Abstract
The DpsA protein plays a dual role in Streptomyces coelicolor, both as part of the stress response and contributing to nucleoid condensation during sporulation. Promoter mapping experiments indicated that dpsA is transcribed from a single, sigB-like dependent promoter. Expression studies implicate SigH and SigB as the sigma factors responsible for dpsA expression while the contribution of other SigB-like factors is indirect by means of controlling sigH expression. The promoter is massively induced in response to osmotic stress, in part due to its sensitivity to changes in DNA supercoiling. In addition, we determined that WhiB is required for dpsA expression, particularly during development. Gel retardation experiments revealed direct interaction between apoWhiB and the dpsA promoter region, providing the first evidence for a direct WhiB target in S. coelicolor.
Introduction
A common mechanism used by bacteria to selectively modulate gene expression in response to stress involves promoter selection by alternative sigma factors. A paradigm of this regulatory strategy is the stress response regulon controlled by the transcription factor SigmaB. Initially described in Bacillus subtilis, where it controls expression of around 200 genes in response to osmotic, ethanol and temperature stresses, SigmaB orthologs have been shown to perform similar roles in other Gram positives like Staphylococcus aureus and Listeria monocytogenes [1].
The soil, a complex environmental niche where most Streptomyces species thrive, poses serious challenges to the cell's metabolic balance. Sudden modifications of salinity, moisture and temperature are only a few of these challenges, leading to the activation of complex regulatory networks controlling a myriad of genes involved in stress responses and ultimately allowing adaptation to the harsh surroundings. The response to stress in Streptomyces coelicolor has been extensively studied and a central role for a SigmaB ortholog has been identified [2]. Furthermore, the S. coelicolor genome encodes 9 SigmaB-like paralogs, probably an indication of the complex stress-response strategies imposed by its natural environment [3], [4]. In contrast with B. subtilis, where various stress conditions induce a single regulon under the control of SigmaB, proteomics studies indicate that in S. coelicolor different regulons are activated in response to specific stresses. This led to the interpretation that independent control mechanisms could govern individual stress responses. Interestingly, numerous stress-induced proteins are also developmentally controlled, suggesting a dual role for regulatory elements involved in both stress responses and development [5].
The multiple SigmaB-like paralogs encoded by S. coelicolor support the idea of ‘one SigmaB-like paralog per stress type’, but this notion has been consistently challenged as further genetic and gene expression data describing SigmaB-like sigma factors has accumulated. The characterisation of sigH expression revealed the presence of several promoters induced by heat, osmotic stress and developmental stage [6]. Moreover, sigH is also under the control of BldD, which represses its expression during vegetative growth [7]. In addition to sigH, several of the S. coelicolor SigmaB-like sigma factors are also induced by osmotic stress (sigB, L, I, K and M) while others are mainly involved in morphogenesis (sigF, sigN). The activation of multiple sigma factors in response to a specific stress suggests the existence of a much more complex and overlapping regulatory network [8]. Experimental evidence resulting from in vitro transcription experiments indicates that members of the S. coelicolor SigmaB family can recognise similar promoters [4], leading to the assumption that they have overlapping promoter specificities. In contrast to this in vitro evidence, most of the SigB-like factors in S. coelicolor are apparently quite specific at recognising promoters and are usually autoregulated, as observed when analysing the expression of target genes in the corresponding sigB-like mutants. SigH has been shown to direct transcription of one of its own promoters as well as ssgB, gltB and sigJ (SCO1276), and in all cases their expression is dramatically reduced in a sigH mutant [9]–[12]. A similar behaviour was observed when analysing the expression of sigB and several of its targets identified from bioinformatics and transcriptomics analyses [13], [2], while SigN is autoregulated and controls the in vivo expression of the morphogenetic protein NepA [14]. Furthermore, a regulatory cascade of SigmaB-like factors has been inferred from microarray experiments. Based on induction timing in response to osmotic stress control hierarchies of sigI→sigB→sigM and sigK→sigH→sigL sequence were proposed [8], although a similar study by Lee and colleagues implicates sigB (referred to as sigJ by some authors) as a master regulator, acting at the beginning of a putative cascade consisting of sigB→sigL→sigM [2]. The latter is further supported by a genome wide search using a consensus SigB-dependent promoter sequence, which identified several putative SigB targets including SigL [13].
We have recently described the functional role of DpsA; a nucleoid associated protein whose expression is strongly induced by osmotic and heat stresses. DpsA also contributes significantly to nucleoid condensation during reproductive growth in S. coelicolor, together with its two paralogs DpsB and DpsC [15]. Our initial expression analyses did not reveal a clear dependence between dpsA expression and SigB, despite the fact that DpsA orthologs are part of stress regulons in other bacteria (E. coli, M. smegmatis) and in Bacillus subtilis its expression is part of the general stress response controlled by SigB [16].
Here we describe how dpsA is regulated in response to stress and during development. Stress-dependent expression of dpsA is dependent on a regulatory cascade involving SigB-like sigma factors in S. coelicolor. A single promoter drives dpsA expression and is a target for both SigB and SigH. We also identify a role for DNA supercoiling and the WhiB transcription factor in regulation of dpsA, indicating how developmental and stress-dependent regulation mediated by these sigma factors can be finessed from a single promoter.
Results
dpsA is transcribed from a single SigB-like dependent promoter
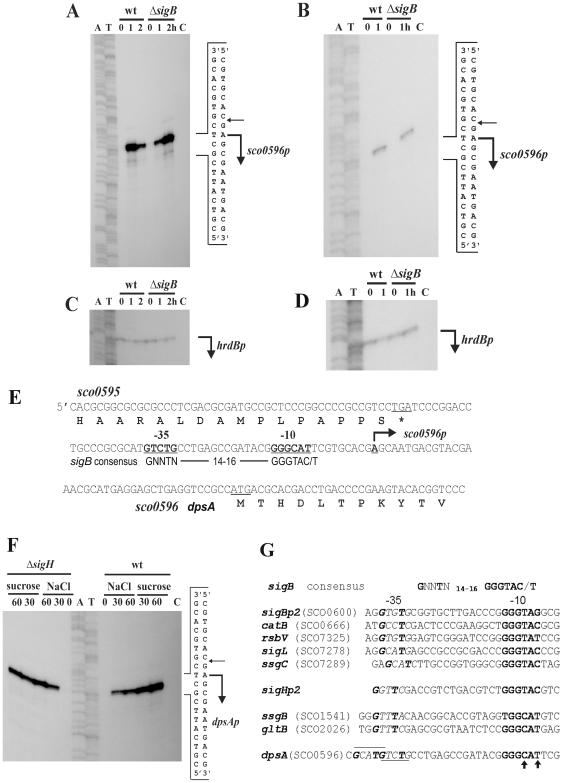
Our initial studies using both Quantitative Real-Time PCR (qRT PCR) and immunoblots revealed that dpsA expression is strongly up-regulated in response to osmotic up-shift and high temperature [15]. High resolution S1 protection assay experiments performed using total RNA extracted from S. coelicolor M145 grown in MS agar and MS agar containing 250 mM KCl confirmed that dpsA expression is strongly induced by osmotic stress from a single transcription start point (Figure 1A and C), while transcripts are almost undetectable in the non-stressed sample. Similar experiments using cells grown under heat shock (42°C) also showed induction of expression from the same transcription start point (Figure 1B and D). In both cases total RNA samples isolated from a sigB mutant grown under the conditions described above were processed in a similar manner, revealing that transcription still proceeds from the same transcription start point (Figure 1A and B). The only noticeable difference between the parental strain and sigB mutant is the apparent delay in dpsA induction by 250 mM KCl in the latter. While in M145 the highest induction is observed after 1 hour of osmotic up-shift, in a sigB mutant comparable induction levels are only reached after 2 hours, which suggests that in the absence of SigB the observed induction could be mediated by another sigma factor which either binds with less affinity to the dpsA promoter or is induced to the required levels after longer exposure to high salt concentration. The putative −10 and −35 sequences were identified and shown to resemble SigmaB-dependent consensus promoters (Figure 1E). We also explored dpsA expression in a sigH mutant, as this SigB-like sigma factor is known to be induced by both osmotic stress and heat [6]. Osmotic stress caused by NaCl and sucrose resulted in an increase of dpsA transcription in a sigH mutant from the single promoter described above (Figure 1F).
Figure 1. High-resolution S1-nuclease mapping the transcription start point (TSP) of S. coelicolor dpsA (SCO0596).
Bent arrows indicate the positions of RNA-protected fragments. A: Total RNA from S. coelicolor M145 and its isogenic sigB mutant grown for 17 h on a cellophane disc on top of MS agar (lane 0), then transferred to MS containing 250 mM KCl for 1 h (lane 1) and 2 hours (lane 2). B: Total RNA from cells grown as above but transferred for 1 h to 42°C (lane 1). C and D: Control S1-nuclease mapping experiments with the same RNA samples using a DNA probe for the hrdBp promoter. E: Nucleotide sequence of S. coelicolor M145 SCO0596 promoter region. The deduced protein product is shown below. The TSP of the SCO0596 promoter is indicated by the bent arrow. The proposed −10 and −35 boxes of the promoter are in bold characters and underlined. F: S1-nuclease protection assay using RNA isolated from S. coelicolor M145 and sigH mutant (as indicated above the figure), grown for 20 h in liquid minimal NMP+0.5% mannitol medium (lane 0) and osmotic stress induced by addition of NaCl (final concentration 0.5 M) or sucrose (final concentration 1 M) and incubated for 30 min (lane 30) and 60 min (lane 60). In all cases lane C is E. coli tRNA, used as negative control G: Promoter sequences of consensus sigB-like, sigBp2, sigHp2 and several promoters known to be controlled by SigB or SigH respectively. The underlined sequence in dpsA promoter indicates putative −35 sequences and arrows indicate modifications from consensus −10 sequence.
Careful examination of the −10 sequence of the dpsA promoter reveals subtle differences when compared to its equivalent in SigB-dependent consensus promoters. The consensus −10 sequence GGGTAC/G changes to GGGCAT (T→C, C/G→T) in dpsAp, similar to −10 sequences from genes known to be transcribed by SigH (ssgB, gltB). This suggests that dpsA could be a direct target for SigH regulation, rather than a member of the SigB regulon (Figure 1G). It is also noticeable that there are two putative −35 sequences in dpsAp, depending on a spacer length of 14 or 18 nucleotides between the −10 and −35 sequences.
Stress-induced control of dpsA expression does not depend on a single SigB-like transcription factor
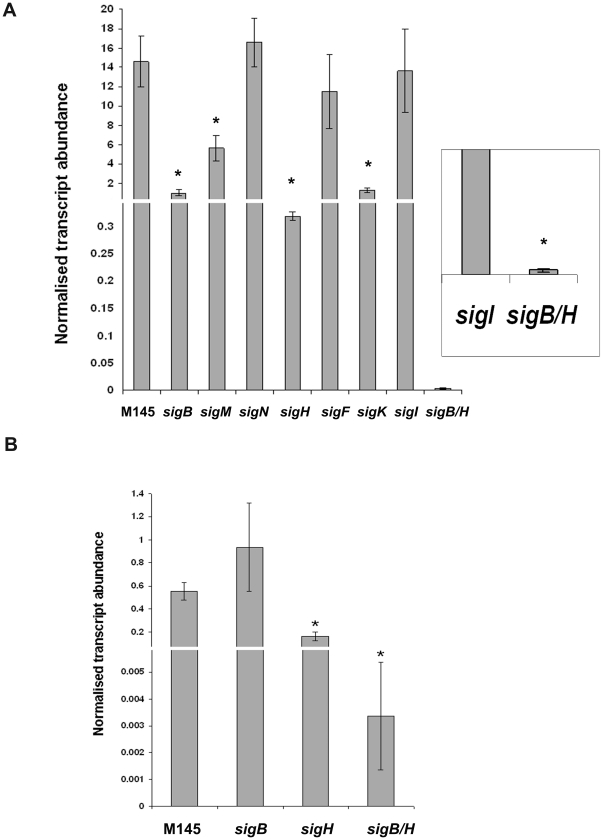
We used qRT PCR in order to determine precisely how expression levels from the single, sigB-like, dpsA promoter are affected in sigB-like mutants. Total RNA was extracted from S. coelicolor M145 (wild type), sigB, sigH, sigI, sigK, sigM, sigN, sigK, sigF and sigB/H mutants grown on cellophane discs placed on the surface of MS plates; incubated for 16–18 hours and then transferred to MS plates containing 250 mM KCL. In all cases a set of MS plates was kept as a non-stressed control. Cells were collected after 1 hour of further incubation, RNA extracted and cDNA synthesised. The relative abundance of dpsA transcripts was determined by qRT PCR as described [15] using dpsA gene specific primers and hrdB as internal, normalising control. Basal, non-induced, dpsA expression levels are very low and upon osmotic shock dpsA transcript abundance dramatically increases in all strains (ranging from 10-fold to 80-fold). The normalised dpsA basal expression level for each strain under study was subtracted from the corresponding induced expression levels detected, in order to ensure that only differences in dpsA expression resulting from osmotic shock were scrutinised. Wild-type dpsA induction levels were observed in sigI, sigN and sigF mutants. Although in both sigB and sigH mutants dpsA was induced by osmotic stress, interestingly transcript abundance never reached the levels attained in the parental strain M145 (Figure 2A) and were particularly low in a sigH mutant. Statistical analyses (one way Anova) revealed that the observed differences between the S. coelicolor M145 strain and the sigB, sigH, sigM, sigK and sigB/H mutants were indeed significant. Remarkably, in a sigB/H mutant dpsA expression remained almost non-induced by osmotic stress, indicating an absolute requirement for both SigB and SigH during dpsA osmotic stress up-regulation. No induction was observed in the sigB/H mutant despite the prolonged incubation under stress (not shown). This result also confirms that both SigB and SigH are responsible for dpsA expression and none of the remaining SigB-like elements can replace them functionally.
Figure 2. Quantification of dpsA transcript abundance in response to osmotic stress.
qRT PCR monitoring dpsA expression levels after induction by 250 mM KCL in S. coelicolor M145 and sigB, sigH, sigM, sigN, sigF, sigK, sigI and sigB/H (inset) mutants (A). dpsA expression in S. coelicolor M145, sigB, sigH and sigB/H mutants after 1 hour of incubations at 42°C (B). * indicates significant differences with equivalent S. coelicolor M145 sample (One Way Anova, P<0.05). Broken Y axis has been used. Error bars indicate standard deviation.
Since dpsA induction levels were significantly affected in a sigH mutant background either as a single or double mutant, we further investigated the role of this sigma factor in dpsA control. Heat-mediated dpsA induction was examined in S. coelicolor M145 parental strain and sigB, sigH and sigB/H mutants. dpsA expression was up-regulated to similar levels by heat in both M145 and sigB mutant strains, although transcript abundance never reached the levels observed during osmotic stress induction. In a sigH mutant there is a three-fold reduction on dpsA induction level after 1 hour at 42°C when compared to the M145 strain, while in a sigB/H mutant dpsA expression is heavily compromised in both control and heat treated samples, reminiscent of the lack of induction by osmotic stress on this double mutant (Figure 2B). It is noticeable that while SigB is required for full dpsA osmotic stress induction, the lack of this sigma factor alone does not affect heat-induced activation of the dpsA promoter, which is dependent on a functional SigH.
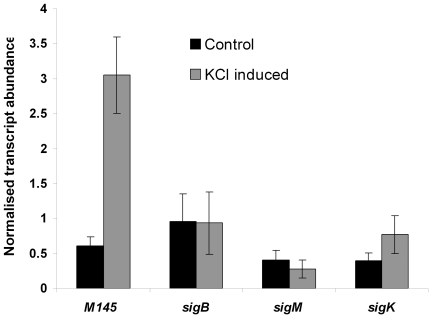
It is evident from the above results that SigH plays a key role in dpsA stress mediated induction, although this role may be modulated by SigB or other SigB-like factors. We used qRT PCR to assess the expression of sigH after osmotic stress in the parental strain and in different mutant strains deficient in SigB-like sigma factors known to be induced by osmotic stress [8]. A marked sigH induction by KCl was observed in the parental M145 strain, while this induction was abolished or reduced in sigB, sigM and sigK mutants (Figure 3), suggesting that these sigma factors influence directly or indirectly sigH osmotic induction. Basal sigH transcript abundance remained within comparable levels, unaffected by the loss of the SigB-like factors. Combined with the dpsA expression studies described earlier (Figure 2A), these results provide an explanation for reduced dpsA osmotic induction observed in the sigM and sigK mutants, due to a reduced induction of sigH, which is required for proper dpsA activation. The observed reduced sigH expression in the sigB mutant is likely both direct and indirect, as the latter is able to drive dpsA induction in the absence of SigH but is also required for sigM expression [2] and putatively for sigH expression as suggested by the presence of a SigB-consensus promoter in sigH (sigHp2, Figure 1G) and our own observations.
Figure 3. sigH osmotic induction depends on SigB-like factors.
qRT PCR monitoring sigH expression levels after induction by 250 mM KCL for 1 hour in S. coelicolor M145, sigB −, sigM −, and sigK − strains. Error bars indicate standard deviation.
This cascade explains the reduced dpsA expression in a sigB mutant, where reduced sigH expression results in diminished dpsA activation. As disruption of sigH does not completely abolish dpsA osmotic induction, an alternative sigma factor functionally replaces SigH. SigB is a plausible candidate, as dpsA induction is totally abolished in a sigB/H double mutant.
Inhibition of DNA gyrase affects dpsA osmotic stress induction
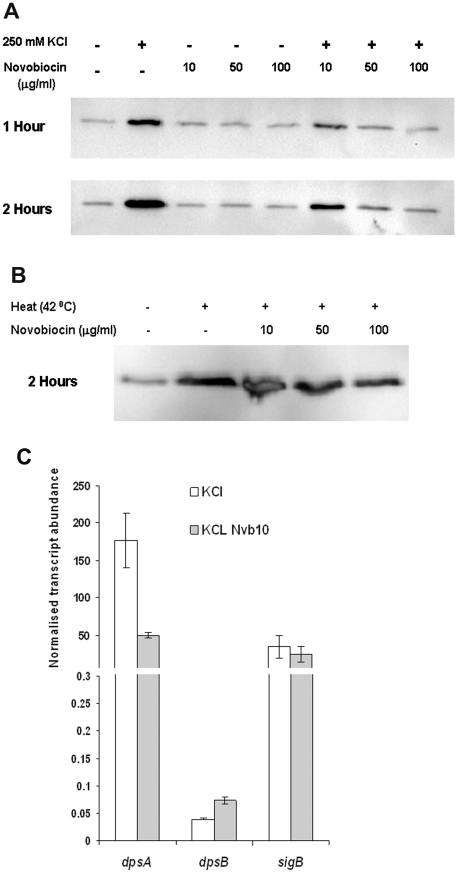
Osmotic stress can result in an increase in negative DNA supercoiling, and the change in DNA topology can directly modify transcription of specific genes [17], [18]. Novobiocin, a gyrase B inhibitor, was used to determine if an increase in negative DNA supercoiling resulting from osmotic stress contributed (directly or indirectly) to DpsA up-regulation, monitored using a C-terminal translational fusion to 6 Histidines under the control of dpsA native promoter. Western blot experiments revealed that DpsA basal expression levels remained unchanged as a result of novobiocin treatment (Figure 4A), indicating that constitutive expression is insensitive to gyrase B inhibition. In contrast, osmotic stress induction was significantly modified by the antibiotic in a dose dependent manner. At the lowest concentration of 10 µg/ml novobiocin some up-regulation of DpsA was observed, although lower than in the untreated control. At higher concentrations DpsA abundance remained similar to non-induced levels (Figure 4A). This indicated a requirement for active gyrase B and hence an increase in negative DNA supercoiling for induction of dpsA expression after osmotic stress. A similar experiment analysing heat-stress induction of DpsA revealed that this up-regulation is independent of DNA negative supercoiling, as novobiocin treatment had no apparent effect even after 2 hour incubation (Figure 4B).
Figure 4. Negative DNA super-coiling contributes to dpsAp induction by osmotic stress.
A: Increasing concentrations of novobiocin abolish DpsAHis induction in the presence of osmotic stress but basal expression levels remain unchanged. B: Heat dependent induction of DpsAHis is independent of novobiocin treatment. S. coelicolor dpsA −/pDpsA7H was used for both experiments. Time indicates incubation period under stress in the presence or absence of novobiocin at concentrations shown. C: qRT PCR showing decrease in salt-induced dpsA transcript abundance in response to novobiocin treatment. dpsB and sigB transcript abundance under the same conditions was also determined. Error bars indicate standard deviation.
To establish that the observed novobiocin effect was specific to dpsA expression and not a consequence of global reorganisation of gene expression due to reduced DNA supercoiling, we grew S. coelicolor M145 on MS agar for 16 h and then transferred it to MS agar/250 mM KCL and MS agar/250 mM KCL/10 µg/ml novobiocin. Total RNA was isolated, converted to cDNA and used as template to perform qRT PCR to quantify dpsA transcript levels. As negative controls we monitored the expression levels of dpsB and sigB. The novobiocin treatment caused a reduction in dpsA transcript abundance by a third as compared with the untreated sample, while expression levels of dpsB and sigB remained unaffected, confirming that dpsA expression is indeed influenced by topological changes in DNA (Figure 4C). As an additional control, we quantify the expression of 16S rRNA transcript, which remained also unaffected by the novobiocin treatment even at the higher concentrations (not shown).
Developmental control of dpsA expression depends on SigH and SigB and requires WhiB
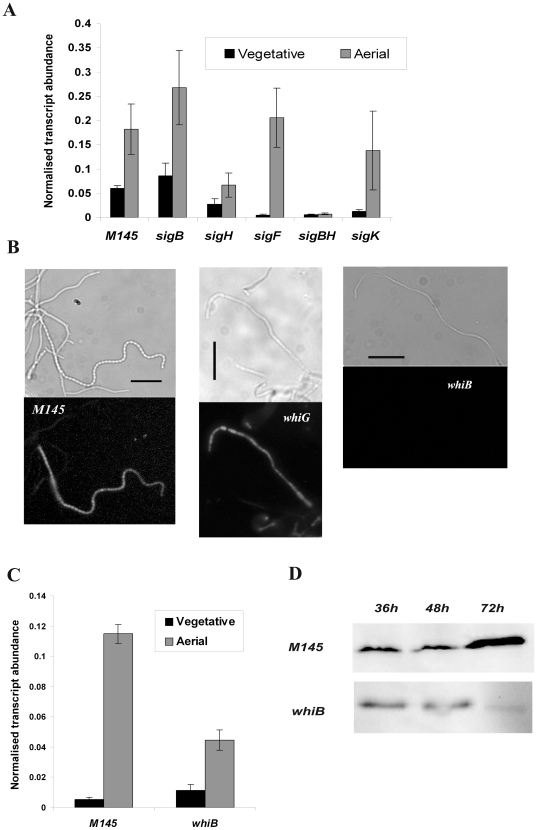
The dpsA gene is developmentally controlled, as its expression is drastically up-regulated during sporulation [15]. SigH is an obvious candidate to modulate such up-regulation as it is known to exert developmental control [10] and is also required for normal aerial development [9]. We performed qRT PCR experiments to monitor and compare dpsA expression in vegetative and aerial hyphae from S. coelicolor M145 and mutant strains (sigH, sigK and sigF, all of which are known to be involved in aerial development). We also included sigB and sigB/H mutants. The sigN mutant was not included in this study because our previous work [15] showed that dpsA expression is not supported in the sub-apical compartment, opposite to what has been described for the SigN target nepA [14]. Developmentally controlled expression of dpsA in sigB, sigK and sigF mutants remained similar to that of the parental M145 strain, while a reduced expression was detected in sigH aerial hyphae. Remarkably, developmental up-regulation of dpsA was abolished in a sigB/H mutant, reminiscent of the absence of stress induction in this mutant strain and indicating that no other sigma factor can replace SigB or SigH (Figure 5A).
Figure 5.
Developmentally controlled expression of dpsA in S. coelicolor M145, sigB −, sigF −, sigK − and sigB/H − strains assessed by qRT PCR (A). Bright field and corresponding fluorescence image showing DpsAmCh expression in aerial hyphae of S. coelicolor M145, whiG − and whiB − strains. Bar: 10 µm (B). qRT PCR showing dpsA transcript abundance in vegetative and aerial hyphae of S. coelicolor M145 and whiB mutant. Error bars indicate standard deviation. (C). Immunoblot comparing DpsAHis abundance in S. coelicolor M145 and whiB − strain throughout the developmental life cycle. Similar amounts of total protein were loaded in each lane (D).
We used a dpsA:mCherry translational fusion (DpsAmCh) under the control of the dpsA native promoter to determine in situ DpsA expression in aerial hyphae from two early whi mutants (whiG and whiB). An integrative PhiC31-derived plasmid (pDpsA6A) carrying the fusion was conjugated into the mutants under study and the expression of DpsAmCh visualised using fluorescence microscopy as described [15]. Interestingly, while a whiG mutant supports normal dpsA expression in aerial hyphae, a requirement for a functional WhiB was detected as we could not visualise red fluorescence due to DpsAmCh in this mutant (Figure 5B). This result was corroborated by a qRT PCR experiment using RNA extracted from whiB aerial hyphae that revealed reduced dpsA expression in this mutant (Figure 5C). Although reduced, dpsA mRNA expression levels in aerial hyphae are higher than those observed in vegetative cells. This suggests that additional mechanisms, perhaps involving post-translational DpsA processing, are in place in a whiB mutant. We further analysed this WhiB dependence by introducing into the whiB mutant an integrative plasmid encoding a dpsAHis translational fusion under the control of the dpsA native promoter (pDpsA7A, [15]). Total proteins were isolated from the resulting strain after growth on MS agar at different time points until aerial development was evident. Similar amounts of total protein were assessed by Western blot using an anti-His antibody in order to monitor the abundance of DpsAHis. A clear reduction of DpsAHis levels was observed after the onset of aerial growth (72 hours), confirming our observation of reduced DpsA levels in whiB aerial hyphae (Figure 5D).
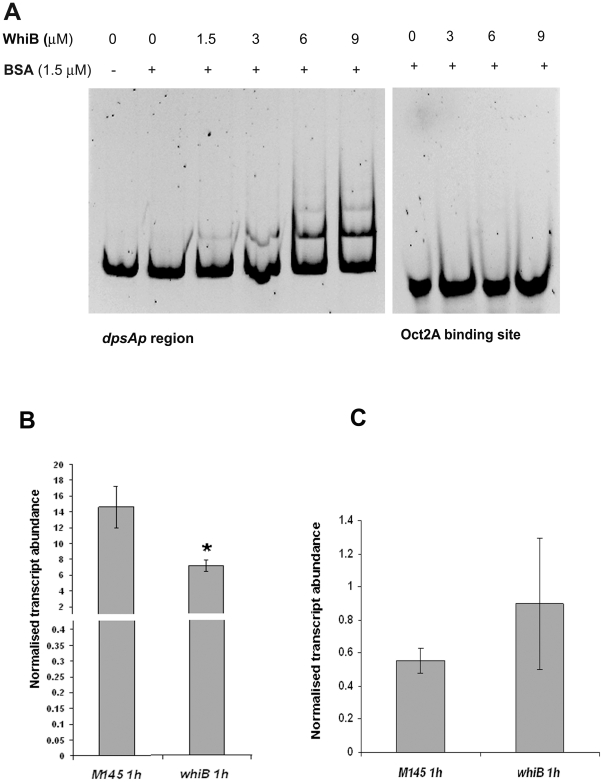
Evidence for direct interaction of WhiB with the dpsA promoter region was provided by gel retardation experiments. A PCR fragment, encompassing a region from the dpsA start codon up to 417 bp upstream DNA sequence, was amplified using primers P1DpsAF1 and P1DpsAR1 and 20 ng were mixed with recombinant apoWhiB at various concentrations (0–9 µM). After 30 minute incubation at room temperature the protein-DNA mix was electrophoresed in a 6% Acrylamide gel, followed by Syto9 staining (Electrophoretic Mobility Shift Assay, Invitrogen). The stained gels were visualised under UV light and an image recorded. As negative control a parallel experiment was carried out using a 39-mer oligonucleotide containing the binding site for Oct2A (Roche). Figure 6A shows the shift in electrophoretic migration of the dpsA promoter region caused by apoWhiB, while the negative control remained unaffected.
Figure 6. Increasing concentrations of apoWhiB causes electrophoretic shift of dpsAp region.
A double-stranded oligonucleotide containing the OctA2 binding site was used as negative control (A). qRT PCR monitoring dpsA expression levels in S. coelicolor M145 and whiB mutant in response to osmotic stress (B) and heat stress (C). Significant differences in dpsA expression levels (One way Anova, P<0.05) were detected between S. coelicolor M145 and whiB − strain after 1 hour of osmotic stress (*). Error bars indicate standard deviation.
We also tested dpsA KCl-mediated induction in a whiB mutant using qRT PCR. The induced levels of dpsA transcript in the whiB − strain are lower (∼two fold reduction) than in the parental M145 strain (Figure 6B). This result indicates that the loss of a functional WhiB protein affects dpsA osmotic induction, albeit mildly. The fact that a noticeable dpsA osmotic induction is still detected suggests that other factors may functionally replace WhiB or that WhiB only has a minor contribution to the dpsA up-regulation by osmotic stress. We failed to detect up-regulation of whiB transcript in response to osmotic stress, or a dependence on SigB or SigH (not shown). This suggests that the contribution of WhiB to dpsA induction during osmotic stress is independent of SigB and SigH control, and constitutes an additional regulatory switch. Similar experiments revealed that WhiB is not required for heat dependent induction of dpsA (Figure 6C), confirming earlier observations that indicate the existence of alternative regulatory strategies to activate dpsA in response to different stresses.
Discussion
The osmotic stress response in Streptomyces coelicolor is a complex process involving numerous regulatory elements among which SigB-like sigma factors play a central role, together with their cognate anti and anti-anti sigma factors. This network is only superficially understood, as the existence of co-regulation and interaction among its components makes it a challenging puzzle to assemble. Available experimental evidence indicates that SigB may act as a ‘master regulator’ of the osmotic stress response while regulating the expression of many genes, among which there are at least two SigB-like sigma factors, SigL and SigM [2].
Our initial studies characterising S. coelicolor Dps paralogs revealed a clear link between dpsA expression and the osmotic stress response [15], corroborating previously published data reporting dpsA as a target for regulation by SigB [2]. Indeed, promoter mapping experiments confirmed the existence of a single promoter driving dpsA expression resembling SigB-like dependent promoters, in particular those transcribed by SigH. Interestingly the dpsA promoter is dependent on both SigB and SigH for full induction by high osmolyte concentration, which in turn is abolished in a sigB/H double mutant. Other SigB-like factors like SigM and SigK are also needed to achieve full dpsAp osmotic induction, as its expression is significantly reduced in the corresponding mutants. Analysis of sigH expression in response to stress revealed a dependence on various sigB-like sigma factors, namely SigB, SigM and SigK. This expression profile supports the proposed cascade governing SigB-like sigma factors in S. coelicolor, where SigB acts early on in response to osmotic stress and regulates the expression of its targets among which is sigM, which in turns controls sigH. The existence of a sigH promoter identical to those recognised by SigB strongly supports the existence of a direct regulatory link between SigB and sigH expression as well, as shown by our experiments. The up-regulation of dpsA expression by heat is mainly dependent on SigH, in contrast with the dual regulation exerted by both SigB and SigH during osmotic stress. This difference can be explained by the presence of a heat inducible promoter of sigH, driving expression independently from the salt stress induced sigHp2 promoter that we propose is controlled by SigB.
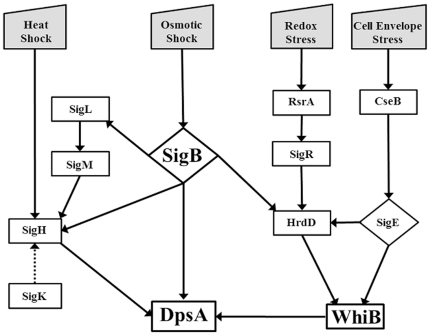
Our data suggests that sigH is also regulated by SigK, although it is not possible to determine if it is a direct or indirect control. Similarly, we cannot overlook the possibility of SigB-dependent control of sigK expression, although the published or available transcriptomics experiments analysing expression in a sigB mutant have so far failed to provide evidence for such connection ([2]; Stanford Microarray Database). We explored this idea by monitoring sigK expression in sigB and sigH mutants using qRT PCR, but failed to detect any differences on sigK expression levels in response to osmotic stress in those strains (not shown). The lack of dpsA induction by high osmolyte in a sigB/H double mutant further reinforces our interpretation that SigH and SigB are the main modulators of dpsA expression. SigH likely regulates dpsA directly, as they are both induced by the same signals (salt stress, heat and development). In contrast to sigH, dpsA has a single promoter, so the only possible way it can share inducing signals (osmotic and heat stresses) with sigH is by being its direct target. SigB is able to replace SigH and drive dpsA induction, although less efficiently possibly due to having less affinity for the promoter sequence. A model for this regulatory cascade places SigB as a main modulator of various sigma factors (SigL, SigM, SigH), which results in indirect regulation of dpsA expression, but also acting directly on the dpsA promoter, although less efficiently. This multilayered regulatory network allows the integration of multiple signals leading to the activation of a specific gene. SigH may act as a node integrating multiple signals and mediating expression of specific genes in response to various stresses (Figure 7).
Figure 7. Model depicting the regulatory network controlling dpsA expression.
Only stress induced elements are shown. Solid lines indicate experimentally verified relationships (direct and indirect) while the dotted line indicates a partially verified one.
Various authors have suggested that SigB-like factors have overlapping promoter specificities, in other terms are able to ‘cross talk’. To our knowledge the experimental evidence for the cross talk is based on in vitro transcription experiments on the B. subtilis promoter Pctc, which can be transcribed by both SigB and SigH [4]. Our experimental work confirms that at least two SigB-like factors (SigB and SigH) modulate dpsA induction directly, offering the first in vivo experimental evidence for this ‘cross talk’.
The need for stimulus-specific responses to an extremely variable and nutrient depleted environment like the soil calls for energy efficient mechanisms of stress response. Individual sigma factors reactive to specific challenges and able to mediate transcription of defined regulons could permit fine-tuning of specific stress responses. On the other hand, diverse stimuli may require the induction of stress response elements able to protect the cell in a variety of ways. DpsA is one such element, able to protect against oxidative stress by preventing free radical formation and shielding DNA from damage, while contributing to DNA condensation during sporulation [15]. Our data supports the idea of dpsA been modulated primarily by SigH, but its activation in response to stress can also be mediated by SigB. Rather than relying on gene expression driven from multiple promoters, each recognised by specific sigma factors and induced by specific stimuli; a sigma factor cascade modulates and integrates various environmental signals. This leads to the induction of well defined and specialised regulons, but is flexible enough to converge in a global stress response element like DpsA. The role of sigma factor antagonists within this model should also be considered, and must be included in future experimentation attempting to offer a more complete view of the regulatory network controlling stress response in Streptomyces.
Although dpsA is induced by both osmotic up-shift and heat in a SigH dependent manner, the resulting expression levels are very different for each stimulus, namely much higher for salt induced expression. We found evidence for a mechanism that contributes to the difference in induction levels, providing an additional layer of gene expression control. Differences between osmotic and heat stress induction of dpsA are in part the result of changes in DNA topology, in particular an increase in negative supercoiling dependent on the activity of gyrase B. An increase in negative supercoiling as a result of osmotic stress is well documented and topoisomerase gene promoters are sensitive to changes in DNA topology. The presence of two potential −35 sequences in dpsAp (separated from the −10 sequence by 14 and 18 nucleotides respectively) may explain such topology sensitivity. An increase in negative supercoiling in response to osmotic stress may bring the closer −35 sequence out of phase from the −10, affecting recognition by the corresponding sigma factor. The existence of a second −35 sequence further apart would compensate for the change in promoter topology, as it will ensure that a suitable −35 is always in phase with the −10 sequence and will ensure a sustained expression of dpsA. Exploring the relative contribution of each −35 sequence to dpsAp activity extends beyond the scopes of this paper, but surely constitutes an exciting proposition. However, the regulation of dpsA expression after heat shock is independent of this DNA topology-dependent mechanism, implicating at least two parallel stress-dependent regulatory systems influencing expression of this gene.
Developmental control of dpsA expression depends primarily on SigH and SigB. An important finding is the dependence of dpsA on WhiB for developmentally controlled expression, the first reported target for this transcription factor in S. coelicolor. dpsA expression in aerial hyphae was heavily compromised in a whiB mutant, and we confirmed in vitro binding of apoWhiB to the dpsA promoter region. Osmotic induction of dpsA is reduced in a whiB mutant, expanding the role of this transcription factor beyond a mere development-related switch and into a modulator of gene expression during stress. There are several whiB paralogs encoded by S. coelicolor genome, and they may also contribute to dpsA control during osmotic stress, similar to the combined action of SigH and SigB. We have performed searches on publicly available microarray data (Stanford Microarray Database) and found two wbl (whiB like) genes whose expression is induced by osmotic stress (SCO5190 and SCO7306) and therefore likely subjects for future studies assessing their potential dpsA regulatory role.
These observations fit with published data investigating in vitro expression of genes regulated by SigB-like sigma factors. The developmentally controlled gene nepA is not transcribed by SigN in vitro despite the uncontroversial evidence for nepAp dependence on SigN in vivo [14]. The whiEVII promoter sequence also fails to be transcribed by SigF in vitro [19], and in both cases the existence of an additional transcriptional activator has been suggested. We propose that WhiB or a WhiB-like paralog is (are) responsible, together with the corresponding SigB-like factor, for the up-regulation of the above genes in aerial hyphae, just as we observed for dpsA. Similarly, the existence of whiB-like (wbl) genes induced by osmotic stress suggests that they may play a similar role during osmotic stress. Moreover, additional stress factors, such as redox stress or cell envelope stress, are likely to occur concomitantly during conditions of osmotic stress (see Figure 7). In this respect we note that whiB can be expressed from one of two promoters [20], [21] and is developmentally regulated by BldD [22]. The first promoter requires HrdD which may be involved in coordinating “cross talk” from osmotic (via SigB), redox (via SigR) and cell envelope (via SigE) stress sensing systems [21], [23], [2]. The second requires SigE, part of a multicomponent system directly involved in monitoring changes in the integrity of the cell envelope [24]. Thus WhiB, like other WhiB-like proteins, presumably senses stress induced changes in the intracellular redox status of the cell via an [FeS] cluster, leading to an enhanced DNA binding affinity [25]–[28]. This would allow fine tuning of dspA gene expression as a result of the dual action of the SigB (or SigB-like) and WhiB (or WhiB-like) proteins in response to varying degrees of osmotic stress. We intend to continue exploring the putative connection between SigB-like and WhiB-like factors, particularly the role played by the former in the expression of the latter, in order to identify dependence on each other while controlling their putative gene targets in response to stress and developmental stage.
In summary, we have dissected the dpsA expression control mechanism and shown that two sigma factors (SigB and SigH) are able to drive dpsA expression in response to stress and during developmental differentiation, and also how expression levels can be modulated by additional transcription factor(s) (WhiB) and DNA topology status. The results described here revealed how a single promoter can be the subject of multiple regulatory factors in response to a variety of stress signals, leading to finely tuned levels of gene expression.
Methods
Bacterial strains and media
Streptomyces coelicolor A3 (2) and E. coli strains are listed and described in Table 1. All cloning procedures were performed in E. coli JM109, while E. coli ET12567/pUZ8002 was used for intergeneric conjugative transfer of plasmid DNA into Streptomyces strains [29]. Gene replacement experiments were performed in BW25113 (pIJ790) strain as described [30]. Culturing of E. coli strains was as recommended [31]. S. coelicolor strains were grown at 30°C on the surface of MS (mannitol soya flour) agar and/or on cellophane discs [29]. For osmotic up-shock, MS agar was supplemented with 250 mM KCl. Streptomyces mutant strains were obtained using Tn5062-mutagenised cosmids as described ([32]; Table 1), the double sigB/sigH mutant was created by disruption of sigB using an apramycin resistant Tn5062 mutagenised cosmid in an existing thiostrepton resistant sigH mutant [9]. The identity of all mutants was confirmed by Southern blot [31].
Table 1. Bacterial strains, plasmids and cosmids.
| Strain or plasmid | Description | Transposon insertiona (genome position), Genbank accession | Reference/Source |
| Strains | |||
| S. coelicolor A3(2) M145 | Prototrophic SCP-1 SCP-2 Pgl+ | [29] | |
| DSCO0600, sigB− | M145 sigB−::Tn5062 (apra) | SC5G5.1.C05 (639940) | [35] |
| K101, sigF− | M145 sigF−::apra | [14] | |
| sigH− | M145 sigH−::thio | [9] | |
| DSC03068, sigI− | M145 sigI−::Tn5062 (apra) | 7F11.01.F06 (3361076) | This study |
| DSCO7314, sigM− | M145 sigM−:: Tn5062 (apra) | SC5F8.2.C11 (8120634) | This study |
| sigN− (K100) | M145 sigN−::apra | [14] | |
| sigK− | M145 sigK−::kan | [36] | |
| DSCO0600/DSCO5243, sigB/H | M145 sigB−::apra, sigH::thio | This study | |
| J2402, whiB− | M145 whiB−::hyg | [37] | |
| J2400, whiG− | M145 whiG−::hyg | [37] | |
| JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14- gyrA96 recA1 relA1endA1 thi hsdR17 | [38] | |
| ET12567 (pUZ8002) | dam13::Tn9 dcm6 hsdM hsdR recF143 16zjj201 ::Tn10 galK2 galT22 ara14 lacY1 xyl5 leuB6 thi1 tonA31 rpsL136 hisG4 tsx78 mtli glnV44, containing the non-transmissibleoriT mobilizing plasmid,pUZ8002 | [39] | |
| BW25113 (pIJ790 | K12 derivative: deltaaraBAD, deltarhaBAD containing lambdaRED recombination plasmid pIJ790 | [30] | |
| Plasmids | |||
| pQM5062 | pMOD+Tn5062, AmpicillinR and AramycinR | AJ566337.1 | [32] |
| pDpsA6A | dpsA::mCherry, ApramycinR | This study | |
| pDpsA6H | dpsA::mCherry, HygromycinR | [15] | |
| pDpsA7 | dpsA::His6, ApramycinR | [15] | |
| pDpsA7H | dpsA::His6, HygromycinR | [15] |
Access http://strepdb.streptomyces.org.uk/ for information about transposon insertion details.
DNA manipulation and plasmid construction
All plasmids are listed in Table 1. DNA manipulation and cloning were carried out following standard protocols [31] using E. coli JM109 as a host. Plasmids were verified by restriction analysis and sequencing, and introduced in Streptomyces strains by intergeneric conjugation.
An apramycin resistant version of plasmid pDpsA6H [15] was created by replacing the hygromycin-resistance marker using the PCR targeted system [30] with the apramycin gene from plasmid pQM5062 digested HindIII. The resultant plasmid, pDpsA6A, was used for conjugal transfer into hygromycin resistant mutant strains. Plasmid pIJ6999 was used for recombinant expression of WhiB (C. den Hengst, JIC, personal communication). Briefly the whiB coding sequence was amplified by PCR using primers WB1 and WB2 (Table 2), which contain NdeI and BamHI recognition sequences respectively. The PCR product was cloned into pET15b, resulting in pIJ6999. PCR amplifications were performed using the high fidelity polymerase Pfu (Promega), following the manufacturer's recommendations.
Table 2. Oligonucleotides.
| Name (target gene) | Sequence (5′ to 3′ direction) |
| P1DpsAF1 | TAGATATCCCATGCTCGGTGAGACCGACG |
| P1DpsAR1 | TACATATGGGACCTCAGCTCCTCATGCG |
| WB1 | GCGCATATGACCGAGCTGGTGCAGC |
| WB2 | GTTGGATCCGCCGCGTGGGGCGGC |
| hrdBFor | CCTCCGCCTGGTGGTCTC |
| hrdBRev | CTTGTAGCCCTTGGTGTAGTC |
| 0596RTF1 (dpsA) | AGCGGAAGTGGGACGACTAC |
| 0596RTR1(dpsA) | TCAGAAGGTCCTCGGTGGC |
| whiBRTF1 | ACCCCGAGTCCTTCTTCC |
| whiBRTR1 | ATTCGGAGCGGACCTCAC |
| sigHQRT2F | CCCTGGACGACCTGACC |
| sigHQRT2R | GGAAGTGCCGCTTGATCTC |
High-resolution S1-nuclease protection assay
Promoter mapping experiments were performed as described [9]. The probe used to analyze dpsA promoter region was amplified by PCR using a 5′-32P-labeled reverse primer SCO0596R (located in the SCO0596 coding region 100 bp downstream the start codon) and the unlabeled forward primer SCO0569F, binding upstream of SCO0595 gene (ca. 70 bp upstream of SCO0595 start codon). The single end-labelled DNA fragment was hybridized with 40 µg total RNA, and treated with 100 U of S1-nuclease. The RNA-protected DNA fragments were analyzed on DNA sequencing gels together with G+A (lane A) and T+C (lane T) sequencing ladders derived from the end-labelled fragments [33].
Purification of apo-WhiB
Soluble apo-WhiB was over produced from plasmid pIJ6999 as a (His)6-tagged protein in aerobic E. coli cultures (BL21 lambdaDE3 Star, Novagen) using Luria-Bertani (LB) medium supplemented with 100 mg/L ampicillin. Cultures were grown according to Rybniker [28] and induced with 0.5 mM IPTG. Briefly, cell pellets were resuspended in binding buffer (50 mM NaH2PO4, 200 mM NaCl, 10% (v/v) Glycerol, pH 7.5), treated with 30 mM Imidazole, lysozyme (0.5 mg/ml), DNaseI (0.125 mg/ml), 1.2 mM PMSF, disrupted by sonication and centrigufed at 40,000×g for 45 min at 2°C. Apo-WhiB was isolated under aerobic conditions, via Ni2+-NTA affinity chromatography (HisTrap FF Crude, GE-Healthcare) [28]. Bound proteins were eluted (1 ml/min) using a 6 ml linear gradient from 0 to 100% (v/v) elution buffer (50 mM NaH2PO4, 200 mM NaCl, 500 mM Imidazole, 10% Glycerol, pH 7.5). Fractions (1 ml) containing apo-WhiB were pooled, diluted 10 fold with binding buffer and concentrated using a 1 ml Ni2+-NTA column [34]. before being exchanged (PD10, GE-Healthcare) into 50 mM Tris, 100 mM, NaCl 10% (v/v) Glycerol, pH 7.5. Apo-WhiB was stored at −20°C until needed. As isolated, aerobically prepared apo-WhiB was devoid of an iron sulfur cluster (not shown), as previously observed for WhiB2 [28].
Other Protein methods
Total protein was used for immunodetection of proteins. Cellophane disc cultures were set up as described previously [15] and incubated overnight (∼16 h). To provide osmotic up-shock, overnight cellophane cultures were transferred to MS agar/250 mM KCl and incubated for the specified time. MS agar plates were used as controls. After incubation, mycelia were scraped from the cellophane discs and suspended in Sonication Buffer [50 mM Tris-HCl, pH 8, 200 mM NaCl, 15 mM EDTA, Complete protease inhibitor cocktail (Roche Diagnostics)]. Cells were disrupted by several burst of sonication on ice (20 s at 30% amplitude). Cell-free extracts were obtained by centrifugation (13 000 r.p.m. for 5 min) and recovery of the supernatant. Total protein concentration was determined using the Bradford method (Bio-Rad). SDS-PAGE was performed as previously described [31], loading 10 µg of total protein per lane in 15% SDS PAGE gels. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Hybond-P, Amersham) using a semi-dry electrophoretic transfer cell (Trans-Blot SD, Bio-Rad). Immunological detection was performed using an ECL Advance Western blotting detection kit (Amersham Pharmacia Biotech). His-tagged proteins were detected with a Penta- His peroxidase conjugate (QIAGEN).
RNA isolation and qRT PCR
Total RNA isolation, reverse transcription and qRT PCR procedures were performed as previously described [15]. Briefly, sterile cellophane cultures were set up as described above. After the required incubation, cells were collected and total RNA isolated with a Qiagen RNeasy mini kit as per the manufacturers' recommendations. cDNAs were obtained from 1 µg of total RNA using a RETROscript reverse transcription kit (Ambion); using the manufacturers recommendations with random decamers in a reaction volume of 20 µl. cDNAs were diluted 1/15 in nuclease free water (Ambion). RT-QPCR was carried out on 5 µl of diluted cDNA with an iCycler iQ real-time PCR detection system (Bio-Rad) using SYBR-Green Supermix 2X containing Thermo-Start DNA Polymerase (ABgene). Gene specific primers used for Quantitative PCR were designed using Beacon Design (Premier Biosoft, USA) and shown in Table 2. The specificity of the reaction was assessed using melt curve analysis. Transcript abundance was determined using the standard curve method against serial dilutions of S. coelicolor genomic DNA. S. coelicolor hrdB was used as internal control to normalise samples.
qRT PCR data analysis
Normalised starting quantities were initially tested for normality using the Kolmogorov Smirnov test. Significant differences between transcript abundance among strains were tested using a one-way ANOVA. Dunnett's T3 Test was used post-hoc (equal variances not assumed) to highlight stains that differed most significantly from each other. All statistical analysis procedures were performed in SPSS version 16 for Windows.
Microscopy
Localisation of dpsA expression in mycelia and aerial hyphae was determined using C-termius fusions to mCherry protein and imaged as described [15]. Briefly, spores or mycelia of S. coelicolor were inoculated into the acute angle between glass coverslips inserted obliquely into an agar plate and the surface of the medium. Coverslips were removed from the agar and placed onto microscope slides with a drop of 20% glycerol. Preparations were sealed with clear nail varnish and images obtained using a Nikon Eclipse E600 epifluorescence microscope fitted with a Coolsnap microscope camera (RS Photometrics., Tucson, AZ).
Electrophoretic Mobility Shift Assays
EMSA was performed using the fluorescence based Electrophoretic Mobility-Shift Assay (EMSA) Kit (Invitrogen) according to the manufacturer's instructions. In brief, binding reactions were prepared using recombinant apoWhiB mixed with dpsA promoter region amplified by PCR. Reactions were incubated at room temperature for 30 min. BSA was included as a non-specific competitor. Binding reactions were mixed with 1X EMSA gel-loading solution and electrophoresed for 2 hours in 6%, pre-run (90 V 30 min) polyacrylamide gels in 0.5× Tris-Borate/EDTA running buffer. Gels were post-stained for 30 min at room temperature in the dark in 1× TBE containing 1X SYBR® Green EMSA staining solution.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a VEGA grant 2/0104/09 from Slovak Academy of Sciences. MDH is supported by a Biotechnology and Biological Sciences Research Council Studentship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–36. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 2.Lee EJ, Karoonuthaisiri N, Kim HS, Park JH, Cha CJ, et al. A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol Microbiol. 2005;57:1252–64. doi: 10.1111/j.1365-2958.2005.04761.x. [DOI] [PubMed] [Google Scholar]
- 3.Cho YH, Lee EJ, Ahn BE, Roe JH. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol Microbiol. 2001;42:205–14. doi: 10.1046/j.1365-2958.2001.02622.x. [DOI] [PubMed] [Google Scholar]
- 4.Viollier PH, Kelemen GH, Dale GE, Nguyen KT, Buttner MJ, et al. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol Microbiol. 2003;47:699–714. doi: 10.1046/j.1365-2958.2003.03302.x. [DOI] [PubMed] [Google Scholar]
- 5.Vohradsky J, Li XM, Dale G, Folcher M, Nguyen L, et al. Developmental control of stress stimulons in Streptomyces coelicolor revealed by statistical analyses of global gene expression patterns. J Bacteriol. 2000;182:4979–86. doi: 10.1128/jb.182.17.4979-4986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kormanec J, Sevcikova B, Halgasova N, Knirschova R, Rezuchova B. Identification and transcriptional characterization of the gene encoding the stress-response sigma factor sigmaH in streptomyces coelicolor A3(2). FEMS Microbiol Lett. 2000;189:31–8. doi: 10.1111/j.1574-6968.2000.tb09202.x. [DOI] [PubMed] [Google Scholar]
- 7.Kelemen GH, Viollier PH, Tenor J, Marri L, Buttner MJ, et al. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol Microbiol. 2001;40:804–14. doi: 10.1046/j.1365-2958.2001.02417.x. [DOI] [PubMed] [Google Scholar]
- 8.Karoonuthaisiri N, Weaver D, Huang J, Cohen SN, Kao CM. Regional organization of gene expression in Streptomyces coelicolor. Gene. 2005;353:53–66. doi: 10.1016/j.gene.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 9.Sevcikova B, Benada O, Kofronova O, Kormanec J. Stress-response sigma factor sigma(H) is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch Microbiol. 2001;177:98–106. doi: 10.1007/s00203-001-0367-1. [DOI] [PubMed] [Google Scholar]
- 10.Kormanec J, Sevcikova B. The stress-response sigma factor sigmaH controls the expression of ssgB, a homologue of the sporulation-specific cell division gene ssgA, in Streptomyces coelicolor A3(2). Mol Genet Genomics. 2002a;267:536–43. doi: 10.1007/s00438-002-0687-0. [DOI] [PubMed] [Google Scholar]
- 11.Kormanec J, Sevcikova B. Stress-response sigma factor sigmaH directs expression of the gltB gene encoding glutamate synthase in Streptomyces coelicolor A3(2). Biochim Biophys Acta. 2002b;1577:149–54. doi: 10.1016/s0167-4781(02)00409-8. [DOI] [PubMed] [Google Scholar]
- 12.Mazurakova V, Sevcikova B, Rezuchova B, Kormanec J. Cascade of sigma factors in streptomycetes: identification of a new extracytoplasmic function sigma factor sigmaJ that is under the control of the stress-response sigma factor sigmaH in Streptomyces coelicolor A3(2). Arch Microbiol. 2006;186:435–46. doi: 10.1007/s00203-006-0158-9. [DOI] [PubMed] [Google Scholar]
- 13.Lee EJ, Cho YH, Kim HS, Roe JH. Identification of sigmaB-dependent promoters using consensus-directed search of Streptomyces coelicolor genome. J Microbiol. 2004;42:147–51. [PubMed] [Google Scholar]
- 14.Dalton KA, Thibessard A, Hunter JI, Kelemen GH. A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol Microbiol. 2007;64:719–37. doi: 10.1111/j.1365-2958.2007.05684.x. [DOI] [PubMed] [Google Scholar]
- 15.Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, et al. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol. 2009;73:1186–202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 16.Antelmann H, Engelmann S, Schmid R, Sorokin A, Lapidus A, et al. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor sigmaB in Bacillus subtilis. J Bacteriol. 1997;179:7251–6. doi: 10.1128/jb.179.23.7251-7256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003;13:206–15. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–5. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelemen GH, Brian P, Flardh K, Chamberlin L, Chater KF, et al. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2). J Bacteriol. 1998;180:2515–21. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soliveri J, Brown KL, Buttner MJ, Chater KF. Two promoters for the whiB sporulation gene of Streptomyces coelicolor A3(2) and their activities in relation to development. J Bacteriol. 1992;174:6215–20. doi: 10.1128/jb.174.19.6215-6220.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang JG, Hahn MY, Ishihama A, Roe JH. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2). Nucleic Acids Res. 1997;25:2566–73. doi: 10.1093/nar/25.13.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, et al. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol Microbiol. 2010;78:361–79. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- 23.Paget MS, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol Microbiol. 2001;42:1007–20. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 24.Paget MS, Leibovitz E, Buttner MJ. A putative two-component signal transduction system regulates sigmaE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2). Mol Microbiol. 1999;33:97–107. doi: 10.1046/j.1365-2958.1999.01452.x. [DOI] [PubMed] [Google Scholar]
- 25.Crack JC, den Hengst CD, Jakimowicz P, Subramanian S, Johnson MK, et al. Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry. 2009;48:12252–64. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J. 2010;432:417–27. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybniker J, Nowag A, van Gumpel E, Nissen N, Robinson N, et al. Insights into the function of the WhiB-like protein of mycobacteriophage TM4–a transcriptional inhibitor of WhiB2. Mol Microbiol. 2010;77:642–57. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- 29.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation 2000 [Google Scholar]
- 30.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–6. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russel DW. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, New York; 1989. [Google Scholar]
- 32.Bishop A, Fielding S, Dyson P, Herron P. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 2004;14:893–900. doi: 10.1101/gr.1710304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 34.Crack JC, Le Brun NE, Thomson AJ, Green J, Jervis AJ. Reactions of nitric oxide and oxygen with the regulator of fumarate and nitrate reduction, a global transcriptional regulator, during anaerobic growth of Escherichia coli. Methods Enzymol. 2008;437:191–209. doi: 10.1016/S0076-6879(07)37011-0. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Martinez L, Bishop A, Parkes L, Del Sol R, Salerno P, Sevcikova B, et al. Osmoregulation in Streptomyces coelicolor: modulation of SigB activity by OsaC. Mol Microbiol. 2009;71:1250–62. doi: 10.1111/j.1365-2958.2009.06599.x. [DOI] [PubMed] [Google Scholar]
- 36.Mao XM, Zhou Z, Hou XP, Guan WJ, Li YQ. Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J Bacteriol. 2009;191:6473–81. doi: 10.1128/JB.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flardh K, Findlay KC, Chater KF. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology. 1999;145(Pt 9):2229–43. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–19. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Flett F, Mersinias V, Smith CP. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–9. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]