Abstract
Posttranslational modification of proteins by poly(ADP-ribosyl)ation regulates many cellular pathways that are critical for genome stability, including DNA repair, chromatin structure, mitosis and apoptosis1. Poly(ADP-ribose) (PAR) is composed of repeating ADP-ribose units linked via a unique glycosidic ribose-ribose bond, and is synthesised from NAD by poly(ADP-ribose) polymerases (PARPs)1,2. Poly(ADP-ribose) glycohydrolase (PARG) is the only protein capable of specific hydrolysis of the ribose-ribose bonds present in PAR chains; its deficiency leads to cell death3,4. Here we show that filamentous fungi and a number of bacteria possess a divergent form of PARG that exhibits all the main characteristics of the human PARG enzyme. We present the first PARG crystal structure (derived from the bacterium Thermomonospora curvata), which reveals that the PARG catalytic domain is a distant member of the ubiquitous ADP-ribose-binding macro domain family5,6. High resolution structures of T. curvata PARG in complexes with ADP-ribose and the PARG inhibitor ADP-HPD, complemented by biochemical studies, allow us to propose a model for PAR binding and catalysis by PARG. Our insights into the PARG structure and catalytic mechanism should greatly improve our understanding of how PARG activity controls reversible protein poly(ADP-ribosyl)ation and potentially of how the defects in this regulation link to human disease.
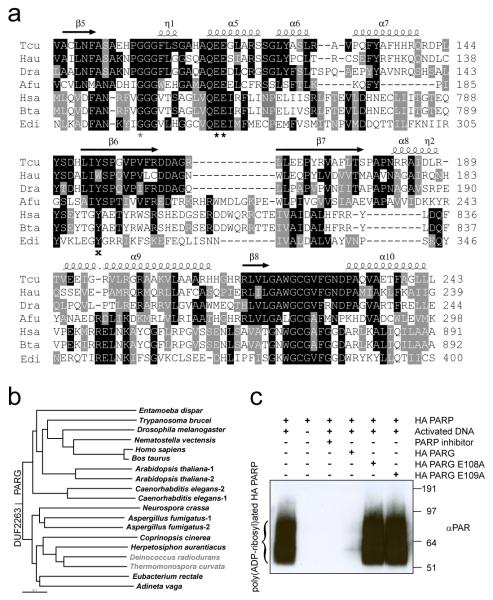
While the mechanism and the functional aspects of poly(ADP-ribose) synthesis have been extensively characterised, the present understanding of the PAR-degradation pathways catalysed by PARG is comparatively poor. To date, the catalytic fold and the mechanism of PARG-mediated hydrolysis remain unknown. Our homology searches revealed that the majority of fungal genomes lack close mammalian/canonical PARG homologues, but instead possess a divergent PARG-like protein, annotated as DUF2263. DUF2263 protein sequences contain the PARG signature (GGG-X6-8-QEE)7, which includes the previously identified key residues: two consecutive glutamates7 (Fig. 1a, black asterisk) and a glycine8 (Fig. 1a, grey asterisk). In addition, a tyrosine residue (Fig. 1a, black cross) implicated in binding to the PARG inhibitor ADP-HPD9 is also present in DUF2263. The DUF2263 orthologue is found also in other eukaryotes lacking canonical PARGs such as the rotifer Adineta vaga (Fig. 1b), where it is fused to several copies of the poly(ADP-ribose)-binding zinc finger10. As both filamentous fungi and A. vaga have functional PARP orthologues and an active poly(ADP-ribose) metabolism11,12, we postulated that DUF2263 was likely to be a functional PARG in these organisms. Surprisingly, although bacteria are historically thought to be devoid of poly(ADP-ribose) metabolism, certain bacterial species possess both PARP (closely related to PARP113) and DUF2263 genes, while some only contain the DUF2263 homologue (Fig. 1a, b). To analyse the biochemical activities of the corresponding bacterial PARP and PARG proteins, we purified His6-tagged Herpetosiphon aurantiacus (HA) PARP and DUF2263. HA PARP was able to synthesise poly(ADP-ribose) from NAD, as revealed by antibodies recognising PAR (Fig. 1c) and by the analysis of PAR ladders using sequencing gels (Supplementary Fig. 1). Analogous to the human PARP1 enzyme, HA PARP was sensitive to the PARP inhibitor KU-0058948 and required DNA for its activation (Fig. 1c). Strikingly, poly(ADP-ribosyl)ation produced by HA PARP is efficiently hydrolysed by the HA DUF2263 protein, demonstrating that DUF2263 is a functional PARG (Fig. 1c) and suggesting the presence of functional PAR metabolism in bacteria.
Figure 1. Phylogeny and functional relationship between DUF2263 and canonical-type PARGs.
a, Multiple sequence alignment of different DUF2263 and PARG proteins from Thermomonospora curvata (Tcu), Herpetosiphon aurantiacus (Hau), Deinococcus radiodurans (Dra), Aspergillus fumigatus (Afu), Homo sapiens (Hsa), Bos taurus (Bta) and Entamoeba dispar (Edi). The two catalytic glutamates, a conserved glycine and tyrosine are marked with black asterisks, grey asterisk and black cross respectively. Secondary structure elements from the Tcu PARG structure are indicated above. b, YmdB-rooted phylogenetic tree of PARGs implied by the neighbour-joining method. Organisms devoid of PARP are marked in grey. c, H. aurantiacus (HA) PARP and PARG enzymes are active as shown by Western blotting with anti-PAR antibodies.
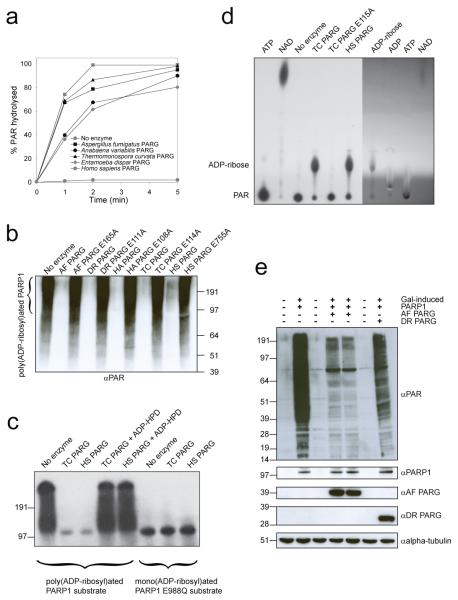
We extended our analyses to other bacterial and fungal PARGs (Fig 1a, b and Supplementary Fig. 2) using established PAR substrates formed by the activity of the well-characterised human PARP1 enzyme. Canonical human and Entamoeba dispar PARGs served as controls. The in vitro activity of the purified PARG enzymes was evaluated in five ways: (i) an assay which measures the loss of PAR from PARP1-modified histones; (ii) a Western-blot analysis of the hydrolysis of PAR from the auto-modified human PARP1 using anti-PAR antibodies; (iii) an SDS-PAGE analysis of the [32P]-PAR auto-modified human PARP1; (iv) an analysis of the release of ADP-ribose from PAR either by thin-layer chromatography (TLC), and (v) liquid chromatography-mass spectrometry (LC/MS) (Fig. 2a-d, Supplementary Fig. 4). The quantification of PAR hydrolysis by the PARG assay shows that bacterial and fungal PARGs are highly active in hydrolysing PAR in vitro; their catalytic potency is comparable to the E. dispar and human PARG enzymes (Fig. 2a). Moreover, the mode of action of these proteins and mutant versions on poly(ADP-ribosyl)ated PARP1 shows the same pattern observed for the canonical PARGs: (i) all are inactivated by the mutation of a key glutamate residue (Fig. 2b, d), (ii) and by the treatment with the PARG inhibitor ADP-HPD (Fig. 2c), and (iii) they are active on ribose-ribose bonds between the two ADP-ribose units, but they are not efficient on mono(ADP-ribosyl)ated protein substrates with the ADP-ribose unit linked directly to the PARP1 (Fig. 2c) or the PARP10 protein (Supplementary Fig. 3). Finally, TLC and LC/MS analyses confirmed that the main product of DUF2263-type PARGs is ADP-ribose (Fig. 2d and Supplementary Fig. 4). Collectively, these results demonstrate that bacterial and fungal PARG proteins possess genuine PARG activity in vitro.
Figure 2. Poly(ADP-ribose) hydrolytic activities of divergent and canonical PARGs.
a, A colorimetric PARG assay. b, Analysis of the hydrolysis of the PARP1-generated PAR substrate by anti-PAR antibodies ‘AF’ stands for A. fumigatus; ‘DR’ D. radiodurans; ‘HA’ H. aurantiacus; ‘TC’ T. curvata; ‘AV’ A. variabilis; ‘HS’ H. sapiens; ‘ED’ E. dispar. c, SDS-PAGE-based assay with [32P]-automodified PARP1 substrate. PARGs are inhibited by ADP-HPD. d, TLC analysis of PARG activity on the [32P]-PAR substrate. The right side of the TLC plate was visualised by shadowing under UV. e, Heterologously expressed A. fumigatus and D. radiodurans PARG hydrolyse PAR in hPARP1-expressing budding yeast cells.
To test whether bacterial and fungal PARG proteins can act as PARGs in vivo, we used the Saccharomyces cerevisiae system expressing human PARP114. Yeast cells, naturally deficient in PAR metabolism, can readily express human PARP1, resulting in poly(ADP-ribosyl)ation of various protein substrates14 (Fig. 2e). When divergent PARG proteins were co-expressed with human PARP1, we observed that the poly(ADP-ribosyl)ation levels in yeast extracts diminished, indicating an efficient hydrolysis of PAR in vivo (Fig. 2e, Supplementary Fig. 5).
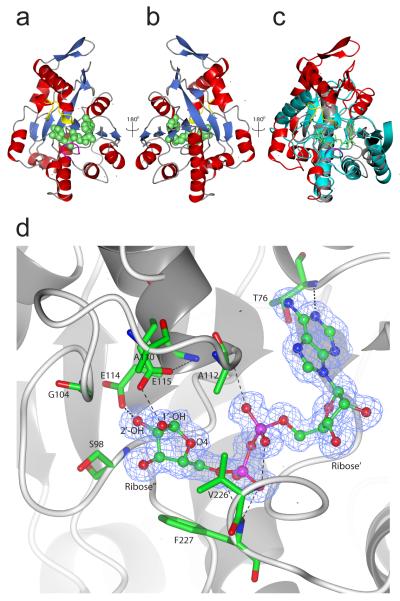
Although the first PARG gene was cloned in 199715, to date there are no structural data available for a PARG enzyme. To gain an insight into the PARG structure and mechanism, we solved the T. curvata PARG ligand-free crystal structure to 1.5 Å in addition to ADP-ribose- and ADP-HPD inhibitor-bound structures (Supplementary Table 1). Comparison with available structures reveals that T. curvata PARG consists of an ADP-ribose-binding macro domain fold with an N-terminal extension (residues 3-67, Fig. 3c). Similar macro domain structures are found in the E. coli YmdB protein (1SPV, Z score 16.2, r.m.s.d. 2.1 Å over 161 Cα atoms, 19% identity), an Archaeoglobus fulgidus macro domain protein (2BFQ, Z score 15.6, r.m.s.d. 2.8 Å over 164 Cα atoms, 16% identity)5 and the histone variant MACROH2A1.1 (1ZR3, Z score 15.0, 2.3 Å r.m.s.d. over 157 Cα atoms, 12% identity)16. The diphosphate-binding loop that flanks one side of the ADP-ribose binding cavity is highly conserved between PARG and other macro domain structures (in magenta, Fig. 3a, c). In contrast, the opposite side of the PARG ADP-ribose binding cavity is lined by a stretch of amino acids corresponding to the PARG-specific GGG-X6–8-QEE signature sequence7 (in yellow, Fig. 3a-c). The structural alignment reveals this PARG-specific loop is inserted into the macro domain fold to accommodate the Glu115 side chain that projects into the PARG active site (Fig. 3c, d). Due to the PARG-specific loop, it appears that only PARGs but not other macro domain proteins can hydrolyse PAR (Supplementary Fig. 6).
Figure 3. T. curvata PARG crystal structure in complex with ADP-ribose.
a, b, Overall fold of PARG with α-helices depicted in red, β-sheets in blue and bound ADP-ribose in green spheres. The PARG-specific catalytic loop is shown in yellow, and the diphosphate-binding loop in magenta. c, Overlay of the T. curvata PARG structure with a representative macro domain structure (PDB code 2BFQ, in cyan). The PARG-specific N-terminal extension and additional PARG structural elements are highlighted in red. Structural features that are similar to 2BFQ and other macro domains are shown in gray. d, Detailed view of ADP-ribose bound in the PARG active site. ADP-ribose is shown with the corresponding 2Fo-Fc omit-map density contoured at 1.2 σ in blue. Key active site residues are represented in atom coloured sticks with hydrogen bonds indicated by black dotted lines.
The ligand-PARG complex structures reveal that ADP-α-ribose occupies a position similar to that observed in other ADP-ribose-macro domain complex structures5 (Fig. 3d and Supplementary Fig. 7). A minor rearrangement can be observed for Val226-Phe227 upon ADP-ribose binding. The Phe227 side chain is in close contact with the ribose” moiety of ADP-ribose, ensuring a close distance between the ribose” O4 and one of the phosphate groups within the constraints of the active site. The ribose” moiety also forms hydrogen bonds with Glu114 (via the 2′-OH group) and Glu115 (via the 1′-OH group) (Fig. 3d). With the exception of water-mediated polar contacts, few direct contacts can be observed with the adenosine moiety. In contrast to the ribose” moiety, the adenosine ribose’ moiety is significantly less accessible. The presence of any substituents on the ribose’ 2′-OH group (such as another ADP-ribose) would require a drastic reorientation of the PARG C-terminal α-helix (see Fig. 4a) with concomitant exposure of the hydrophobic core of the macro domain. This suggests that bacterial PARG binds the PAR terminus and acts as an exo-glycohydrolase.
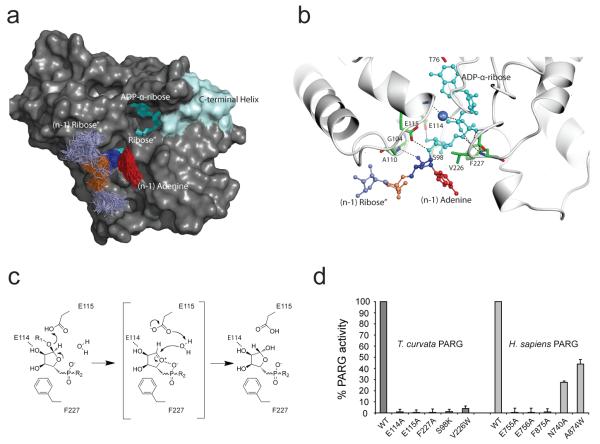
Figure 4. Structural basis of poly(ADP-ribose) glycohydrolysis.
a, The PARG solvent accessible surface derived from molecular dynamic simulations b, A detailed view of the lowest energy PARG:ADP-ribose-dimer model obtained from the 100 snapshot structures. Key active site residues are highlighted in green with PAR structural elements coloured as in 4a. c, Proposed mechanism for PAR glycohydrolysis. R1 and R2 represent (n-1) PAR and terminal adenosine moieties respectively. d, PARG activities of the T. curvata PARG mutants and the corresponding H. sapiens PARG mutants. Error bars represent s.d. (n=3).
The steric restraints imposed by the PARG active site form a basis for the modelling of an α(1″→2′) O-glycosidic linkage with an additional ADP-ribose group to provide further insight into PAR binding (Fig. 4a, b). Our models suggest that the (n-1) adenine moiety is partially enclosed by Ala110, Ala112 and Val226 while the (n-1) ribose’ is in close contact with Ser98 and Gly104 (Fig. 4b). However, MD simulations suggest that the enzyme lacks any obvious binding sites for the (n-1) phosphate and ribose” moieties, as exemplified by significant conformational freedom of the latter with respect to the terminal ADP-ribose and the associated (n-1) ribose’. This suggests that bacterial PARG does not specifically bind additional PAR elements.
The T. curvata PARG complex with ADP-ribose (Fig. 3d) and the corresponding PAR-PARG model (Fig. 4b) allow us to propose a mechanism for PARG catalysis (Fig. 4c). Binding of the PAR terminus positions the key ribose-ribose O-glycosidic linkage in direct hydrogen bonding contact with Glu115, while constraining the conformation of the terminal ribose”. Formation of a putative oxocarbenium intermediate is supported by the protonation of the (n-1) PAR ribose’ 2′-OH leaving group via Glu115, and by the stabilization of the positively charged oxocarbenium through close proximity with the terminal diphosphate group. A water molecule is ideally positioned to attack the oxocarbenium intermediate, activated through concomitant deprotonation by Glu115. This leads to the release of ADP-β-ribose” and (n-1) PAR.
This mechanism is supported by mutagenesis and binding studies. Although mutations of both Glu114 and Glu115 render the enzyme inactive without disruption to the overall PARG fold (Fig. 4d, Supplementary Fig. 8), ITC ADP-ribose binding studies reveal that Glu115A does not affect binding, while Glu114A leads to an approximate 10-fold decrease in binding affinity (Supplementary Fig. 9). This supports the notion that the role of Glu114 is confined to substrate binding/orientation as opposed to the acid-base catalysis proposed for Glu115. The mutation of Phe227 implicated in positioning the terminal ribose” also renders the enzyme inactive (Fig. 4d). Mutations of Ser98 and Val226, implicated by our model in binding the (n-1) ADP-ribose, greatly diminish T. curvata PARG activity (Fig. 4d). Importantly, mutations of the corresponding catalytic residues in the human PARG have a similar effect on the enzymatic activity, which suggests a universal catalytic mechanism for bacterial and mammalian PARGs (Fig. 4d).
These data provide the first insight into PARG structure and mechanism, and improve our understanding of the biochemical strategies regulating reversible poly(ADP-ribosyl)ation. Our results define existence of bacterial PAR metabolism and show that PARG is a new member of the widespread macro domain protein family5,6, which is known to be implicated in genome stability and regulated by poly(ADP-ribosyl)ation17-19. Our data confirm that PARGs are evolutionary unrelated to ARH3 proteins, another class of enzymes with the ability to cleave PAR20,21 (Supplementary Information). We believe that our findings will provide the groundwork for future studies that might ultimately lead to the development of small, cell-permeable PARG inhibitors and the potential to manipulate physiology of health and disease by interfering with PAR metabolism.
METHODS SUMMARY
Proteins with His6-tags were purified from E. coli. A colorimetric PARG Assay Kit (Trevigen) was used to measure the loss of biotinylated PAR from histones. Auto-modified poly(ADP-ribosyl)ated human and bacterial PARP substrates were made by incubation with NAD10. In vivo activity of PARGs was tested in a yeast system with inducible expression of human PARP1. The T. curvata PARG structure was solved by means of single-wavelength anomalous diffraction on Pt-soaked crystals. Ligand-enzyme complexes were obtained by the soaking of PARG crystals. The PAR-PARG model was created using MD simulations of the ADP-α-ribose dimer in complex with PARG.
METHODS
Plasmids and proteins
Bacterial and fungal PARG proteins and the E. dispar PARG were expressed from the pET28a vector (Novagen). Human PARG with an N-terminal truncation (Δ1-455) was expressed from the pColdTF vector (Takara). The A. fumigatus Af293 gene (AFUA_4G11940), the D. radiodurans R1 ATCC13939 gene (DR_B0099) and the E. dispar SAW760 gene (EDI_110590) were amplified from genomic DNA, the human PARP1 and PARG gene (Q86W56) were amplified from HeLa cDNA, while the T. curvata DSM 43183 gene (Tcur_1721) and the A. variabilis ATCC 29413 gene (Ava_4013) were synthesised according to the database sequence (GenScript USA Inc.). The H. aurantiacus ATCC 23779 PARG (Haur_1618) and PARP (Haur_4763) genes were amplified from genomic DNA extracted from a dried culture purchased from DSMZ. All proteins bear an N-terminal his tag with the exception of the A. variabilis PARG, which has a C-terminal his tag. Mutations were introduced using the QuickChange II site-directed mutagenesis kit (Stratagene). Proteins were expressed in E. coli Rosetta2(DE3) cells (Novagen). Recombinant proteins were purified on Ni-NTA beads according to standard procedure. For crystallisation studies, the T. curvata PARG was purified by FPLC on a HisTrap HP column followed by Superdex 200 (GE Healthcare). The catalytic fragment of human PARP10 (amino acids 818–1025) was expressed as a GST-tagged protein and purified on Glutathione Sepharose columns.
PARP activity assay
PARP auto-modification activity of the recombinant H. aurantiacus (HA) PARP was assayed in a 30-minute room-temperature reaction containing 100 nM HA PARP, 200 μM NAD (Trevigen), 50 mM Tris pH 7.5 and 50 mM NaCl with or without activated DNA (Trevigen), and with or without the PARP inhibitor KU-0058948 (10 μM). After the reactions were stopped with the PARP inhibitor, 500 nM HA PARG and its catalytic glutamate mutants were added to the reaction for 30 min. The reactions were analysed by Western blotting with rabbit anti-PAR antibodies as described under ‘PARG activity assays’.
PARG activity assays
PARG activity of recombinant proteins was quantified with a colorimetric PARG assay kit (Trevigen), which measured the loss of biotinylated PAR from histones generated by PARP1. 3 nM PARG enzymes were used for this assay. For Western-blot analysis of PARG activity, poly(ADP-ribose) was synthesised by the automodification of PARP1 in a reaction mixture containing 2 units of PARP1 (Trevigen), 200 μM NAD (Trevigen), activated DNA (Trevigen), 50 mM Tris pH 7.5 and 50 mM NaCl at room temperature. Reactions were stopped after 30 min by the addition of the PARP inhibitor KU-0058948. 500 nM PARGs were added to the reaction and incubated for another 30 min. The reactions were run on 4-12% SDS-PAGE gels and blotted onto a nitrocellulose membrane. PAR hydrolysis was detected by rabbit polyclonal anti-PAR antibodies (1:1000; Trevigen). Western blots were analysed densitometrically by GeneTools (SynGene). For a direct visualisation of PARG activity via SDS-PAGE, [32P]-labelled NAD (GE Healthcare) was added to the above reaction to generate radioactively labelled poly(ADP-ribosyl)ated or mono(ADP-ribosyl)ated PARP substrates. 500 nM PARGs were subsequently added with or without the PARG inhibitor ADP-HPD (50 μM). The reaction products were analysed by SDS-PAGE and visualised by autoradiography. For thin layer chromatography (TLC) analysis, the reactions were stopped by the addition of 50 mM ADP-ribose, spotted onto polyethyleneimine (PEI)-cellulose plates (Macherey-Nagel, Polygram CEL 300 PEI/UV254) and developed in 0.15 M LiCl and 0.15 M formic acid. Dried plates were exposed on X-ray film or visualised by UV254 shadowing.
Expression of heterologous proteins in S. cerevisiae
Plasmids used to introduce human PARP1, A. fumigatus PARG and D. radiodurans PARG into S. cerevisiae were created via the Gateway (Invitrogen) site-specific recombination system using yeast-specific destination vectors (Addgene)22. hPARP1 was introduced into pAG303GAL-ccdB (HIS3), while AF PARG and DR PARG were introduced into pAG423GAL-ccdB (URA3). The host strain W303-1a (MATa, ade2-1, ura3-1, his3-11,15, trp1-1, leu2-3,112, can1-100) was grown in rich YPD medium at 30°C. The strain was first transformed with the hPARP1-harboring plasmid resulting in his+ transformants with chromosomally integrated hPARP1. Subsequent transformation with the PARG-harboring plasmids yielded his+ura+ transformants with episomally maintained PARG. To induce the expression of PARP1 and PARG from the GAL1 promoter, the strains were grown overnight in minimal SC medium with 2% glucose. The overnight cultures were washed, resuspended in SC+2% raffinose at 7×106 cells/ml and grown for one cell generation. Cells were centrifuged before resuspending in SC+2% galactose medium and grown overnight to induce the expression of hPARP1 and PARG from the GAL1 promoter. Yeast protein extracts were analysed by immunoblotting. Rabbit polyclonal anti-PAR antibody (Trevigen) and rabbit anti-PARP1 antibody (Abcam) were used at 1:1000 dilution, while rat monoclonal anti-alpha-tubulin antibody (Abcam) was used at 1:5000 dilution. Rabbit anti-DRPARG and anti-AFPARG polyclonal antibodies were raised against the respective full-length proteins purified from E. coli (Eurogentec) and applied at 1:1000 dilution.
Crystallisation, refinement and model building
Initial T. curvata PARG crystallisation conditions were identified using the JCSG+ matrix screen (Molecular dimensions). Crystals suitable for diffraction experiments were obtained by sitting drop vapour diffusion in 4 μL drops containing equal volumes of protein and a solution containing 10% PEG 6K and 0.1 M Bicine buffer (pH 9.0). The crystals were derivatised by soaking with the same solution supplemented with 1 mM potassium tetracyanoplatinate (II) trihydrate for 10 minutes. For the ADP-ribose and ADP-HPD complexes, native crystals were soaked with 1 mM ligand for 1-2 hrs. The crystals were cryoprotected with the addition of 15% PEG 200 to the mother liquor and flash cooled in liquid nitrogen. Data was collected on beamline I04 at the Diamond Light Source Facility and reduced and scaled with the X-ray Detector Software suite (XDS)23.
The ligand-free PARG crystal structure was determined by single-wavelength anomalous diffraction on a Pt-soaked crystal using Solve and Resolve as implemented in Phenix24. The initial electron density map was of sufficient quality for Resolve to build 70% of the model. Further automated model building with ARPwARP25 successfully constructed the entire model, with the exception of a small loop region incorporating residues 58-66, for which no corresponding density could be observed. The model was completed by iterative cycles of manual model building and real space refinement using the program Coot26 and crystallographic refinement using Phenix.refine24. Validation of the structure was performed with Molprobity. The processing, phasing and final refinement statistics are presented in Supplementary Table 1.
Isothermal Titration Calorimetry
The thermodynamic ligand binding properties of wild-type T. curvata and two mutant proteins (E114A and E115A) were measured using a VP-ITC microcalorimeter. Protein and ligand concentrations were 20 μM and 200 μM respectively in 50 mM Tris (pH 7.5), 100 mM NaCl. Titration curves were fitted using a non-linear least squares method in Microcal Origin software. A model that was indicative of a single binding site was found to give the best fit in each case; this model was used to obtain the thermodynamic parameters shown in Supplementary Fig. 9.
Thermal shift assay
In each 50 μL reaction 15 μL of 300× Sypro Orange (Sigma), 2 μL of water and 33 μL of protein (4 mg/ml) in 25 mm Tris pH 7.5, 50 mM NaCL, 10% glycerol and 1 mM DTT were added to the wells of a 96-well thin-wall PCR plate (Bio-Rad). For thermal shift assays in the presence of ADP-ribose, 2 μL of 25 mM ADP-ribose was added instead of water. Samples were placed in a CFX96 Real Time PCR thermal cycler (Bio-Rad) and slowly heated from 15 to 95 °C with sample fluorescence recorded every 0.2 °C. Fluorescence was monitored at 575 nm (emission), using 490 nm for excitation.
Analysis of poly(ADP-ribose) chains by sequencing gels
[32P]-labelled poly(ADP-ribose) chains produced by PARPs were resolved in 10% sequencing gels in TBE buffer. Reactions were stopped by the addition of 600uL 20% ice-cold trichloroacetic acid (TCA). The precipitated material was washed twice with 5% TCA. The pellet was then incubated with 70 μL of 10 mM Tris and 1mM EDTA buffer (pH 12) for 2 hours at 60 oC to detach PAR chains. PAR was phenol-chloroform extracted, dried and resuspended in 2 mM EDTA-formamide buffer before loading. The products were visualised by autoradiography.
Computational simulations
NAMD software1 was used to perform all molecular dynamics (MD) simulations of the ADP-α-ribose dimer in complex with PARG. The topology and parameters files for the ligand were obtained using the Antechamber program2 and AM1-BCC charges3. The additional ADP-α-ribose monomer was added to the ADP- α -ribose bound to PARG by creating an α (1″→2′) O-glycosidic bond. Five conformers of the added monomer were taken as starting points for MD simulations. The PARG:ADP-α-ribose dimer complex model was then immersed in a periodic water box (TIP3), as half of the ligand was in contact with the bulk solvent. Then, for each conformer, several simulations (0.5 ns) were run with variable parameters (200 K or 310 K, peptidic backbone fixed, restrained or free). During these MD simulations, snapshots (100 in total for clarity) were periodically extracted and energy-minimized, to represent the conformational flexibility of the ADP-α-ribose dimer.
Analyses by ultrahigh-performance liquid chromatography (UHPLC) coupled to quadruple-time-of-flight mass spectrometry (QTOFMS)
The poly(ADP-ribosyl)ated PARP1 was treated with T. curvata PARG, and the mixture was filtered using centricons (30 kD cutoff). The analysis of the filtrate was performed using a modified procedure by Coulier et al.4, which employed ion-pair chromatography for the separation of nucleotides and negative electrospray ionization for mass spectrometric detection. All analyses were performed using a Waters Acquity UPLC system (Waters Corp., Milford, MA, USA), equipped with a binary solvent delivery system and autosampler. The chromatographic separations employed a column (50 mm × 2.1 mm) filled with a 1.7 μm BEH C18 stationary phase (Waters Corp., Milford, MA, USA). Binary gradients at a flow rate of 0.4 mL/min were applied for the elution. The eluent A was water containing 5 mmol/L of pentylamine, while the pH value was adjusted to 6.5 using acetic acid. A fast elution gradient was applied, which allowed an efficient separation of NAD+ and ADP-ribose within 5 min. The gradient started at 2 % B and then the percentage of B linearly increased to 25 % in 5 min.
The mass spectrometry was performed on a QTOF Premier instrument (Waters Micromass, Manchester, UK) using an orthogonal Z-spray-electrospray interface. The drying gas and the nebulizing gas was nitrogen. The desolvation gas flow was set to 700 L/h at a temperature of 300 °C. The cone gas flow was adjusted to 25 L/h, and the source temperature to 120 °C. The capillary and cone voltages were 3200 V and 30 V respectively. The instrument was operated in V mode with TOFMS data being collected between m/z 100–1000, applying a collision energy of 4 eV. All spectra were recorded using the extended dynamic range (DRE) option in order to correct for possible peak saturations, the data were collected in the centroid mode with a scan time of 0.08 s and interscan time of 0.02 s. In order to ensure maximum accuracy and reproducibility of the system, all acquisitions were carried out using an independent reference spray via the lock spray interface. Leucine enkephalin was applied as a lock mass in negative ionization mode (m/z 554.2615).
The chromatograms, recorded in the total ion current (TIC) mode, were systematically examined by manually generating mass spectra of each visible individual peak using the background-subtraction option and searching for target components expected to occur in the mixture, in particular ADP-ribose and NAD+, using extracted ion chromatograms. The target analyses were based on the accurate mass feature of the instrument, applying a mass window of 50 mDa. Elemental composition of the selected ions was calculated using the MassLynx software incorporated in the instrument. The main criterion for selecting the elements to be included in search parameters for the calculation of the molecular formula was the elemental composition of the presumed parent compounds. According to the instrument specifications, the acceptable deviation from the theoretical m/z values was set at 5 mDa.
Supplementary methods references
- 27.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornell WD, et al. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 29.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23:1623–41. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 30.Coulier L, et al. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal Chem. 2006;78:6573–82. doi: 10.1021/ac0607616. [DOI] [PubMed] [Google Scholar]
Supplementary Material
Acknowledgements
We thank G. Clark for genomic DNA from E. dispar, G. Smith for PARP inhibitor, M. Rossi for purified proteins, and R. Thorough for editing English. We thank D. Ahel, A. Jordan, D. Ogilvie, S. Terzic, D. Neuhaus and S. Eustermann for helpful discussions. We are grateful to B. Lüscher for the gift of PARP10 expression plasmid, and K. Labib and the members of his lab for advice with yeast work. This work was funded by Cancer Research UK. D.S. holds an AXA Research Fund post-doctoral fellowship. D.L. is a Royal Society University Research Fellow. Access to Diamond beamlines is gratefully acknowledged.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Atomic coordinates and structure factors have been deposited with the Protein Data Bank (PDB ID codes 3SIG, 3SIH, 3SII and 3SIJ).
References
- 1.Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008;9:1094–100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- 3.Koh DW, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanai S, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101:82–6. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karras GI, et al. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–20. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Front Biosci. 2009;14:3246–58. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 7.Patel CN, Koh DW, Jacobson MK, Oliveira MA. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev Cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 9.Koh DW, et al. Identification of an inhibitor binding site of poly(ADP-ribose) glycohydrolase. Biochemistry. 2003;42:4855–63. doi: 10.1021/bi0272048. [DOI] [PubMed] [Google Scholar]
- 10.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 11.Kothe GO, Kitamura M, Masutani M, Selker EU, Inoue H. PARP is involved in replicative aging in Neurospora crassa. Fungal Genet Biol. 2010;47:297–309. doi: 10.1016/j.fgb.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semighini CP, Savoldi M, Goldman GH, Harris SD. Functional characterization of the putative Aspergillus nidulans poly(ADP-ribose) polymerase homolog PrpA. Genetics. 2006;173:87–98. doi: 10.1534/genetics.105.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–82. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 14.Tao Z, Gao P, Liu HW. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry. 2009;48:11745–54. doi: 10.1021/bi901387k. [DOI] [PubMed] [Google Scholar]
- 15.Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272:11895–901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 16.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–5. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 17.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk AJ, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–4. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–9. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 20.Mueller-Dieckmann C, et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci U S A. 2006;103:15026–31. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–13. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 22.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–9. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Cryst. 1988;21:916–24. [Google Scholar]
- 24.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.