Abstract
RNA polymerase III (pol III) transcription of transfer RNA (tRNA) genes is essential for generating the tRNA adapter molecules that link genetic sequence and protein translation. By mapping pol III occupancy genome-wide in the livers of mouse, rat, human, macaque, dog and opossum, we found that pol III binding to individual tRNA genes varies substantially in strength and location. However, taking into account tRNA redundancies by grouping pol III occupancy into 46 anticodon isoacceptor families or 21 amino acid-based isotype classes shows strong conservation. Similarly, pol III occupancy of amino-acid isotypes is almost invariant among transcriptionally and evolutionarily diverse tissues in mouse. Thus, synthesis of functional tRNA isotypes has been highly constrained, though the usage of individual tRNA genes has evolved rapidly.
Introduction
Tissue-specific gene expression patterns can be highly conserved among vertebrates, and organismal divergences up to 500 million years can leave a core set of highly transcribed tissue-specific genes intact from chicken to fish to human1. Furthermore, non-coding RNAs, such as lincRNAs, may also show elevated purifying selection in location and expression between human and mouse cells2,3. Perhaps surprisingly, the proteins that bind DNA to direct these tissue-specific gene expression programs in mammals can diverge rapidly and dramatically in their genome-wide binding, even between evolutionarily close species4-7. The ability of divergent protein-DNA contacts to direct highly conserved gene expression is not understood, both because mammalian transcription factors (TF) operate in cross-regulatory and functionally redundant modules, and because of the difficulty of disentangling the specific regulatory contributions of the thousands of TF binding events in a mammalian cell.
The synthesis of transfer RNA (tRNA) by RNA polymerase III (pol III) is an ideal system to explore the evolution and function of mammalian transcriptional regulation, because pol III binds to only a few hundred tRNA genes in mammals8-12, and because translation of mRNA into polypeptides is highly conserved13. Each tRNA molecule can be attached to a single amino acid that it couples to a growing polypeptide chain by selectively base-pairing its three-base anticodon to a complementary three-base codon sequence within a messenger RNA (mRNA).
tRNA genes that encode the same anticodon are referred to as isoacceptor classes; there are about 48 in mammals, depending on the species14,15. Isoacceptor families that translate to the same amino acid are known as amino acid isotypes, of which there are 21. In addition to the redundancies arising from multiple genes encoding the same anticodon, and multiple anticodons translating to the same amino acid, tRNAs can use the third base of the anticodon in a wobble pair with a closely-related codon sequence, thereby allowing one tRNA anticodon to pair with multiple codon triplets. These redundancies have been well characterized biochemically16; grouping tRNA genes into isoacceptor families and isotypes can simplify analysis, bypassing redundancy complications15.
Although tRNA biology has been extensively studied, much of the work was based on biochemical assays, and was performed before genome sequences were available for any mammal. More recent work has been mainly computational, using predictive methods to identify possible tRNA genes14,17. The experimental analyses that have been carried out so far have focussed only on single species8-12.
In this study, we have identified the location of, and analyzed the evolutionary stability of, the binding of pol III to tRNA genes in six species from four mammalian orders: mouse and rat (rodents), human and macaque (primates), dog (carnivores), and opossum (marsupials) as a non-eutherian mammalian outgroup. Our results demonstrate how the binding of the basal transcriptional machinery to individual tRNA genes can rapidly diverge while still maintaining highly constrained expression of functional amino acid isotypes.
Results
Pol III binds hundreds of expressed tRNA genes in mice
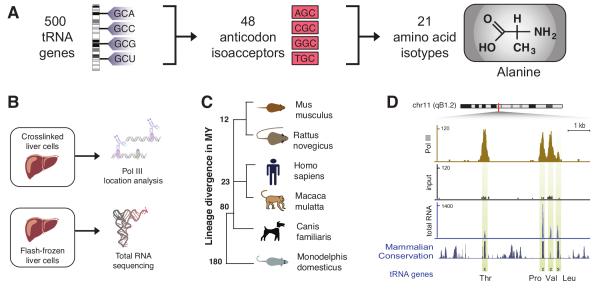
Our analysis focused on actively transcribed nuclear tRNA genes, which we collapsed into anticodon isoacceptors that translate synonymous codons, and further into amino acid isotypes based on their aminoacylation identity (Figure 1A). We first identified the tRNA genes occupied by pol III in mouse liver by performing chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) and validated their expression by sequencing of cDNA from total RNA (RNA-seq) (Figure 1B) (Methods). We identified candidate loci using tRNAscan-SE, mapped ChIP-seq reads to the appropriate reference genome, and counted reads that align to predicted tRNA loci or within 100 bp upstream or downstream (Methods). In tissues from mouse (as well as five other mammalian species) (Figure 1C), different biological replicates for pol III occupancy were found to be highly similar (Figure 2). As expected from prior studies in cultured cells8-12, pol III binding was found primarily at known pol III targets18 such as transposons, other repeat elements, and tRNA genes (Figure S1).
Figure 1. Pol III occupies and transcribes tRNA genes in mouse liver.
(A) tRNA genes are present in hundreds of copies in a mammalian genome, but collapse into 62 isoacceptor families (48 are present in mammals), and 21 amino acid isotypes. (B) Separate portions of each species’ tissues were flash frozen to permit direct RNA sequencing, and treated with formaldehyde to cross link protein-DNA contacts to allow chromatin immunoprecipitation reactions. (C) Primary tissues were isolated from six mammals varying in evolutionary distance to mouse, from 12 million (rat) to over 180 million years (opossum). (D) Typical pol III-bound tRNA loci on murine chromosome 11. Pol III binding shown as a beige enrichment track (top), the input DNA as a black track (middle), and total RNA as blue track (bottom), indicating the pol III bound regions are transcriptionally active. The y-axis of each track specifies read density. The mammalian conservation track, obtained from UCSC, displays the degree of placental mammal basewise conservation (20 species) and sequence constraint. Beneath is the genome annotation in this region obtained for repeats, non-coding RNAs, and tRNAs (UCSC genome browser).
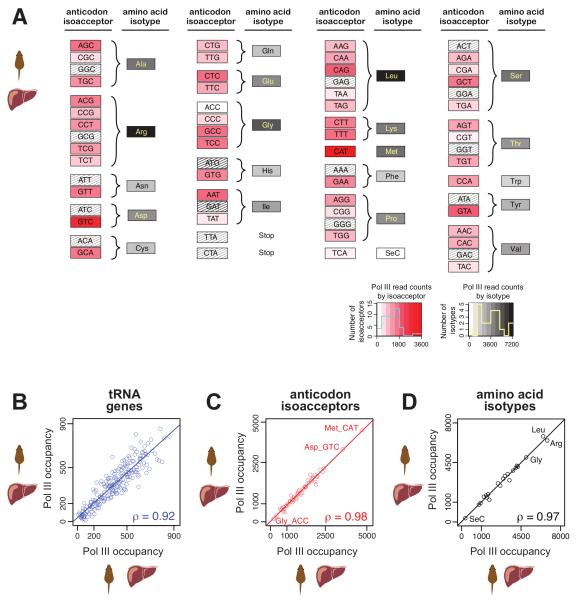
Figure 2. tRNA isoacceptors and isotypes are differentially bound by pol III in mouse liver.
(A) Pol III binding varies widely among isoacceptor families (red), which collapse in number as well as binding variability in the 21 amino acid isotypes (grey). Areas are shaded according to number of pol III read counts (dendrogram shows values white: low to red/black: high). Hatched boxes represent isoacceptors that are absent in mouse, or that do not encode for an amino acid. (B-D) Pol III binding between replicates of mouse liver compared at (B) tRNA gene loci (blue), (C) anticodon isoacceptors (red), and (D) amino acid isotypes (black). Spearman’s rank correlation coefficients (ρ) are shown.
RNA polymerase II (pol II), which is responsible for protein-coding mRNA synthesis, has been well-characterized to bind in vivo to many regions not actively transcribed due to stalled polymerase activity19,20, alternate transcription start sites21, and regulated alternative splicing22. However, sequencing of transcripts in mouse liver indicated that every tRNA gene bound by pol III is expressed (Figure 1D). Conversely, very few RNA transcripts aligned uniquely to predicted tRNA loci not bound by pol III (Figure S2), confirming prior reports that pol III binding can be used as a proxy for tRNA transcription8-12.
Pol III occupies tRNA genes in anticodon- and isotype-specific distributions
Mammals share a set of tRNAs carrying 41-55 anticodons15. In mouse liver, we identified 223 distinct tRNA genes bound by pol III. Almost all of the 223 mouse tRNA genes experimentally determined to be bound by pol III were computationally predicted previously by Lowe et al. 14 (98%) or Coughlin et al.23 (94%) (Table S1). About 70% of the transcribed tRNA genes are located in clusters, indicating that the majority of tRNA gene transcription is concentrated in distinct chromosomal domains (Figure S3)24.
The mouse tRNA genes we identify as pol III bound in primary liver tissue represent 47 of the 62 possible isoacceptor families that encode the 20 standard amino acids, plus selenocysteine (Figure 1A, Figure 2A). One of these 47 isoacceptor families is found only in rodents [tRNAGly(ACC)], and therefore, the remaining 46, which are shared among all interrogated mammals, are predominantly used in our analyses.
Redundancies in the translational code are reflected in the distribution of pol III binding among the different isoacceptor families. For instance, methionine (Met) has only a single anticodon tRNAMet(CAT), which is highly occupied by pol III. In contrast, the five different and partially redundant arginine (Arg) anticodons tRNAArg(ACG), (CCG), (CCT), (TCG), and (TCT) each show intermediate pol III occupancy. Collapsed to the amino acid level, however, the collective amount of pol III at all tRNA genes for arginine is substantially higher than the pol III occupancy of methionine tRNA genes (Figure 2A).
The binding of pol III among these 223 distinct mouse tRNA genes varied by up to two orders of magnitude (Figure 2B). Analysing pol III binding by isoacceptor family showed only one order of magnitude difference. For instance, the highest occupied isoacceptor families, tRNAMet(CAT) and tRNAAsp(GTC), show over 4000 reads versus just over 300 reads at the lowest, tRNAGly(ACC) (Figure 2C). Although the amount of pol III at each isoacceptor family largely correlated with the number of copies of mouse tRNA genes in the family, there were many specific tRNA genes for the same isoacceptor that showed considerable variation (Figure S4).
These observations support prior reports of active regulation of tRNA transcription in a site-specific manner8-12. The amino acid isotypes had a similar distribution of total pol III enrichment as their component isoacceptors (Figure 2D). Pol III occupancy at tRNA genes, isoacceptor anticodon families, and amino acid isotypes classes was highly correlated between different biological replicates (Figure 2B-D).
Tissue-independent expression of tRNA isotypes
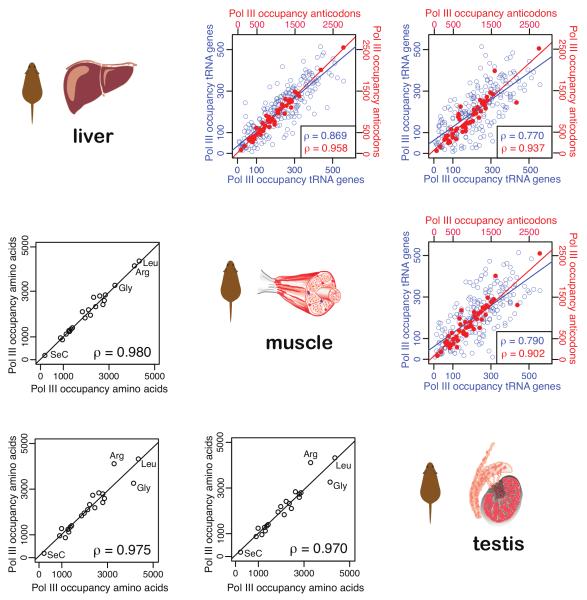
There are large differences in pol III binding between different human cultured cell types8,11 and between different human tissues in tRNA expression25. We asked whether tRNA expression was similarly divergent in phenotypically diverse primary tissues by determining pol III occupancy in mouse muscle, testes, and liver. Our data confirmed prior reports that different cellular types have differences in pol III occupancy on a gene-by-gene basis (Figure 3, top right, blue circles). The collections of tRNA genes active in different mouse tissues overlap almost entirely (Figure S5); the observed differences between these transcriptionally diverse tissues within one species appear instead to be due to differential pol III binding over the same set of tRNA genes.
Figure 3. Amino acid isotypes are bound by pol III in a tissue-independent manner.
Pol III occupancy at tRNA genes was determined in mouse liver, muscle, and testes. The intersection of the row-column for each tissue combination in the upper-right triangle shows the correlation for all pol III bound tRNA gene loci (blue) and isoacceptor (red) and in the lower-left triangle of the binding of pol III to isotypes (black). The Spearman’s rank correlation coefficients (ρ) are reported correspondingly by colour in the lower right of each inter-tissue panel.
When pol III binding is considered by isoacceptor family (Figure 3, top right, red circles) or by amino acid isotype classes (Figure 3, bottom left, black circles), the relative proportion of tRNAs bound by pol III in muscle, testes, and liver is highly correlated (Spearman’s rank correlation coefficient ρ=0.90-0.96 on the isoacceptor and ρ=0.97-0.98 on the isotype level).
In bacteria and yeast, relative levels of tRNA isoacceptors correlate with codon usage in genes encoding highly translated proteins, and can themselves influence protein coding gene expression26. However, the role of codon bias in mammals remains debated, with different studies providing contradictory results in inter-tissue codon biases27,28. Our data show that pol III binding to classes of tRNA genes is surprisingly stable, even among transcriptionally divergent mouse tissues, despite substantial tissue-specific differences in pol III occupancy at individual tRNA genes. This observation provides support for the functional importance of selective codon usage in mammals16. We also note more differential use of certain isotypes in testis relative to muscle or liver, perhaps reflecting optimisation in reproductive tissues for tissue-specific gene products.
Pol III binds to hundreds of tRNA genes in six mammalian species
To explore the evolutionary dynamics of tRNA gene transcription, we experimentally determined pol III occupancy in livers of four additional eutherian and one marsupial mammal. We compared our mouse pol III occupancy data to rat (diverged from mouse 12 million years [MY] ago), human (80 MY), macaque (80 MY), dog (85 MY), and the non-eutherian opossum (180 MY) (Figure 4A). We chose liver, which is primarily composed of a single cell type, hepatocytes29 as a representative vertebrate tissue for interspecies comparisons, because of its conserved tissue structure30, function, and gene expression1.
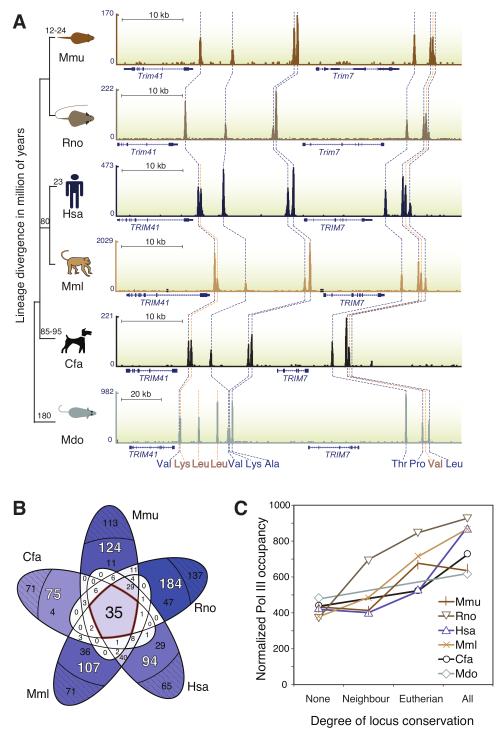
Figure 4. The tRNA genes bound by pol III diverge in genomic location and functional usage among mammals.
(A) Two clusters of multiple tRNA genes flanking the Trim41 and Trim7 genes are bound by pol III in multiple species (shown as genome tracks). The numbers on the left of each track specify read density for pol III ChIP experiments. The syntenic position of each tRNA gene is traced between species with a dashed line. A blue dashed line represents conserved binding in all six mammals and a yellow line indicates species-specific innovation. The cluster upstream of the Trim41 gene shows four tRNA gene loci (two valines, a lysine, and an alanine) with conserved pol III binding. The lysine gene locus does not exist in the rodents, and two leucines are only present in opossum. An additional cluster 3′ downstream of Trim7 demonstrates two tRNA loci (threonine and proline) bound by pol III in all six species. The valine tRNA gene only evolved in the eutherians, and the leucine tRNA gene is not bound in dog. (B) A 5-way Venn diagram intersects the tRNA genes bound by pol III for each possible combination of eutherian mammals used in this study. The total number of species-specific tRNA genes (white) is subdivided into tRNA genes present (inner black numbers) and absent (outer black numbers in dashed subdivision) in Ensembl’s 16 amniote alignment. 24 tRNA genes are bound in all 6 species. Area colourings are shaded according to number of tRNA genes (white: low to blue: high). (C) tRNA loci bound by pol III from panel (B) were sorted by the number of species in which they were bound, and compared to the level of pol III occupancy in each of the study species; demonstrating that higher degree of tRNA gene conservation correlates with higher pol III occupancy (Methods).
In addition to the 223 tRNA genes bound by pol III in mouse, we identified 282 in rat, 224 in human, 233 in macaque, 135 in dog, and 216 in opossum (Tables S2-7). The number of genes corresponding with each isotype is roughly similar across the six species (Table S8). In every species, a substantial fraction (50-70%) of the pol III bound tRNA loci were found in clusters (Figure S3). The higher number of tRNA genes in rat and lower number in dog could be explained by: repeat-driven expansion of tRNA genes, segmental duplications, and genome assembly errors31,32. Regardless, our data captured a set of 46 isoacceptors found in every mammal. Rodents and opossum each have one additional isoacceptor, tRNAGly(ACC) and tRNALeu(GAG), respectively, totalling 48 for all profiled mammals. A similar number of pol III occupied tRNA loci were found using phenotypically and transcriptionally distinct human cell lines8,10-12. Between 61% and 92% of our 224 human tRNA genes overlapped with the tRNA loci identified as bound in other recent studies (Table S1).
We noted a good correlation between the number of tRNA genes and overall pol III occupancy among different species, both by isotype and anticodon, similar to that seen in mouse tissues (Table S9). However, there are significant differences between the occupancy levels of specific tRNA genes, indicating an additional level of per-locus regulation beyond the number of genes.
Publicly available databases predict the existence of 500 to 1000 possible tRNA genes in the genomes that we experimentally profiled for pol III binding1. The hundreds of predicted tRNA genes not occupied by pol III in primary liver tissue must therefore represent a combination of (i) genes actively transcribed in tissues or conditions not profiled in our study and (ii) inactive tRNA pseudogenes. Precedent for the first scenario comes from comparisons between human cell lines. For example, 26% of tRNA loci were occupied by pol III in either HeLa cells or CD4+ T cells, but not in both8. Examples of the latter became apparent when we compared pol III occupancy in livers from different mammals.
The location of Pol III bound tRNAs diverges among mammals
We asked whether the genomic location of pol III-associated tRNA genes is conserved throughout mammalian evolution (Figure 4). tRNA gene synteny is broadly maintained in 12 Drosophila genomes spanning about 40 MY of lineage divergence33. However, our data showed that the specific set of pol III-bound tRNA genes varied considerably between species.
For example, on mouse chromosome 14, a set of four clustered tRNA genes (tRNAThr, tRNAPro, tRNAVal, tRNALeu) is encoded downstream of the murine Trim7 locus, which has conserved synteny in mammals (Figure 4A). Two loci (tRNAThr and tRNAPro) are pol III bound in all interrogated species, and all four loci are pol III-bound in the rodents and primates. Lineage specific loss of pol III occupancy can also be observed in this example, as the tRNALeu gene in dog does not show detectable pol III occupancy. The A- and B-box motifs, which are the type II tRNA gene internal pol III binding motifs34, have degenerated in dog, precluding transcription. The tRNAVal gene in this example is eutherian-specific and either evolved after the eutherian-marsupial divergence or was lost in opossum. Figure S6 is an example of a primate-specific loss in pol III binding of a tRNAArg(ACG) gene. Our data also show lineage-specific gains: in the same figure, a tRNALeu(TAG) gene is bound by pol III in rodents, yet shows no similar evidence of transcription in the other mammals.
To determine the genome-wide evolution of actively transcribed tRNA genes in placental mammals, we compared all six species’ liver pol III binding data using the Ensembl 16-amniota vertebrate PECAN alignment, which contains approximately half of each study species’ genome35,36. Only, 35 of all tRNA genes bound by pol III (ca. 16%) can be aligned among eutherians (Figure 4B), consistent with previous reports that a relatively small amount of functional sequence is alignable across eutherian genomes37. The 35 tRNA genes bound by pol III in all eutherians include at least one anticodon for 18 of the 21 amino acids. Incorporating opposum, 24 tRNA genes are bound by pol III across 180 MY of evolution, and therefore were likely present in the common mammalian ancestor.
Disregarding pol III-occupancy on tRNA genes, 55 predicted tRNA genes (11%) are aligned by PECAN, indicating a slight evolutionary constraint on actively transcribed tRNA genes (Figure S7).
Orthologous tRNA loci bound by pol III in all six species tend to have high pol III occupancy (Figure 4C). For example, among the many tRNAArg(TCG) genes in mouse, the single tRNAArg(TCG) gene on chromosome 7 (Table S2) with orthologues in the other five species has the strongest pol III occupancy (Table S3-7). An additional example is tRNALeu, which is the most highly expressed isotype, and which also has two distinct gene loci strongly occupied by pol III in eutherian mammals.
The number of loci conserved between two species generally reflects evolutionary distance: humans and macaques (23 MY) share 124 loci, humans and mice (80 MY) share 79, while mice and rats (12 MY) share 99 (Figure 4B). The majority of tRNA loci are not alignable to other species within the 16-way alignment; they would be by this definition occupied species-uniquely (Figure 4B, S5 and S8).
We then asked whether pairwise (as opposed to 16-way) alignments of the study species’ genomes would substantially increase the observed overlap in pol III binding. Using synteny nets from UCSC38, we identified the tRNA genes in each species that do not have an ortholog in the syntenic block in a second species. This calculation provides a minimum estimate on the number of tRNA genes that must occur in non-syntenic locations. Mouse and human have the best-annotated genomes, with 93.3% and 93.4% of the genomes, respectively, in synteny blocks; more generally, a typical pairwise synteny map aligns each genome with more than 90% of the other genome39. However, 34% of mouse tRNA genes do not align with human homologs, well over twice as many as would be expected were they randomly distributed in the genome. Similarly, we found 83% of mouse tRNA genes were in the same synteny block in rat, which decreases to 49% in dog, and 43% in opossum (Table S10). It is possible that this reflects a modest preference for tRNA genes to be duplicated in evolutionarily active regions of the genome. In summary, a substantial fraction (ca. 14-55%) of mouse tRNA genes cannot be found in a syntenic location in a second species; this result was unaffected by which species was used to anchor the pairwise comparison.
We conclude that even by relaxed measures of homology, a substantial fraction of pol III binding at tRNA genes occurs in species-specific location. Our results indicate that, in contrast to the apparent high conservation in drosophila tRNA gene evolution33, both tRNA gene location as well as tRNA transcription can evolve rapidly along mammalian lineages.
Pol III occupancy of tRNA isotypes is highly conserved across mammals
We asked whether the observed rapid gain and loss in transcription of particular tRNA genes could be compensated for by simultaneous losses or gains in transcription from other tRNA genes within the same anticodon families. To test this, we began by comparing the total pol III occupancy among the 21 specific amino acid isotypes in all six mammalian species. We reasoned that if there were successful compensation, then the total quantity of pol III at the complete collection of tRNA genes coding for a specific amino acid isotype would not vary between species, despite the divergence we could observe for specific gene loci. We found that the distribution of pol III occupancy among the amino acid isotypes is highly conserved among mammals (Figure 5A), showing correlations uniformly around Spearman’s ρ=0.9 between species.
Figure 5. Isotypes are highly conserved in pol III occupancy and codon usage across mammalian evolution, though the isoacceptors they consist of are only moderately well conserved.
We experimentally determined the strength of pol III binding using ChIP-seq (A-C) and transcript abundance using mRNA-seq (D-F) in livers of six mammals. (A) Proportional frequency of pol III binding by isotype is similar among all six mammals. (B) Proportional frequency of pol III binding to all possible arginine tRNA isoacceptors demonstrate differences among mammals. (C) Pol III binding to tRNA anticodons diverges in correspondence to evolutionary distance (red circles), but pol III binding to isotypes remains largely conserved out to 180 MY of evolution (hollow circles). (D) The frequency of each amino acid weighted by transcript expression was highly conserved between species. (E) Proportional frequencies of codon usage for arginine indicate that mammalian species have little divergence in their codon biases. (F) Amino acids and to a large extent their encoding triplet codon are equally abundant in transcripts across 180 MY of mammalian evolution. Coloured lines within the radial plots (A, B, D, and E) present the data values of each species (purple: mouse, pink: rat, green: human, orange: macaque, black: dog, and yellow: opossum). Labels around the plot indicate amino acids (A and D), anticodons (B) and triplet codons (E). Labels within the grid of the radial plots (A, B, D, and E) describe percentages.
Similarly, we determined the correlation of pol III occupancy among the 46 common anticodon isoacceptors in mouse compared with the same isoacceptor family from each of the other mammals; Figure 5B shows an example of the six arginine anticodons. Figure S6 shows a similar analysis for three orthologous tRNALys genes. These examples demonstrate that increased pol III occupancy of orthologous tRNA genes can be balanced by decreased pol III occupancy of other tRNA genes within the same anticodon isoacceptor family. In general, we found that the anticodon isoacceptor correlation with mouse declined steadily with evolutionary distance from approximately ρ=0.88 in rat (12 MY) to ρ=0.69 in opossum (180 MY) (Figure 5C).
Taken together, in contrast to the rapid gain and loss of pol III activity at specific tRNA genes, the overall expression of tRNA amino acid isotypes has remained constant during 180 MY of evolution.
Transcriptome usage of codon triplets is highly conserved across mammals
Transcription of amino acid isotypes, and to a lesser extent anticodon isoacceptors, is highly conserved in livers of six mammals. We asked whether the usage of codons in the corresponding mRNA transcriptomes was similarly conserved. Prior studies in prokaryotes and simple eukaryotes have found that codon usage can vary considerably among species, although rarely between closely related species16. In addition, gene expression is strongly influenced by tRNA availability in non-mammalian systems40. Other findings suggest that codon usage biases are correlated to tRNA abundance41. To determine whether mammals have species-specific codon biases, we sequenced mRNA from liver of the six mammals used in this work, then counted the occurrences of triplet codons and amino acids necessary for mRNA decoding in each species’ liver transcriptome, weighted by transcript abundance. We found that the frequencies of different encoded amino acids were almost invariant among mammals as diverse as opossum and mouse, with uniformly high correlation values (ρ >0.95) (Figure 5D-F). Codon usage was well conserved across the profiled species (Figure 5E-F, S9), though not as well as amino acid usage. We observed a similar high correlation in codon usage and weighted amino acid frequency in different mouse tissues (Figures S10, S11 and S12).
Finally, we asked whether pol III occupancy of tRNA isoacceptor classes correlated with the weighted codon distribution found in the transcriptome of the same tissue (Figure S13). For liver in 6 species, we found reasonably strong correlation (R≈0.7) between the amount of pol III at the 46 common anticodons, and the weighted occurrence of the corresponding codons in mRNAs. This correlation is particularly surprising, given the number of confounding factors in correlating such disparate data, such as the previously discussed redundancies within the genetic code and post-transcriptional regulation of mRNA causing variations in mRNA longevity.
In sum, we found remarkable conservation in pol III occupancy and transcription of tRNA genes at the amino acid level, and a corresponding conservation among the usage of amino acids in transcriptomes from matched tissues in diverse mammals.
Discussion
tRNA gene binding and transcription by pol III is absolutely required for cellular function, yet has only recently been analysed experimentally in single species on a genome-wide scale. Our global binding data in primary mouse tissues confirm reports based on cultured cell lines that pol III occupancy of tRNA genes varies considerably between cell types8,11. Nevertheless, despite the variable use of each tRNA locus, conserved pol III occupancy becomes apparent on taking into account the redundancies within the genetic code. This analysis shows that within a single species, pol III binding at the isotype level is very similar in tissues as diverse as muscle, liver, and testis. Similar correlations are found across 180 million years of evolution to those found between replicate experiments within a single species. By conducting RNA-seq in all six mammals, we discovered that a similar type of conservation appears to exist within the amino acid usage across mammals. Our data show that pol III binding at the anticodon isoacceptor level and the component triplet codon within transcriptomes must be under strong constraint at the amino acid isotype level, that is, although individual genes and isoacceptor classes vary in expression, the overall quantity of tRNAs for a given isotype is highly conserved. To our knowledge, this type of conservation has not been reported previously.
Genomic binding of pol III showed a remarkably rapid rate of divergence in mammals, largely due to genomic rearrangements, co-option, and gene duplication. We identified 24 genes that are in locations of conserved synteny and therefore likely existed in the early mammalian ancestor. Even between closely related species, such as mouse/rat or human/macaque, 33 to 58% of transcribed tRNA genes appear unique to each species. Proportionally at least, this appears to broadly reflect divergence of the tRNA genes themselves. However, the rapid changes found in functional pol III binding to tRNA genes is similar to site-specific transcriptional regulators like Oct4 and CEBPA, where mouse to human comparisons show that only a minority of binding events are conserved in orthologous locations6,7.
Our data provide evidence for regulatory crosstalk among the tRNA genes corresponding with each isotype, allowing collective output to be coordinated at the transcriptional level. It is possible that distinct regulatory sequence motifs are employed10 or local chromatin state helps direct pol III binding8,10,11. Such highly specific trans-acting communication cannot be explained by the established mechanisms of pol III regulation, which have focused on controls that are mediated through changes to TFs shared by all tRNA genes17. One possibility is that tRNA genes cluster in space according to isotype, despite their dispersal across chromosomes, allowing output from the amino acid group to be controlled independently of usage of individual group members. Spatial clustering of tRNA genes has been observed in yeast42. However, it is unclear how such clustering might overcome chromatin topological constraints or distinguish among isotypes.
Many transcription factors bind the genome tens of thousands of times; Schmidt et al. suggest that many of these binding events are functionally neutral or redundant6. Even well characterized, directly regulated TF targets can have clustered TF binding. These factors can contribute in varying degrees towards controlling transcription, which greatly complicates our understanding of the relationship between binding location and regulator function43-46. In contrast, tRNA expression originates from only a few hundred distinct locations, in genomes as diverse as drosophila33 and mammals. The limited number of actively transcribed tRNA genes, determined by pol III occupancy, allowed us to analyse the otherwise complex relationship between pol III binding and its functional role in translation. Despite conservation of the overall tRNA transcriptional program, our analysis showed that pol III occupancy and tRNA genes themselves can evolve rapidly. Thus, purifying selective pressure in tRNA expression must be operating at the amino acid isotype level, rather than at individual genetic loci.
Supplementary Material
Acknowledgements
We thank James Hadfield, Nik Matthews, Sarah Aldridge, Sara Sayalero and Claire Fielding at the Cambridge Research Institute (CRI) Genomics Core; Ben Davis, Kevin Howe and Rory Stark at the CRI Bioinformatics Core; Tony Davidge (CRI), Selina Ballantyne (CRI) and Mellissa Nixon (CFM). This work was supported by the European Research Council Starting Grant, the European Molecular Biology Organization Young Investigator Award, Hutchinson Whampoa (D.T.O.); Swiss National Science Foundation (C.K.); University of Cambridge (C.K., M.D.W. and D.T.O.); Cancer Research UK (C.K., G.D.B., S.W., M.D.W., R.J.W., and D.T.O.); and European Molecular Biology Laboratory (A.G.).
Appendix
Methods
Tissue preparation
The experiments were performed on liver material isolated from six mammals: human (Hsa; primate), macaque (Mml; primate), dog (Cfa; carnivores), mouse (Mmu; rodent), rat (Rno; rodent) and short-tailed opossum (Mdo; marsupials). For each ChIP and mRNA experiment, at least two independent biological replicates from different animals were performed. Healthy human hepatocytes (2 males, unknown age) were obtained from the Liver Tissue Distribution Program (NIDDK Contract #N01-DK-9-2310; consent forms included in contract) at the University of Pittsburgh. Macaques (2 males, 17 and 18 years old) were obtained from CFM. The dogs (2 males; 14 months of age) were obtained from Harlan, UK and rats (2 males, 9 weeks old) were obtained from Charles River, UK. Mice (2 male adult C57BL6 males, 2.5 months of age) were obtained from CRI under Home Office license PPL 80/2197. Opossums (2 adult males, 17 months of age) were obtained from the University of Glasgow, UK. All tissues were either treated post-mortem with 1% formaldehyde for ChIP experiments or flash-frozen in liquid N2 for RNA experiments.
ChIP sequencing library preparation
ChIP sequencing experiments were performed as described previously47. We used pol III antibody 190048 recognizing antigen POLR3A, the RPC1/155 subunit of pol III. Figure S14 shows this antibody’s binding of pol III around tRNA gene loci. In short, the immunoprecipitated material was end-repaired, A-tailed, ligated to the sequencing adapters, amplified by 18 cycles of PCR and size selected (200-300 bp).
Total RNA and polyA+ sequencing library preparation
Total RNA was extracted using Qiazol reagents (Qiagen) and DNase-treated (Turbo DNase, Ambion). Ribosomal RNA was depleted from total RNA using RiboMinus (Invitrogen). The remaining RNA fraction was fragmented (RNA fragmentation reagent, Ambion), polydenylated (Poly(A) tailing kit, Ambion), reversed transcribed and converted into double-stranded cDNA (Smarter PCR cDNA synthesis kit, Clontech). Adapters suitable for RNA sequencing were ligated to fragments obtained after restriction with RsaI according to the manufacturer’s protocol. Similarly, polyA+ RNA was enriched using the polyATtract mRNA isolation system protocol (Promega), fragmented, reversed transcribed and converted into double-stranded cDNA (SuperScript cDNA synthesis kit, Invitrogen), followed by paired end adapter (Illumina) ligation.
Illumina sequencing
After passing quality control on a Bioanalyzer 1000 DNA chip (Agilent), libraries were sequenced on the Illumina Genome Analyzer II (single-ended) and post-processed using the standard GA pipeline software v1.4 (Illumina).
Read mapping and NGS data analysis
Pol III reads were aligned to their reference genomes (Table S11) with BWA49 version 0.5.7, using default parameters. Reads that aligned equally well to multiple loci in the genome were assigned to a particular locus probabilistically, according to the occurrence of nearby uniquely mapped reads, as follows: If a read aligns to k loci L1 … Lk, and there are Mi uniquely mapped reads in a 200bp region around locus Li, then the read is assigned to locus Li with probability unless , in which case the read is assigned to locus Li with probability pi = 1/k.
Reads that aligned equally well to more than 20 loci were discarded. It is common to discard reads that map to multiple loci. However, since tRNA genes are frequently duplicated in mammals, in this case discarding multiply mapped reads would imply discarding most of the reads, which align to tRNA genes. Allocating them proportionally based on the relative level of nearby uniquely mapped reads more accurately reflects true pol III abundance. This method is also used by Mortazavi et al. 50.
Candidate tRNA genes were identified with tRNAscan-SE14 version 1.21. We excluded tRNA genes mapping to mitochondrial sequences from our analysis. Loci marked as pseudogenes, and those with an “undetermined” isoacceptor family were discarded. (Some loci marked as “undetermined” are highly occupied by pol III. Future work may reveal their family and/or other functions.)
Loci with at least 10 pol III reads in each of a) the mature tRNA sequence, b) 100bp upstream of the gene, and c) 100bp downstream of the gene, in at least one replicate, were considered to be actively transcribed. Flanking regions were included because they discriminate more clearly between loci that are transcribed and those which are not than do the tRNA genes themselves, due to the high similarity of multiple copies of the genes, as compared to the relatively low similarity of the flanking regions.
RNA-seq reads were aligned to their reference genomes using BWA version 0.5.7. Many reads map to multiple loci, due to the multiple copies of most tRNA genes in mammalian genomes. They were also assigned to particular loci based on the occurrence of uniquely mapped pol III in the regions around the equally best-matching loci, as described above for multiply-mapping pol III reads.
tRNA clustering was computed by counting transcribed tRNA genes within 7.5 kb of another transcribed tRNA gene. 7.5 kb was chosen because there is a natural break in the inter-tRNA distance histogram between about 5 kb and 10 kb (that is, there are many tRNA pairs closer than 7.5 kb, many further apart than 7.5 kb, but relatively few precisely around 7.5 kb).
Pol III occupancies between mouse tissues were compared by computing Spearman’s correlation coefficient for the read counts at each transcribed locus (plus 100 bp upstream and downstream) between the two liver replicates, and between liver, muscle, and testes replicates. Then pol III counts for all loci from each isoacceptor family were summed, and the summed values correlated between replicates and between tissues. Likewise, pol III counts for all loci of each isotype were summed, and correlations between those values computed.
Mapping of orthologous tRNAs
Cross-species comparisons were performed by first combining the liver pol III replicates for each species by quantile normalizing the replicates, then computing the average between the replicates, for each species. Loci were grouped by isoacceptor family and isotype class, and the corresponding sums computed for each. Then Spearman correlations were computed for anticodon families and isotypes, between each pair of species. The cross-species radar plots were made from loci grouped by isotype, then values for the isotypes for the six species were quantile normalized and the results plotted using the “radial.plot” function from the “plotrix” package of R.
To determine genomic conservation of tRNA loci, we used Ensembl’s PECAN multiple genome alignment of 16 amniotes, from Ensembl version 58, because it is the current best available (the only available) multiple species alignment which includes all the species described in this report 35. Using each species as a starting point, we retrieved via the Ensembl API the regions of each other species that aligned to that region (if any), and recorded which have a transcribed tRNA gene at that position, using our data to identify transcribed genes. By grouping these results, we computed the loci that were occupied in 1, 2, 3, 4, 5 and 6 species. The Venn diagram in Figure 4B was generated in the same manner, but without opossum.
Pol III occupancy by locus conservation was computed using the PECAN multi-genome alignments described in previous paragraphs. Each locus was classified as being species-specific, shared only with a near neighbour (i.e. human with macaque, and mouse with rat), with all five eutherians, or with all six mammals. Pol III occupancy was normalized by scaling according to total tRNA-aligned read count, on the assumption that mammals have roughly the same total pol III-occupancy of tRNA genes. Then the read counts for each species and group (species-unique, shared-with-neighbour, shared-with-eutherians, shared-with-mammals) were plotted.
UCSC BlastZ pairwise determination of conserved pol III occupancy
Because the evolutionary results we report are dependent upon the accuracy of the multi-genome alignment, we sought to validate our results by independent means. We used UCSC’s BlastZ-based pairwise nets to identify synteny blocks between pairs of species38,39,51. Top-level blocks of length at least 300Kb were considered; shorter blocks were discarded. We discarded shorter blocks to conform to the definition of sytenic blocks from the mouse-human synteny map derived from the draft mouse genome in 200252, in which 300 kb was used as the minimum block length. Then we computed, for each species, the list of tRNA loci on each block, in order of genomic position. If the syntenic blocks from the two species contained the same tRNA isotypes, in the same order, then they were considered to be unchanged, whether they appeared alignable or not. In blocks where there were changes, the longest common subsequence of isotypes from the two blocks was counted as the number of loci that had not changed, taking into account whether the entire block was reversed in one genome.
Codon usage and codon bias analysis
Raw reads of RNAseq libraries from mouse tissues50 (SRA accession SRA001030) and each species (generated in this study) were truncated to 35-mers and mapped to the corresponding transcripts sequences (cDNA sequences from Ensembl 58) using default options of Bowtie version 0.12.753. Transcripts expression estimates were obtained with MMSEQ54 and normalized using the TMM method55. Possible GC content biases were loess corrected, subtracting from the real value the difference between this and the predicted value. Codon usage and codon bias were obtained by multiplying the mean expression of each transcript with its number of anticodon isoacceptors and amino acid isotypes and summed across all transcripts.
Footnotes
Data Access
RNA-seq and Pol III ChIP-seq sequencing data are available from ArrayExpress under accession E-MTAB-424.
Input libraries used for assessing the relative efficiency of the pol III antibody are available from ArrayExpress under accesion E-MTAB-442.
The authors declare that no competing financial interests exist.
References
- 1.Chan ET, et al. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33.1–17. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124.1–.12. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–2. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourque G, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–62. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–40. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 8.Barski A, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–34. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–21. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–40. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–8. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raha D, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–44. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–45. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–46. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011 doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 18.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–77. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–55. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 23.Coughlin DJ, Babak T, Nihranz C, Hughes TR, Engelke DR. Prediction and verification of mouse tRNA gene families. RNA Biol. 2009;6:195–202. doi: 10.4161/rna.6.2.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–14. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221, 2107–15. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–55. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- 27.Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 2004;101:12588–91. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semont A, et al. Mesenchymal stem cells increase self-renewal of small intestinal epithelium and accelerate structural recovery after radiation injury. Adv Exp Med Biol. 2006;585:19–30. doi: 10.1007/978-0-387-34133-0_2. [DOI] [PubMed] [Google Scholar]
- 29.Blouin A, Bolender RP, Weibel ER. Distribution of organelles and membranes between hepatocytes and nonhepatocytes in the rat liver parenchyma. A stereological study. J Cell Biol. 1977;72:441–55. doi: 10.1083/jcb.72.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romer AS, Parsons TS. The vertebrate body. Saunders College Pub.; Philadelphia: 1986. [Google Scholar]
- 31.Gibbs RA, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 32.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 33.Rogers HH, Bergman CM, Griffiths-Jones S. The evolution of tRNA genes in Drosophila. Genome Biol Evol. 2010;2:467–77. doi: 10.1093/gbe/evq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 35.Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 2008;18:1814–28. doi: 10.1101/gr.076554.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paten B, Herrero J, Beal K, Birney E. Sequence progressive alignment, a framework for practical large-scale probabilistic consistency alignment. Bioinformatics. 2009;25:295–301. doi: 10.1093/bioinformatics/btn630. [DOI] [PubMed] [Google Scholar]
- 37.Meader S, Ponting CP, Lunter G. Massive turnover of functional sequence in human and other mammalian genomes. Genome Res. 2010;20:1335–43. doi: 10.1101/gr.108795.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiaromonte F, Yap VB, Miller W. Scoring pairwise genomic sequence alignments. Pac Symp Biocomput. 2002:115–26. doi: 10.1142/9789812799623_0012. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100:11484–9. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Man O, Pilpel Y. Differential translation efficiency of orthologous genes is involved in phenotypic divergence of yeast species. Nat Genet. 2007;39:415–21. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- 41.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 42.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstein MB, et al. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science. 2010;330:1775–87. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–5. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy S, et al. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011:27–35. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt D, et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–8. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fairley JA, Scott PH, White RJ. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003;22:5841–50. doi: 10.1093/emboj/cdg544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turro E, et al. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.