the xenografts. Although it has been shown that xenografting can alter the expression of miRNAs [52], our study focused primarily on differences in miRNAs between matched samples and these differences are therefore likely to be real. Taken together, the data obtained in this study should be useful for the delineation of miRNAs with oncogenic properties that are involved in the development of prostate cancer metastasis.
The highest reads in the two RNA libraries were observed for miR-148a. The expression of this miRNA is androgen-inducible in LNCaP cells [53]. This suggests that the relatively high expression of miR-148a found in the two libraries is a result of the testosterone supplementation of the animals.
Of the 104 miRNAs that were found to be down- or up-regulated in the metastatic prostate cancer xenografts, relative to their non-metastatic counterparts, 39 had previously been reported to be involved in prostate cancer (Table 5,6,7). These reports were mostly based on comparisons of miRNA expressions in prostate cancer tissues versus normal prostate tissues without defining the metastatic ability of the malignant samples. It is of interest that 21 of the 39 miRNAs showed down- or up-regulations in the metastatic xenografts which matched those reported for the prostate cancer tissues (relative to benign tissues), suggesting that these prostate cancer tissues may have had metastatic ability. Of the miRNAs found to be down-regulated in the metastatic xenografts, miR-16, showing a >17-fold decrease in expression, has been reported to be down-regulated in prostate cancer [23], [24] and to have a metastasis-suppressing function. Moreover, metastatic prostate tumor growth in vivo could be inhibited by systemic delivery of synthetic miRNA-16 [25]. The reduced expression of miR-34a in the metastatic xenograft line is consistent with its reported inhibition of prostate cancer metastasis [33]. The lower expression of miR-126* is in agreement with reports that this miRNA is down-regulated in prostate cancer metastasis [34] and that ectopic expression of miR-126* inhibited the migration and invasiveness of prostate cancer cells [34]. The latter being an example of a miRNA* playing a role as a tumor suppressor. Interestingly, miR-126, a miRNA reported as down-regulated in prostate cancer relative to normal prostate tissue [54], was up-regulated in the metastatic xenograft line. In this context it is of interest that whereas in LNCaP cells, the antagomir of miR-126 did not affect cell migration, the antagomir of miR-126* induced cell migration with up-regulation of prostein [34], suggesting that miR-126* affects cell migration more than miR-126. This raises the possibility that alternative strand selection as a mechanism for changing the expression of either arm is involved in the development of cancer or cancer metastasis.
The down-regulation of miR-145 in the metastatic xenograft line is in agreement with many reports identifying it as down-regulated in prostate tumors [23], [24], [27], [29], [30]. As well, a role for miR-145 in prostate cancer metastasis is suggested by its down-regulation observed in clinical samples of metastatic prostate cancer relative to localized high grade prostate cancer [35]; furthermore, miR-145 is considered a putative tumor suppressor in colon cancer cells [55] and can reduce breast cancer cell motility [56]. The expression of miR-205 was also reported to be down-regulated in prostate cancer cells, and ectopically expressed miR-205 showed a tumor-suppressive effect, including reduction of cell migration and tissue invasion [36].
Of the miRNAs found to be up-regulated in the metastatic xenografts, miR-125b has been reported to be up-regulated in prostate cancer and shown to be oncogenic [24], [29], [40]; other studies, however, have reported miR-125b to be down-regulated in prostate cancer [23], [24], [27]–[29]. miR-125b-2, a component of the miR-125b cluster, has been identified as part of an androgen receptor-mediated transcriptional network [57]. In a number of single studies, miRNAs such as let-7d [26], let-7i [26] and miR-210 [23] were also found to be up-regulated in prostate cancer, in contrast to let-7g [23], miR-27b [28], miR-99a [23], miR-126 [54], miR-128 [26], miR-152 [28], miR-200a [58] and miR-449a [59] which were down-regulated in prostate cancer samples. Both up- and down-regulation in prostate cancer was reported for a number of miRNAs. The reason for these discrepancies is not clear. Our finding indicating that upregulation of miR-486 is coupled to increased tissue invasiveness, as found with 22Rv1 human prostate cancer cells (Fig. 1b), supports the biological significance of the present study.
It is apparent from the above discussion that a number of the differentially expressed miRNAs identified in this study probably have a significant role in prostate cancer metastasis. Thus some of the miRNAs have already been linked to this phenomenon, in particular down-regulated miRNAs such as miR-16, miR-34a, miR-126*, miR-145 and miR-205, supporting the validity of our analytical approach. However, the prostate cancer xenografts did not show significant differential expression for miRNAs such as miR-221, whose down-regulation in a study using prostate cancer samples from a large number of patients was reported to be a hallmark in human prostate cancer metastasis [37]. This deficiency likely stems from the tumor heterogeneity of prostate cancers and illustrates the need for using a larger number of matched metastatic and non-metastatic xenografts and also clinical samples.
The present study has also identified differentially expressed miRNAs that have not previously been linked to prostate cancer, but to metastasis of other types of cancer (Table 5,6,7). Of the miRNAs down-regulated in the metastatic xenografts, miR-185 has been shown to suppress growth and progression of certain human cancers (e.g., breast, ovary) by targeting the Six1 oncogene which regulates c-myc expression [60]. The miR146b-5p [61] and miR-335 [62] miRNAs have been shown to be metastasis suppressors in breast and colon cancers, facilitating the metastatic phenotype at reduced levels [63]. Of the up-regulated miRNAs in the metastatic line, miR-9 has been reported to target E-cadherin [64] and CDH1, the E-cadherin-encoding messenger RNA [65]; overexpression of miR-9 in non-metastatic breast tumor cells enables such cells to form pulmonary micrometastases in mice [65]. The miR-30a, miR-142-5p and miR-450a have roles in metastatic breast and colon cancer [61] and the miR-151-3p can enhance hepatocellular carcinoma cell mobility [66]. The upregulation of miR-31 is consistent with its ability to induce migration and tissue invasion of colon cancer cells via targeting of T-cell lymphoma invasion and metastasis 1 (TIAM1) [67]. It appears likely that these miRNAs also have a critical role in the development of prostate cancer metastasis on the basis of their role in the metastasis of other cancers, but further validation is needed.
The identification of novel putative miRNAs (Table 4) is of major interest for follow-up studies. In advanced prostate cancer, DNA copy number gain is commonly observed in the chromosome 8q arm [68], and the LTL-313 xenograft lines that were used in the present study also show an 8q arm copy number gain (data not shown). The finding that five of the 46 novel miRNAs were located on chromosome 8q, including the most abundantly expressed candidates (Table 4), suggests that there is a correlation between tissue-specific expression of an miRNA and its DNA copy number.
As found in the present study, some of the miRNA*s (identified by miRBase) are more highly expressed than their corresponding miRNAs (Table 2). This is likely a result of cancer-induced changes in miRNA processing and stability, including strand selection [69]. The latter is thought to normally involve an RNA-Induced Silencing Complex (RISC), a multiprotein complex that can incorporate one strand of an miRNA for subsequent silencing of the complementary mRNA [70]. Switching of a strand should lead to activation/inhibition of a different set of target genes and may underlie oncogenic properties of miRNA*s. There are several reports of miRNA* contributions in cancer progression [34], [71], [72].
The occurrence of miRNA isoforms (isomiRs) in the present study has also been observed in other studies using miRNA deep sequencing [16]. The variability in isomiR expression during Drosophila melanogaster development, generally thought to result from inexact Dicer processing and RNA editing, may not be arbitrary and in fact be regulated and biologically meaningful [73]. At present, the contribution of isomiRs in target recognition is not clear. However, it appears likely that the seed sequence of mature miRNA, i.e. the first 2–8 nucleotides at the 5′ end, is a key of the miRNA's target recognition and a variation of the 5′ end could readily alter the group of target genes of the miRNA. Likewise, the role of isomiRs in cancer has to be elucidated. Lee et al. [74] have constructed a database cataloguing an entire repertoire of miRNA sequences that can be useful for showing isomiR expression pattern differences in various cell types and conditions. The role of the differentially expressed miRNAs, including miRNA*, isomiR and novel miRNA (candidates), in the development of metastatic prostate cancer remains to be shown. Use of patient-derived prostate cancer xenograft models in conjunction with clinical sample analysis and in vitro models may bring unique perspectives to translational research.
It is of interest that many of the genes associated with the differentially expressed mRNAs in the xenografts were found to be identical to predicted target genes of differentially expressed miRNAs, and that they were related to cancer and metastasis, e.g., FSCN1 [42], CCL2 [46], ADAMTS1 [45], FGFR1 [44], CTGF [48] and SERPINB5 [49]. However, while an miRNA potentially has hundreds of target genes, relatively few targets have been experimentally validated and few miRNA loss-of-function phenotypes have been assigned [75]. More research is required into the effect of specifically inhibiting/enhancing the function of miRNAs on the activity of their putative target genes, gene translation and the various stages of cancer development.
In summary, we have utilized next generation sequencing to identify differentially expressed known and novel miRNAs in a pair of metastatic and non-metastatic prostate cancer xenografts derived from one patient's primary cancer. The use of xenografts generated by subrenal capsule grafting of cancer tissue, a technique that tends to preserve properties of the original cancers, coupled to the finding that a substantial number of the differentially expressed genes have previously been linked to metastasis of prostate cancer or other types of cancer, makes it likely that the identified miRNAs include potential biomarkers and/or therapeutic targets for prostate cancer metastasis.
Supporting Information
Novel miRNA candidates.
(XLS)
List of target genes of up-regulated and down-regulated miRNAs as predicted via the Microcosm database.
(XLS)
Enriched KEGG pathways using the DAVID analysis of up-regulated and down-regulated miRNA target genes.
(XLS)
List of up/down-regulated mRNAs identified via microarray analysis.
(XLS)
Lists of genes associated with (i) up-regulated mRNAs/down-regulated miRNAs and (ii) down-regulated mRNAs/up-regulated miRNAs.
(XLS)
Acknowledgments
The authors wish to acknowledge the BC Cancer Agency Genome Sciences Centre, Vancouver, Canada for the small-RNA library construction and sequencing and the Vancouver Prostate Centre, Vancouver, Canada for microarray data production.
Footnotes
Competing Interests: The authors declare that no competing interests exist.
Funding: This study was supported by the Canadian Institutes of Health Research (YZW/MG). YZW is a recipient of an Overseas Chinese Scholar Award from the National Natural Science Foundation of China (No 30928027), and a recipient of an Innovative Scholar Award from International Cancer Alliance for Cancer Research and Education and the Fibrolamellar Cancer Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for prostate cancer. Annu Rev Med. 2009;60:139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 3.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandellini P, Folini M, Zaffaroni N. Emerging role of microRNAs in prostate cancer: implications for personalized medicine. Discov Med. 2010;9:212–218. [PubMed] [Google Scholar]
- 7.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 8.Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89–96. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Xue H, Cutz JC, Bayani J, Mawji NR, et al. An orthotopic metastatic prostate cancer model in SCID mice via grafting of a transplantable human prostate tumor line. Lab Invest. 2005;85:1392–1404. doi: 10.1038/labinvest.3700335. [DOI] [PubMed] [Google Scholar]
- 10.Lee CH, Xue H, Sutcliffe M, Gout PW, Huntsman DG, et al. Establishment of subrenal capsule xenografts of primary human ovarian tumors in SCID mice: potential models. Gynecol Oncol. 2005;96:48–55. doi: 10.1016/j.ygyno.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 11.Cutz JC, Guan J, Bayani J, Yoshimoto M, Xue H, et al. Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: potential models for studying tumor progression-related changes. Clin Cancer Res. 2006;12:4043–4054. doi: 10.1158/1078-0432.CCR-06-0252. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Revelo MP, Sudilovsky D, Cao M, Chen WG, et al. Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate. 2005;64:149–159. doi: 10.1002/pros.20225. [DOI] [PubMed] [Google Scholar]
- 13.Lin D, Watahiki A, Bayani J, Zhang F, Liu L, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–4359. doi: 10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Grant SF. Advances in Whole Genome Sequencing Technology. Curr Pharm Biotechnol. 2011;12:293–305. doi: 10.2174/138920111794295729. [DOI] [PubMed] [Google Scholar]
- 15.Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010;39:D876–82. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 21.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang P, Wu H, Wang W, Ma W, Sun X, et al. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, et al. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 24.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 25.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 28.Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 29.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 30.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandellini P, Folini M, Zaffaroni N. Towards the definition of prostate cancer-related microRNAs: where are we now? Trends Mol Med. 2009;15:381–390. doi: 10.1016/j.molmed.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Manzur M, Fleming P, Huang DC, Degli-Esposti MA, Andoniou CE. Virally mediated inhibition of Bax in leukocytes promotes dissemination of murine cytomegalovirus. Cell Death Differ. 2009;16:312–320. doi: 10.1038/cdd.2008.152. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leite KR, Sousa-Canavez JM, Reis ST, Tomiyama AH, Camara-Lopes LH, et al. Change in expression of miR-let7c, miR-100, and miR-218 from high grade localized prostate cancer to metastasis. Urol Oncol. 2009 doi: 10.1016/j.urolonc.2009.02.002. 2009 Apr 15. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–2295. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 37.Spahn M, Kneitz S, Scholz CJ, Stenger N, Rudiger T, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2010;127:394–403. doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 38.Shi XB, Xue L, Yang J, Ma AH, Zhao J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 40.Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, et al. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2011;71:538–549. doi: 10.1002/pros.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu H, Hu Z, Wen J, Wang K, Liu Y. TGF-beta promotes invasion and metastasis of gastric cancer cells by increasing fascin1 expression via ERK and JNK signal pathways. Acta Biochim Biophys Sin (Shanghai) 2009;41:648–656. doi: 10.1093/abbs/gmp053. [DOI] [PubMed] [Google Scholar]
- 43.Kusters B, Kats G, Roodink I, Verrijp K, Wesseling P, et al. Micronodular transformation as a novel mechanism of VEGF-A-induced metastasis. Oncogene. 2007;26:5808–5815. doi: 10.1038/sj.onc.1210360. [DOI] [PubMed] [Google Scholar]
- 44.Sato T, Oshima T, Yoshihara K, Yamamoto N, Yamada R, et al. Overexpression of the fibroblast growth factor receptor-1 gene correlates with liver metastasis in colorectal cancer. Oncol Rep. 2009;21:211–216. [PubMed] [Google Scholar]
- 45.Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, et al. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–1894. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Patel L, Pienta KJ. CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev. 2010;21:41–48. doi: 10.1016/j.cytogfr.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethi S, Macoska J, Chen W, Sarkar FH. Molecular signature of epithelial-mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am J Transl Res. 2010;3:90–99. [PMC free article] [PubMed] [Google Scholar]
- 48.Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008;15:675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 49.Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, et al. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- 50.Kortmann U, McAlpine JN, Xue H, Guan J, Ha G, et al. Tumor Growth Inhibition by Olaparib in BRCA2 Germline-Mutated Patient-Derived Ovarian Cancer Tissue Xenografts. Clin Cancer Res. 2011;17:783–791. doi: 10.1158/1078-0432.CCR-10-1382. [DOI] [PubMed] [Google Scholar]
- 51.Karlou M, Tzelepi V, Efstathiou E. Therapeutic targeting of the prostate cancer microenvironment. Nat Rev Urol. 2010;7:494–509. doi: 10.1038/nrurol.2010.134. [DOI] [PubMed] [Google Scholar]
- 52.Bogner PN, Patnaik SK, Pitoniak R, Kannisto E, Repasky E, et al. Lung cancer xenografting alters microRNA profile but not immunophenotype. Biochem Biophys Res Commun. 2009;386:305–310. doi: 10.1016/j.bbrc.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murata T, Takayama K, Katayama S, Urano T, Horie-Inoue K, et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13:356–361. doi: 10.1038/pcan.2010.32. [DOI] [PubMed] [Google Scholar]
- 54.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, et al. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379:726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Guo H, Zhang H, Wang H, Qian G, et al. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gotte M, Mohr C, Koo CY, Stock C, Vaske AK, et al. miR-145-dependent targeting of junctional adhesion molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene. 2010;29:6569–6580. doi: 10.1038/onc.2010.386. [DOI] [PubMed] [Google Scholar]
- 57.Takayama K, Tsutsumi S, Katayama S, Okayama T, Horie-Inoue K, et al. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene. 2011;30:619–630. doi: 10.1038/onc.2010.436. [DOI] [PubMed] [Google Scholar]
- 58.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 60.Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29:4971–4979. doi: 10.1038/onc.2010.233. [DOI] [PubMed] [Google Scholar]
- 61.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 62.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khew-Goodall Y, Goodall GJ. Myc-modulated miR-9 makes more metastases. Nat Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 65.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding J, Huang S, Wu S, Zhao Y, Liang L, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 67.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293–35302. doi: 10.1074/jbc.M110.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol. 1998;153:141–148. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 70.Murchison EP, Hannon GJ. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol. 2004;16:223–229. doi: 10.1016/j.ceb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 71.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis. 2009;30:953–959. doi: 10.1093/carcin/bgp094. [DOI] [PubMed] [Google Scholar]
- 72.Pass HI, Goparaju C, Ivanov S, Donington J, Carbone M, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70:1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qin W, Shi Y, Zhao B, Yao C, Jin L, et al. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. doi: 10.1371/journal.pone.0009429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 78.Guessous F, Zhang Y, Kofman A, Catania A, Li Y, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9 2010 Mar 18;9(6) doi: 10.4161/cc.9.6.10987. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, et al. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Epis MR, Giles KM, Barker A, Kendrick TS, Leedman PJ. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J Biol Chem. 2009;284:24696–24704. doi: 10.1074/jbc.M109.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Novel miRNA candidates.
(XLS)
List of target genes of up-regulated and down-regulated miRNAs as predicted via the Microcosm database.
(XLS)
Enriched KEGG pathways using the DAVID analysis of up-regulated and down-regulated miRNA target genes.
(XLS)
List of up/down-regulated mRNAs identified via microarray analysis.
(XLS)
Lists of genes associated with (i) up-regulated mRNAs/down-regulated miRNAs and (ii) down-regulated mRNAs/up-regulated miRNAs.
(XLS)