Abstract
The ends of eukaryotic chromosomes consist of long tracts of repetitive GT-rich DNA with variable sequence homogeneity between and within organisms. Telomeres terminate in a conserved 3'-ssDNA overhang that, regardless of sequence variability, is specifically and tightly bound by proteins of the telomere-end protection family. The high affinity ssDNA-binding activity of S. pombe Pot1 protein (SpPot1) is conferred by a DNA-binding domain consisting of two subdomains, Pot1pN and Pot1pC. Previous work has shown that Pot1pN binds a single repeat of the core telomere sequence (GGTTAC) with exquisite specificity, while Pot1pC binds an extended sequence of nine nucleotides (GGTTACGGT) with modest specificity requirements. We find that full-length SpPot1 binds the composite 15mer, (GGTTAC)2GGT, and a shorter two-repeat 12mer, (GGTTAC)2, with equally high affinity (< 3 pM), but with substantially different kinetic and thermodynamic properties. The binding mode of the SpPot1/15mer complex is more stable than that of the 12mer complex, with a 2-fold longer half-life and increased tolerance to nucleotide and amino acid substitutions. Our data suggest that SpPot1 protection of heterogeneous telomeres is mediated through 5'-sequence recognition and the use of alternate binding modes to maintain high affinity interaction with the G-strand, while simultaneously discriminating against the complementary strand.
Keywords: telomeres, ssDNA-binding domain, end-protection, OB-fold, Pot1, ssDNA recognition
Many critical cellular functions, including telomere maintenance and DNA replication, repair, and recombination, involve the management of single-stranded DNA (ssDNA) and depend on the recognition of ssDNA by OB-fold-containing proteins (1–4). While many of these interactions are not sequence specific, proteins of the telomere-end protection (TEP) family tightly and specifically bind to and protect the GT-rich 3'-ssDNA overhang present at the end of eukaryotic chromosomes (1). In the absence of this telomere cap, the free ssDNA end is subject to recognition by DNA damage machinery leading to chromosomal-end fusions, DNA resection, and chromosomal rearrangements (5–10). In addition to this critical protective function, TEP proteins interact with a suite of other telomere-associated proteins to regulate telomere homeostasis, including defining telomere structure and coordinating telomerase access to telomeric ssDNA (5, 8, 11–16). In fission yeast and humans, the TEP protein Pot1 associates with five other proteins forming a core telomere-interaction complex termed shelterin (17, 15, 18). The Tpz1 protein (TPP1 in humans) specifically interacts with Pot1 and is responsible for Pot1 recruitment to the telomere bridging Pot1 interaction with other shelterin components (19, 17). Cdc13, the budding yeast TEP protein, coordinates with a variety of other telomere-associated protein to affect telomere protection and regulation (11, 20, 21); however, to date a budding yeast shelterin complex has not been identified.
While telomere structure and TEP protein interactions vary between organisms, specific recognition of telomeric ssDNA by TEP proteins is key to their function. Analysis of high-resolution structures of TEP proteins in complex with ssDNA has helped define common mechanisms of recognition, as well as those unique to each system (1). Structures of the DNA-binding domains of the TEP proteins from the yeasts Saccharyomyces cerevisiae and Schizosaccharomyces pombe, and humans reveal the common use of aromatic stacking and H-bonding networks for the specific recognition of cognate telomere sequences (22–26). Human Pot1 (hPOT1) utilizes two OB-folds (hOB1 and hOB2) to bind two repeats of the human telomere sequence, (GGTTAG)2 (27), with a binding interface characterized by four pairwise nucleotide stacks and 31 protein-ssDNA H-bonds, including 13 base-specific H-bonds located in hOB1 (25, 26). The corresponding domain in S. pombe Pot1, Pot1pN, in isolation binds a single telomere repeat (GGTTAC) (6, 28, 29) in a more compact configuration than that observed for the homologous hPOT1 OB1 (24). While the presence of the second domain in the hPOT1 structure may contribute to the differences in DNA structure, the extensive array of base-specific interactions within the Pot1pN structure indicate the compact arrangement is involved in specific recognition. The six nucleotides participate in three sets of stacking interactions, and, in addition to 15 sequence-specific protein-ssDNA H-bonds, several intraoligonucleotide H-bonds contribute to specificity. The single OB-fold of S. cerevisiae Cdc13-DBD is distinct from those of the Pot1 proteins and binds 11 nucleotides (GTGTGGGTGTG) in an extended conformation, resulting in an uncharacteristic lack of inter-nucleotide stacking (23). Additionally, the binding interface contains relatively few H-bonds; however, as S. cerevisiae telomeres do not consist of regular repeating sequence, this scarcity of H-bonding likely plays a role in accommodation of degenerate telomere sequence (30–33). These structures demonstrate the complexity of interactions contributing to recognition and highlight the adaptability of TEP OB-folds to varying specificity requirements.
Pot1pN and hOB1 exhibit a high degree of structural similarity, which likely extends beyond the first OB-fold (24, 25). In the absence of any significant sequence similarity, secondary structure prediction analysis of SpPot1 supports presence of a second OB-fold (Pot1pC) (34). Biochemical studies revealed that Pot1pC has telomere-specific DNA-binding activity (35); however, a structure of Pot1pC has not been determined. The independent nature of Pot1pN and Pot1pC differs from that of hPOT1, in that hOB1 and hOB2 cannot be expressed individually (25). While Pot1pN binds a single repeat GGTTAC (6mer) (24, 29) Pot1pC minimally requires a 9mer (GGTTACGGT), representing 1.5 telomere repeats. We previously found that in addition to the composite 15mer sequence (GGTTACGGTTACGGT), Pot1-DBD, encompassing Pot1pN and Pot1pC, binds two telomere repeats, (GGTTAC)2 (12mer), with high affinity (34, 35). This ability to accommodate multiple sequence lengths contrasts with hPOT1 specificity for the precise 3'-end of the GGTTAG telomere repeat (25).
The differences in Pot1 activity between homologues likely arise from the divergent nature of S. pombe and human telomeres. While the core 6-nucleotide telomere repeat sequences are highly similar to each other, differing only at position 6, S. pombe repeats are interrupted by spacer elements of variable length and sequence, yielding the general sequence GGTTAC(A/AC)0–1(G)0–7 (36, 37). To function properly at these telomeres, SpPot1 must be able to accommodate these spacers, most likely through more flexible interdomain interactions and increased malleability within the ssDNA-binding interface. Additionally, the CTD of both SpPot1 and hPOT1 contains a third putative OB-fold (38). While this domain is known to participate in protein-protein interactions (17, 19, 39), the S. pombe CTD may provide additional nucleotide contacts or allosteric effects that alter interaction with different telomeric sequences.
Here we present quantitative binding studies of full-length SpPot1 and confirm that Pot1-DBD represents the complete DNA-binding domain. As 12mer and 15mer respectively represent the minimal binding and Pot1pN/Pot1pC composite sequences, we examined these two oligonucleotides for differences in binding characteristics. We find that, despite equivalent binding affinities in the low picomolar range, SpPot1 interactions with 12mer and 15mer are characterized by distinct thermodynamic and kinetic properties. Our results suggest that the molecular recognition elements in this system are exceptionally flexible and that alterations in binding mode allow for accommodation of changes in length and sequence arising from the inherent heterogeneity of S. pombe telomeres.
Experimental Procedures
Chemicals, Reagents and Proteins
All chemicals and reagents were obtained from Fisher Scientific unless otherwise indicated. Oligonucleotides were commercially synthesized by Integrated DNA Technologies and [γ–32P]ATP was purchased from Perkin Elmer. Professor Thomas Cech graciously provided the plasmid encoding FL-Pot1.
Expression and Purification of Full-length S. pombe Pot1 (FL-Pot1)
FL-Pot1 (full-length, residues 1–]555) was expressed as a His10-SUMO fusion protein using the pET-His-Smt3 expression vector (40), modified to encode His10, rather than His6. Cells were grown at 37°C in LB containing 100 μg/mL kanamycin to an OD600 of 0.5 – 0.6. Cells were then placed on ice for 1 h, after which protein production was induced by addition of 0.5 mM IPTG and continued shaking for 15 h at 15°C.
For purification of FL-Pot1, cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris, pH 8.4, 500 mM NaCl, 20 mM imidizole, 3 mM βME, 5% glycerol) and lysed by sonication. Cellular debris was removed by centrifugation and the soluble fraction was passed over pre-equilibrated, packed Ni-NTA superflow resin (Qiagen). The column was washed with wash buffer (20 mM Tris, pH 8.4, 1 M NaCl, 50 mM imidizole, 3 mM βME, 5% glycerol) and eluted with lysis buffer containing 300 mM imidizole. FL-Pot1 was concentrated to ~5 mg/mL and injected onto a Superdex S200 16/60 size exclusion chromatography column (GE Healthcare) pre-equilibrated with column buffer ((20 mM Tris, pH 8.4, 500 mM NaCl, 3mM βME, 5% glycerol). Monomeric protein was concentrated to ~5 mg/mL and cleaved with Ubl-specific protease 1 (ULP1) (40) at a ratio of 1:100 mg/mg while spinning at 4°C. The final step in purification was gel filtration, performed with a Superdex S200 16/60 size exclusion chromatography column. Purified monomeric protein was concentrated to 60 μM, flash frozen in liquid nitrogen and stored at −80°C; no loss of activity was observed upon freezing. Typical protein yields were ~2 mg/L.
Expression and Purification of S. pombe Pot-DBD Constructs
Pot1-DBD (residues 1–389) was cloned into the pTXB1 vector (New England BioLabs), generating a C-terminally tagged intein/chitin binding domain fusion protein, and expressed in BL21(DE3) E. coli. Cells were grown at 37°C in LB containing 100 μg/mL ampicillin to an OD600 of 0.5 – 0.6. Cells were then placed on ice for 1 h, after which protein production was induced by addition of 0.5 mM IPTG and continued shaking for 20 h at 18°C. Cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris, pH 8.5, 500 mM NaCl) and lysed by sonication. Cellular debris was removed by centrifugation and the soluble fraction was slowly passed over a packed column of chitin beads (New England BioLabs). Columns were washed with 20 column volumes of lysis buffer and resuspended in lysis buffer as a 50% slurry. Intein self-cleavage was induced by the addition of 100 mM βME and overnight incubation at 4°C. Fully cleaved Pot1 protein was eluted, concentrated and injected onto a Superdex S200 16/60 size exclusion chromatography column (GE Healthcare) pre-equilibrated with column buffer (50 mM potassium phosphate, pH 8.0, 150 mM NaCl, 3mM βME). Monomeric protein was concentrated to 200 – 500 μM, flash frozen in liquid nitrogen and stored at −80°C. Typical protein yields were ~6 mg/L.
Pot1-DBD alanine mutants, previously generated by site-directed mutagenesis, were expressed and purified as N-terminal His6-tagged proteins, as previously described (35). Briefly, proteins were expressed in BL21(DE3)C41 E. coli, purified via Ni-NTA affinity and size exclusion chromatography, flash frozen in liquid N2, and stored at −80°C with final yields of ~2 mg/L.
Determination of FL-Pot1/ssDNA Off-rates
ssDNA-binding off-rates for 12mer and 15mer were determined using a competition double-filter binding assay as previously described for Pot1-DBD (34). Briefly, FL-Pot1/ssDNA complexes were formed in binding buffer (20 mM Tris, pH 8.4, 50 mM NaCl, 5 mM DTT, 15% glycerol, 1 mg/mL BSA) at concentrations well above the KD (FL-Pot1 = 20 nM, [32P]ssDNA = 2 nM) at 4°C for 60 min. Following complex formation, unlabeled competitor ssDNA (5 μM) was added at 1440, 960, 720, 600, 480, 360, 240, 120, 60, 50, 40, 30, 20, 10, 7, 5, 3, 1 and 0.5 min. FL-Pot1-bound [32P]ssDNA was quantified and data were plotted as Nintensity versus time and were fit to a single-state exponential decay model.
Electrophoretic Mobility Shift Assays (EMSA)
All oligonucleotides were 5'-end labeled with [γ-32P]ATP using T4 polynucleotide kinase according to manufacturer's protocol (New England Biolabs). Labeled oligonucleotides were separated from unincorporated ATP using G25 spin columns (GE Healthcare). To prevent degradation, labeled oligonucleotides were stored at −20°C and subjected to no more than 3 freeze/thaw cycles. Active concentrations of FL-Pot1 and Pot1-DBD were determined as previously described (34) using 200 nM 15mer (25,000-fold above estimated KD). Proteins were serially diluted over a range of 5 fM to 2 μM in binding buffer, and complexes were formed with radiolabeled ssDNA for 30 min at 4°C. Protein/DNA complexes at a volume of 30 μL were loaded onto 1.5 mm thick, 6.7% nondenaturing polyacrylamide gels containing 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) and 5% glycerol and run in running buffer (1× TBE, 5% glycerol) at 200 V for 20 min at 4°C. Gels were dried on filter paper (Whatman), exposed to phosphorimaging screens (GE Healthcare) and visualized on a Typhoon Imager (GE Healthcare). Data were quantified using ImageQuant version 5.1 (GE Healthcare) to determine fraction bound and plotted (KaleidaGraph version 4.0; Synergy Software) to determine protein concentration at which saturation is reached.
EMSA experiments used to determine binding constants were performed as above with the following modifications. The concentration of DNA (1 pM) was at least 5-fold below measured binding affinities and the protein concentration range was typically between 5 fM and 1 μM (up to 4 μM for oligonucleotides in Table 3). Lower concentrations of oligonucleotide were tested with no change in measured binding affinity; 1 pM was chosen in order to increase signal and remain below the titration range. Binding affinities were determined using triplicate or quadruplicate EMSA experiments performed with different protein batches. Data were quantified to determine fraction bound and globally fit using nonlinear curve regression to a standard two-state binding model to calculate KD. All reported KD values represent apparent KDs. Errors in the reported KD values are the standard error of the mean extracted from the global fits. We conservatively define KD changes of less than 2-fold insignificant.
Table 3.
KD and free energy of binding values for FL-Potl binding to partial and non-telomeric ssDNA oligonucleotides.
| Oligonucleotide Name | Oligonucleotide Sequencea | KD (pM)b | ΔΔG°' (kcal/mol)c | Fold Changed |
|---|---|---|---|---|
| T12 | TTTTTTTTTTTT | > 250000 | > 16 | > 100000 |
| T15 | TTTTTTTTTTTTTTT | 8600 ± 1900 | 5 ± 1 | 3900 |
| 12mer_comp | CCAATGCCAATG | NDBe | ||
| 15mer_comp | CCAATGCCAATGCCA | NDBe | ||
| 6mer_T6 | GG11ACTTTTTT | 3.2 ± 0.3 | 0.21 ± 0.02 | 1.5 |
| 6mer_T9 | GGTTACTTTTTTTTT | 2.6 ± 0.2 | 0.092 ± 0.007 | 1.2 |
| 6mer_6 comp | GGTTACCCAATG | 9.5 ± 0.3 | 0.81 ± 0.03 | 4.3 |
| 6mer_9 comp | GGTTACCCAATGCCA | 3.8 ± 0.1 | 0.031 ± 0.008 | 1.7 |
| 15 comp GTT | CGTTTGCCAATGCCA | 10600 ± 300 | 4.7 ± 0.1 | 4800 |
All oligonucleotides are deoxynucleotide sequences; wild-type portions are underlined
Apparent KD values were obtained from at least three independent EMS A experiments conducted in 50 mM sodium chloride, 20 mM Tris, 5 mM DTT, pH 8.4 at 4° C. Errors represent the standard error of the mean equilibrium dissociation constant obtained from three independent binding experiments.
Changes in free energy were calculated as follows: ΔΔG°' = RT ln(KDalternate/KDwt), where R = 1.9872 cal/(mol•K), T = 277.15 K.
Fold difference is relative to the respective wild-type.
NDB = no detectable binding up to 4 μM protein.
EMSA experiments to determine the oligomeric state of FL-Pot1 and Pot1-DBD were modified as follows. Oligonucleotide concentration was held at 50 pm. Pot1pN concentration was kept at 1 μM; Pot1-DBD and FL-Pot1 concentrations were 50 nM for individual binding and the protein mixture binding was performed at 0, 0.005, 0.5, and 50 nM. Gels used consisted of 12% polyacrylamide layered on 25% polyacrylamide, allowing for complex resolution and retention of free ssDNA oligonucleotides. Gels were run at 250 volts for 2 hr 45 min and visualized as described above.
Results
Pot1-DBD Fully Recapitulates FL-Pot1 ssDNA-binding Activity
Full-length S. pombe Pot1 (FL-Pot1) (6), is a 555-residue protein consisting of three domains: the structurally characterized N-terminal OB-fold (Pot1pN) (24, 29), a second domain predicted to be an OB-fold (Pot1pC) (34, 35, 41), and a C-terminal domain that interacts with other telomere-associated proteins (17) (Figure 1A). Pot1pN and Pot1pC represent independently folded domains, each displaying DNA-binding activity, with Pot1pN minimally binding a single telomere repeat sequence (6mer; GGTTAC) and Pot1pC binding 1.5 repeats (9mer; GGTTACGGT) (29, 35). We previously identified Pot1-DBD, which encompasses the Pot1pN and Pot1pC subdomains, as an extended DNA-binding domain of FL-Pot1 (Figure 1A). As expected, Pot1-DBD binds the composite Pot1pN and Pot1pC 15-nucleotide sequence, representing 2.5 telomere repeats (15mer; GGTTACGGTTACGGT), with high affinity (35). Unexpectedly, however, Pot1-DBD also binds two telomere repeats (12mer; GGTTACGGTTAC) with an affinity similar to 15mer (34, 35). To determine whether the CTD modulates the DNA-binding activity between these two substrates, we compared binding of FL-Pot1 and Pot1-DBD to 12mer and 15mer. FL-Pot1 and Pot1-DBD bind 12mer equivalently, with < 2-fold difference in KD (2.8 ± 0.4 pM and 4.0 ± 0.3 pM, respectively) (Table 1; Supporting Information Table S1). Additionally, FL-Pot1 binding to 15mer exhibits no change from that of Pot1-DBD (2.2 ± 0.2 pM and 2.9 ± 0.3 pM, respectively) (Figure 1B,C; Table 1; Supporting Information Table S1). These data strongly indicate equivalent activities for FL-Pot1 and Pot1-DBD binding to 12mer and 15mer.
Figure 1.
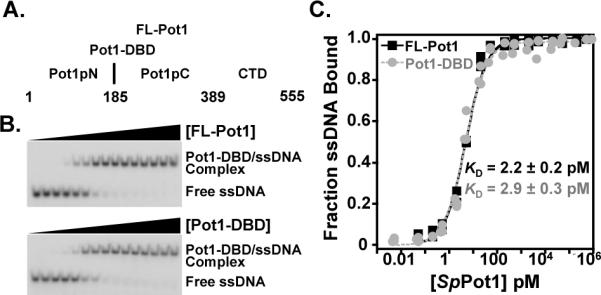
Domain organization and representative EMSA binding and analysis. (A) Domain boundaries are indicated for FL-Pot1, Pot1-DBD, Pot1pN, and Pot1pC. (B) Representative EMSA gels showing FL-Pot1 (top panel) and Pot1-DBD (lower panel) binding to 15mer ssDNA. (C) Normalized data were collected for triplicate EMSA experiments for FL-Pot1 (black squares) and Pot1-DBD (gray circles) binding to 15mer. Data are plotted as a function of fraction ssDNA bound versus Pot1 concentration (pM).
Table 1.
KD and free energy of binding values for SpPotl binding to telomeric ssDNA oligonucleotides of increasing length.
| Oligonucleotide Name | Oligonucleotide Sequencea | KD (pM)b | ΔΔG°' (kcal/mol)c | Fold Changed |
|---|---|---|---|---|
| 12mer | GGTTACGGTTAC | 2.8 ± 0.4 | 0.13 ± 0.02 | 1.3 |
| 13mer | GGTTACGGTTACG | 2.3 ± 0.3 | 0.012 ± 0.002 | 1.0 |
| 14mer | GGTTACGGTTACGG | 2.4 ± 0.4 | 0.042 ± 0.007 | 1.1 |
| 15mer | GGTTACGGTTACGGT | 2.2 ± 0.1 | --- | --- |
| 16mer | GGTTACGGTTACGGTT | 2.20 ± 0.04 | 0.0 | 1.0 |
| 17mer | GGTTACGGTTACGGTTA | 2.3 ± 0.1 | 0.0124 ± 0.0007 | 1.0 |
| 18mer | GGTTACGGTTACGGTTAC | 3.8 ± 0.3 | 0.31 ± 0.03 | 1.7 |
| 24mer | (GGTTACGGTTAC)2 | 3.4 ± 0.3 | 0.24 ± 0.02 | 1.5 |
| 12mer_AC | GGTTACACGGTTAC | 7.7 ± 0.8 | 0.69 ± 0.07 | 35 |
| 15mer_2xAC | GGTTACACGGTTACACGGT | 3.2 ± 0.3 | 0.21 ± 0.02 | 1.5 |
All oligonucleotides are deoxynucleotide sequences.
Apparent KD values were obtained from at least three independent EMSA experiments conducted in 50 mM sodium chloride, 20 mM Tris, 5 mM DTT, pH 8.4 at 4° C. Errors represent the standard error of the mean equilibrium dissociation constant obtained from three independent bindin experiments.
Changes in free energy were calculated as follows: ΔΔG°' = RT ln(KDalternate/KD15mer), where R = 1.9872 cal/(mol•K), T = 277.15 K.
Fold difference is relative to the 15mer sequence.
We next tested whether the CTD participates in DNA binding to longer oligonucleotides. Consistent with previous data for Pot1-DBD (34), FL-Pot1 exhibits no significant change in binding affinity between oligonucleotides ranging in length from 12mer (2.8 ± 0.4 pM) to the 4-telomere repeat 24mer (3.4 ± 0.3 pM) (Table 1). To understand how Pot1 binds representative S. pombe telomere sequences, we tested the effect of the addition of spacer sequences between the core GGTTAC repeats on binding affinity. Inclusion of a two-nucleotide spacer within 12mer (GGTTACACGGTTAC) reduces the binding affinity by 3.5-fold (7.7 ± 0.8 pM). However, no significant reduction in affinity (3.2 ± 0.3 pM) was observed when a two-nucleotide spacer was inserted between the repeats of 15mer (GGTTACACGGTTACACGGT). Thus, FL-Pot1 maintains high affinity binding to representative S. pombe telomere sequences of varied length.
The CTD of SpPot1 promotes interaction with at least one other telomere-associated protein (Tpz1) (17). As a protein-protein interaction domain, the CTD may mediate homodimerization or other higher order oligomerization. In order to probe the effect of the CTD on oligomeric states, we examined Pot1-DBD and FL-Pot1 15mer binding separately and in a mixture by gel electrophoresis (Figure 2). Individually, FL-Pot1 and Pot1-DBD migrate as single bands (Figure 2, lanes 5–6). A mixture of Pot1-DBD and FL-Pot1 migrates as two distinct bands with the same migration patterns as the individual complexes (compare Figure 2, lanes 2–4 and lanes 5–6). We used Pot1pN complexes as benchmarks for Pot1-DBD and FL-Pot1 complex mobility. Pot1pN binds a single repeat of GGTTAC as a monomer (29) and oligonucleotides containing additional repeats are bound cooperatively and with increasing stability in the gel (29). Thus, the Pot1pN/15mer complex represents a dimer (Figure 2, lane 7) with a mass to charge ratio comparable to that of the Pot1-DBD/15mer complex and the Pot1pN/18mer complex represents a trimer (Figure 2, lane 8) with a mass to charge ratio comparable to that of the FL-Pot1/15mer complex. On the time scale of this experiment, Pot1pN/6mer interaction is not observable (data not shown), the Pot1pN/15mer complex partially dissociates (Figure 2, lane 7), while Pot1pN remains fully bound to 18mer (Figure 2, lane 8). Dimerization of Pot1-DBD and/or FL-Pot1 would be evident by a significant shift in migration relative to that of the Pot1pN trimer. Additionally, we independently confirmed the monomeric nature of the FL-Pot1 and Pot1-DBD interactions with 15mer via multiangle light scattering (Supporting Information Figure S1). These data demonstrate that both Pot1-DBD and FL-Pot1 bind as monomers, indicating that the CTD of SpPot1 does not promote protein-protein interactions that lead to homo-oligomerization.
Figure 2.
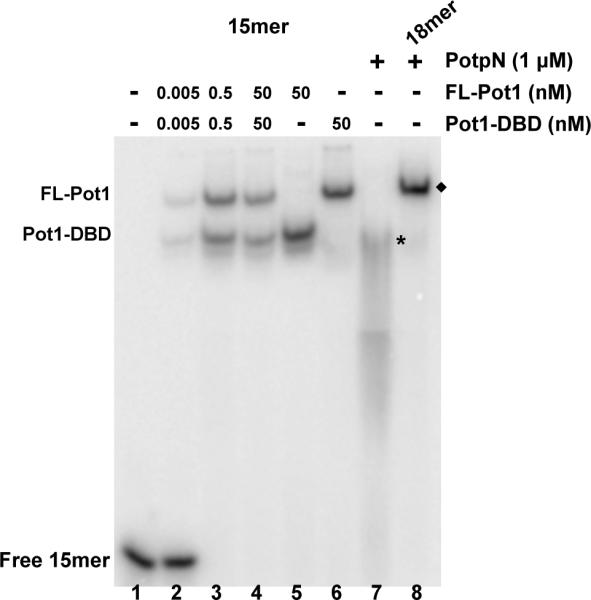
SpPot1 binds 15mer as a monomer. Increasing concentrations of a mixture of FL-Pot1 and Pot1-DBD (0, 0.005, 0.5, 50 nM; lanes 1–4) or 50 nM FL-Pot1 or Pot1-DBD alone (lanes 5–6) were added to 50 pM radiolabeled 15mer. For comparison, lane 7 shows 2× Pot1pN + 15mer (*) and lane 8 shows 3× Pot1pN + 18mer (◆). Total molecular weight of each complex: Pot1-DBD/15mer = 50 kDa; FL-Pot1/15mer = 69 kDa: Pot1pN/15mer = 50 kDa; Pot1pN/18mer = 73 kDa. The Pot1pN/15mer complex dissociates on the time-scale of the experiment.
FL-Pot1 Binds 12mer and 15mer with the Same Affinity but Different Off-rates
In order to address possible kinetic differences between oligonucleotide binding, we measured the dissociation rates of the FL-Pot1/12mer and FL-Pot1/15mer complexes using a competition experiment. FL-Pot1/12mer exhibits an off-rate (koff) of 2.4 × 10−4 ± 3 × 10−6 s −1, corresponding to a half-life (t1/2) of 48 min (Figure 3). This value is consistent with the previously reported half-life of the Pot1-DBD/12mer complex (34). Interestingly, we found that the FL-Pot1/15mer complex displays a slower dissociation rate (1.1 × 10−4 ± 1 × 10−5 s−1), with a half-life approximately twice that of FL-Pot1/12mer (t1/2 = 105 min; Figure 3). This result was unexpected considering the affinities of FL-Pot1 for the two sequences are identical. The observed difference in the dissociation rates suggests changes in binding mode between 12mer and 15mer not manifest in the equilibrium binding constants.
Figure 3.
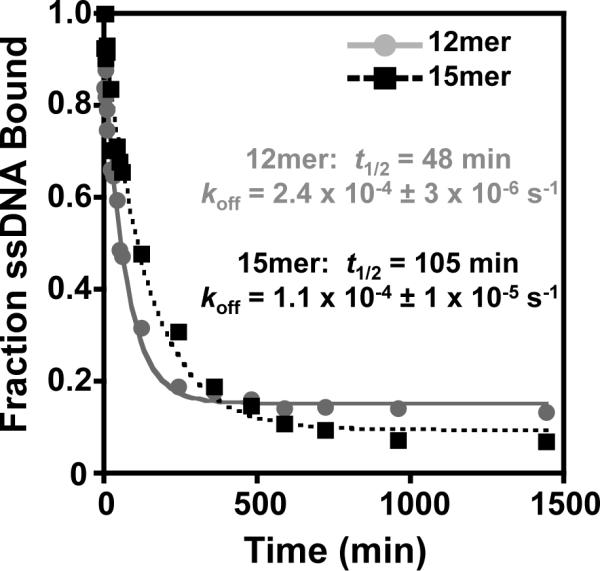
The FL-Pot1/15mer complex displays a longer half-life than the FLPot1/12mer complex. FL-Potl/32 P-12mer complexes (grey circles) and FL-Pot1/32P-15mer complexes (black squares) were competed with a large molar excess of unlabeled 12mer or 15mer, respectively, (5 μM, 2500-fold excess over 32P-ssDNA) at various time points. Data are plotted as 32P-ssDNA bound versus time (min) and globally fit to a single-exponential decay model. Dissociation rate constant (koff) and half-life (t1/2) for each complex are indicated.
FL-Pot1 and Pot1-DBD Have Different Specificity Profiles for 12mer and 15mer
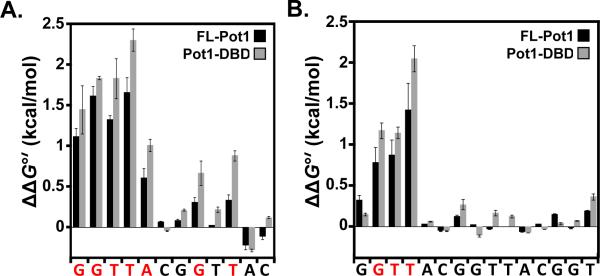
In order to examine potential alterations in binding mode between these two similar substrates, we performed in-depth specificity profiling for both 12mer and 15mer. By making single complementary nucleotide substitutions at each position, we created a panel of “complemers” to probe the specific recognition of SpPot1 for each site. We measured apparent KDs for each complemer via gel shift and calculated the change in the free energy of binding compared to wild-type binding for each position. All binding performed within in this study was conducted at 50 mM NaCl, and thus these data can be directly compared to that of Pot1pN and Pot1pC obtained under the same conditions (29, 35). Consistent with their behavior in other binding assays, Pot1-DBD and FL-Pot1 exhibit similar specificity profiles (Figure 4; Supporting Information Table S1). We set a threshold for significance at ΔΔG > 0.4 kcal/mol, corresponding to a 2-fold difference in binding affinity. Using this threshold, we found that complementary changes within 12mer at positions 1–5, 8 and 10 result in significant changes in the free energy of binding compared to 12mer (Figure 4A). These data designate a recognition profile for SpPot1 binding to 12mer of GGTTACGGTTAC, with specifically recognized nucleotides in bold. The discrimination observed here within the 5'-repeat is consistent with the GGTTAC (all nucleotides are specifically recognized) specificity profile for Pot1pN binding to 6mer, differing only at position 6 (29). SpPot1 specificity for the second repeat of 12mer is not directly comparable to the recognition of 9mer (GGTTACGGT) by Pot1pC due to the lack of a full Pot1pC binding site (35). These data suggest that recognition of the 5'-end contributes the vast majority of the specificity within the SpPot1/12mer interaction.
Figure 4.
The specificity profiles determined by complementary base substitution in 12mer and 15mer on FL-Pot1 and Pot1-DBD reveal alternate modes of binding. (A) ΔΔG°' values for each 12mer complemer binding to FL-Pot1 (black bars) and Pot1-DBD (gray bars) were calculated as follows: ΔΔG°' = RT ln(KDcomplemer/KD12mer), where R = 1.9872 cal/(mol•K), T = 277.15 K, and KD12mer = 4.0 pM. (B) ΔΔG°' values for each 15mer complemer binding to FL-Pot1 (black bars) and Pot1-DBD (gray bars) were calculated as in (A): ΔΔG°' = RT ln(KDcomplemer/KD15mer), where R = 1.9872 cal/(mol•K), T = 277.15 K, and KD15mer = 2.2 pM.
The specificity profile of SpPot1 binding to 15mer is strikingly different from that of 12mer (Figure 4). Significantly fewer positions are specifically recognized, with the specificity pattern mapped in bold as GGTTACGGTTACGGT. These data reveal, as with 12mer, a preference for the 5'-end of the oligonucleotide. However, unlike 12mer, there is a complete lack of nucleotide discrimination within the second telomeric repeat. In addition, positions 1 and 5 are no longer specified and recognition of G2 and T3 is significantly diminished compared to the SpPot1/12mer interaction (Figure 4B). This profile is also notably and unexpectedly different from the combined profiles of Pot1pN and Pot1pC (35), suggesting that while the two domains are separable, interaction between the two permits broadened sequence accommodation. These data demonstrate an overall relaxation of specific nucleotide recognition across the entire 15mer sequence as compared to 12mer and indicate significant differences between the two binding modes.
Non-canonical Nucleotide Substitutions within 12mer and 15mer Reveal Further Differences between Binding Modes
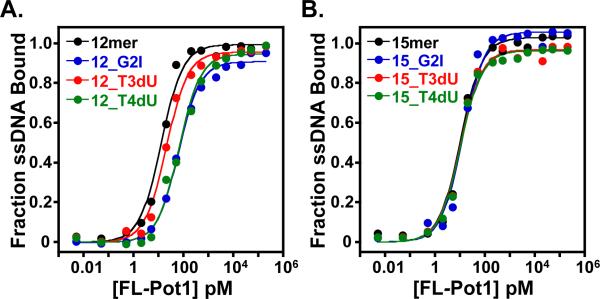
Although full binding activity is only conferred by the DBD, some potential insights into recognition are provided by the Pot1pN/6mer crystal structure. In addition to aromatic and H-bonding DNA/protein contacts, other specific hydrophobic (e.g., burial of the T4 methyl group) and unusual intra-oligonucleotide H-bonding interactions are present (24). Substantial reductions in binding affinity to 6mer were observed with the subtle base alterations of G2 to inosine (I) and T4 to deoxyuracil (dU), but not T3dU, establishing the specific requirement for some of these interactions (24). We examined the significance of specific ssDNA functional groups in the context of FL-Pot1 binding utilizing 12mer and 15mer oligonucleotides containing the nucleotide changes G2I, T3dU, and T4dU. FL-Pot1 binds G2I and T4dU substituted 12mer with KDs of 23.6 ± 3.6 pM and 30.6 ± 1.3 pM, respectively (Figure 5A, Table 2), representing an ~10-fold weakening in binding affinity for each sequence. The T3dU substitution results in a more modest 2-fold increase in KD to 5.6 ± 0.2 pM. These binding trends parallel those of the Pot1pN/6mer interaction; G2I and T4dU affect FL-Pot1 binding to 12mer, whereas T3dU incorporation has little effect on affinity (Figure 5A, Table 2). In contrast to 12mer substitutions, the G2I, T3dU, and T4dU changes made within 15mer have absolutely no effect on binding, exhibiting wild-type affinities of 2.8 ± 0.1, 2.6 ± 0.2, and 3.0 ± 0.4 pM, respectively (Figure 4B, Table 2). These results indicate that specific functional groups participate in promoting recognition of 12mer, but do not contribute specifically to the 15mer binding mode.
Figure 5.
Comparison of binding of non-canonical single nucleotide substituted ssDNA to FL-Pot1 reveals differential binding between 12mer and 15mer. (A) Binding curves for FL-Pot1 binding to 12mer (black) and the single non-canonical substitutions G2I (blue), T3dU (red) and T4dU (green) within 12mer. (B) Binding curves for FL-Pot1 binding to 15mer (black) and the single non-canonical substitutions G2I (blue), T3dU (red) and T4dU (green) within 15mer. Normalized data were collected for triplicate EMSA experiments and plotted as a function of fraction ssDNA bound versus FL-Pot1 concentration (pM).
Table 2.
KD values for FL-Potl binding to telomenc ssDNA oligonucleotides containing non-canonical nucleotide substitutions.
| Oligonucleotide Name | Oligonucleotide Sequencea | KD (pM)b | ΔΔG°' (kcal/mol)c | Fold Changed |
|---|---|---|---|---|
| 12_G2I | GITTACGGTTAC | 23.6 ± 3.6 | 1.2 ± 0.2 | 8.4 |
| 12_T3U | GGdUTACGGTTAC | 5.6 ± 0.2 | 0.38 ± 0.01 | 20 |
| 12_T4U | GGTdUACGGTTAC | 30.6 ± 1.3 | 1.3 ± 0.4 | 10.9 |
| 15_G2I | GITTACGGTTACGGT | 2.8 ± 0.1 | 0.133 ± 0.004 | 1.3 |
| 15_T3U | GGdUTACGGTTACGGT | 2.6 ± 0.2 | 0.092 ± 0.007 | 1.2 |
| 15_T4U | GGTdUACGGTTACGGT | 3.0 ± 0.4 | 0.17 ± 0.02 | 1.4 |
All oligonucleotides are deoxynucleotide sequences.
Apparent KD values were obtained from at least three independent EMSA experiments conducted in 50 mM sodium chloride, 20 mM Tris, 5 mM DTT, pH 8.4 at 4° C. Errors represent the standard error of the mean equilibrium dissociation constant obtained from three independent binding experiments.
Changes in free energy were calculated as follows: ΔΔG°' = RT ln(KDalternate/KDwt, where R = 1.9872 cal/(mol•K), T = 277.15 K.
Fold difference is relative to the wild-type oligonucleotide.
FL-Pot1 Accommodates Sequence Alteration of the 3' Portion of Telomeric Oligonucleotides
As we found that SpPot1 is tolerant of the majority of single base changes, particularly in the 3'-end of the ligand, we tested binding to various oligonucleotides with more extensive sequence changes (Table 3). Not unexpectedly, FL-Pot1 displays markedly reduced binding affinities for polyT oligonucleotides. The affinity for FL-Pot1 binding to a 12-nucleotide polyT sequence (T12, KD > 250 nM) is diminished by at least five orders of magnitude from wild-type binding. Interestingly, the binding affinity for FL-Pot1 binding to T15 (8.6 ± 1.9 nM) is significantly tighter than that of T12. We observed no detectable binding to the complementary sequences of 12mer and 15mer with oligonucleotide concentrations up to 4 μM, demonstrating that binding is not strictly non-specific. As the majority of the specificity determining positions are within the first telomeric repeat, we tested the effects of altering only the 3'-portion of 12mer and 15mer. FL-Pot1 binds GGTTACTTTTTT (6mer_T6) or GGTTACTTTTTTTTT (6mer_T9) with equal affinities to wild-type sequence, exhibiting KDs of 3.2 ± 0.3 and 2.6 ± 0.2 pM, respectively. Intriguingly, we saw no change in affinity for 6mer_9 comp (GGTTAC CCAATGCCA; 3.8 ± 0.1 pM) relative to the telomeric 15mer and a less than 5-fold decrease in affinity for 6mer_6 comp (GGTTAC CCAATG; 9.5 ± 0.3). Considering the high affinity binding observed for these oligonucleotides, we tested the effect of including only the three specifically recognized nucleotides in the 15mer_comp sequence (CGTTTGCCAATGCCA), and observed substantial restoration of binding to 10.6 ± 0.3 nM. These results demonstrate that while SpPot1 binding is specific for telomeric sequence and discriminates completely against its complement, it is also remarkably robust to large alterations in nucleotide sequence, both individually and in block sequence replacement.
Mutation of Residues within the ssDNA-binding Cleft of Pot1-DBD Show a Greater Effect on 12mer Binding than on 15mer Binding
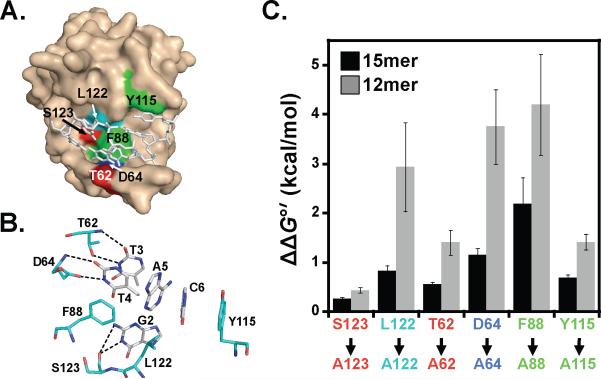
While there is no structure of Pot1-DBD, insights into the DNA-contacting residues have been obtained through examination of the Pot1pN/6mer crystal structure (24) and solution NMR chemical shift indexing of the free and 6mer-bound Pot1pN complexes (42, 43). Chemically diverse residues make a variety of contacts with the 6mer DNA, including aromatic stacking, hydrogen bonding and van der Waals interactions. We previously characterized the effects on Pot1pN binding utilizing several mutants containing alanine substitutions for residues within the DNA binding cleft (Figure 6AB) (35). To further probe the basis for the differential binding modes observed for 12mer and 15mer, we examined the effects of these residue alterations on Pot1-DBD binding to 12mer and 15mer.
Figure 6.
Analysis of Pot1-DBD containing single alanine substitutions within ssDNA-binding interface of the N-terminal OB-fold reveals alternate binding modes for 12mer and 15mer. (A) Surface representation of Pot1pN/6mer crystal complex highlighting ssDNA-binding interface residues mutated to alanine. 6mer ssDNA ligand (sticks) is shown within the ssDNA-binding cleft for reference. (B) Hydrogen bonding and aromatic stacking interactions between protein residues (cyan) and nucleotides (white) within the ssDNA-binding interface. (C) ΔΔG°' values for 12mer (gray bars) and 15mer (black bars) binding to each Pot1-DBD alanine mutant were calculated as follows: ΔΔG°' = RT ln(KDala/KDwt), where R = 1.9872 cal/(mol•K), T = 277.15 K. Images in (A) and (B) were generated using PyMOL version 1.3 (53).
We first examined S123 and L122, which interact with nucleotide G2 via hydrogen bonding and van der Waals interactions, respectively, in the context of the Pot1pN/6mer complex (Figures 6A and 6B). Mutation of S123 to alanine causes a modest 3-fold loss in affinity compared to 12mer (8.2 ± 0.9 pM; Figure 6C, Table 4). Similarly, binding of Pot1-DBD_S123A to 15mer results in < 2-fold change in affinity (3.7 ± 0.3 pM; Figure 6C, Table 4). In contrast, mutation of L122 to alanine has a large effect on binding to 12mer, with a KD of 770 ± 230 pM, corresponding to a 280-fold reduction in binding affinity (Figure 6C, Table 4). Interestingly, Pot1-DBD_L122A binds 15mer with much greater affinity than 12mer (10 ± 1.4 pM; Figure 6C, Table 4), representing a 77-fold difference in binding. Taken together, these data demonstrate that S123 is dispensable for high affinity binding of Pot1-DBD to both sequences, while only 12mer binding is profoundly affected by the loss of L122.
Table 4.
KD values for Potl-DBD N-terminal ssDNA-binding cleft mutants bound to 12mer and 15mer.
| Potl-DBD construct | 12mer KD(pM)a | Fold Changeb | 15mer KD(pM)a | Fold Changec |
|---|---|---|---|---|
| Potl-DBD_WT | 2.8 ± 0.4 | --- | 2.2 ± 0.1 | --- |
| Potl-DBD_S123A | 8.2 ± 0.9 | 2.9 | 3.7 ± 0.3 | 1.7 |
| Potl-DBD_L122A | 770 ± 230 | 20 | 10 ± 1.4 | 4.7 |
| Potl-DBD_T62A | 48 ± 8 | 17 | 6.2 ± 0.6 | 2.8 |
| Potl-DBD_D64A | 3400 ± 670 | 1200 | 18 ± 2.4 | 8.2 |
| Potl-DBD_F88A | 7700 ± 1800 | 2700 | 120 ± 30 | 26 |
| Potl-DBD_Y115A | 49 ± 5 | 17 | 8.0 ± 0.7 | 3.6 |
Apparent KD values were obtained from at least three independent EMSA experiments conducted in 50 mM sodium chloride, 20 mM Tris, 5 mM DTT, pH 8.4 at 4° C. Errors represent the standard error of the mean equilibrium dissociation constant obtained from three independent binding experiments.
Fold difference is relative to the 12mer sequence.
Fold difference is relative to the 15mer sequence.
Two hydrogen bonds are inferred between nucleotide T3 and residue Thr62, and T4 stacks with F88 and hydrogen bonds to D64 (Figure 6B). Alteration of Thr62 leads to an intermediate change in binding to 12mer (48 ± 8 pM) and little change in binding affinity to 15mer sequences (6.2 ± 0.6 pM), as compared to wild-type Pot1-DBD binding (Table 4, Figure 6C). This represents an approximate 10-fold difference in binding between the 12mer and 15mer interactions. These data suggest that Thr62 interactions have moderate influence on both the 12mer or 15mer binding modes, but again show a differential effect between the two modes.
The largest discriminatory mutation is at residue D64, with mutation at this position resulting in an ~200-fold difference in affinity between 12mer and 15mer (Table 4, Figure 6C). Mutation of D64 to alanine causes a 1200-fold reduction in binding to 12mer (3400 ± 670 pM), while conferring only a modest change in KD to 18 ± 2.4 pM for binding 15mer. Mutation of F88 to alanine drastically weakens both 12mer and 15mer binding (7700 ± 1800 pM and 120 ± 30pM, respectively), but once again has a larger effect on 12mer. These data indicate that the contribution of the T4/D64 interaction is greatly reduced for 15mer binding and that the T4/F88 stack is critical for both binding modes, however, has a larger impact upon interaction with 12mer.
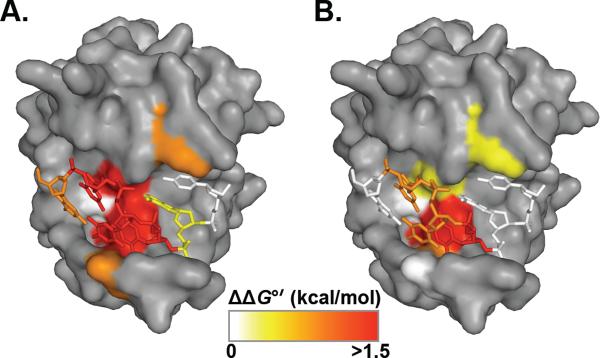
The final residue change that we investigated was Y115A. In the Pot1pN complex structure, Y115 forms a three-ring stack with nucleotides C6 and A5 (Figure 6B). Pot1-DBD_Y115A binds 12mer with an ~20-fold reduction in binding affinity (49 ± 5 pM) and binds 15mer with an ~4-fold reduction in binding affinity (8.0 ± 0.7 pM) (Table 4, Figure 6C). These data follow the pattern of Pot1-DBD_T62A, with Y115 making very little contribution to 15mer binding and playing a modest role in 12mer binding. The effects of the six alanine mutations on 12mer and 15mer binding reveal significant differences in the accommodation of ssDNA-binding cleft alterations within the different binding modes. Together the nucleotide and alanine substitution data reveal a substantially smaller hot-spot of binding affinity and specificity in the 15mer binding mode (Figure 7).
Figure 7.
SpPot1 hot-spot of binding specificity. Representations of Pot1pN (surface) and bound 6mer (sticks) with the ssDNA-binding interface colored according to effects of substitution on 12mer (A) and 15mer (B) binding modes. ΔΔG°' < 0.6 kcal/mol (white), 0.6 – 1 kcal/mol (yellow), 1 – 1.5 kcal/mol (orange) > 1.5 kcal/mol (red). All images were generated using PyMOL version 1.3 (53).
Discussion
The C-terminal Domain of FL-Pot1 Does Not Alter the Binding Activity of SpPot1
We find that FL-Pot1 binds S. pombe telomeric sequences ranging from 12 nucleotides (two telomeric repeats) to 24 nucleotides (four telomeric repeats) with no change in KD (Table 1). Additionally, there is little to no change in binding affinity with the inclusion of small sequences between telomeric repeats (Table 1). These data are consistent with our previous Pot1-DBD binding studies (34). Furthermore, we show that FL-Pot1 and Pot1-DBD bind telomeric DNA sequences equivalently (Figure 1; Supporting Information Table S1), and display the same nucleotide recognition profiles (Figure 4). We also observe that, like Pot1-DBD, FL-Pot1 binds 15mer as a monomer (Figure 2; Supporting Information Figure S1). These data support the established model, in which Pot1-DBD confers the full DNA binding activity of FL-Pot1, and indicate that there is no modulation of affinity or oligomerization state by the C-terminal domain. These observations are also consistent with the hPOT1 system, in which binding of the 2-OB fold DBD recapitulates that of full-length hPOT1 (6, 25).
Alternate Binding Modes Confer Differential Tolerance to Substitutions within the SpPot1/ssDNA Interface
Several lines of evidence indicate marked biochemical differences between the 12mer and 15mer binding modes. First, we observe a 2-fold difference in the half-lives of the FL-Pot1/12mer and FL-Pot1/15mer complexes (Figure 3). This result is intriguing in light of the nearly identical affinities for SpPot1 binding to the two oligonucleotides. These data indicate that 15mer forms a more temporally stable complex with SpPot1. Indeed, this increased stability is consistent with data from exonuclease protection assays. Investigating long oligonucleotide sequences (> 5 telomeric ssDNA repeats), Trujillo, et al. observed a 2–4× increase in end protection with sequences terminating GTT (corresponding to 15mer) compared to those ending in TAC (36).
Second, we observe significant relaxation of the specificity requirements in the context of 15mer binding, from seven specifically recognized nucleotides for 12mer (GGTTACGGTTAC) to three positions for 15mer (GGTTACGTTACGGT) (Figure 4; Supporting Information Table S1). Unexpectedly, while 15mer is the composite oligonucleotide of the Pot1pN and Pot1pC minimal binding sequences, neither the net binding affinity nor the 15mer specificity pattern recapitulates those of Pot1pN and Pot1pC. Interestingly, mutation of the telomere RNA template such that a G is incorporated into the C6 position has no effect on telomere maintenance in vivo (37), which is in agreement with both the 12mer and 15mer nucleotide specificity profiles. Despite the difference in sequence length, the 12mer specificity profile more closely resembles those of Pot1pN and Pot1pC, with the majority of the 5'-recognition intact, and a limited amount of specificity maintained for the 3'-end of the oligonucleotide. Consistent with loss of discrimination against base substitutions in the 15mer binding mode, non-canonical nucleotide substitutions (G2I, T3dU and T4dU) have no effect upon SpPot1 binding to 15mer (Table 2; Figure 5B). Complementing the effects of nucleotide alterations, alanine substitutions for amino acids in the ssDNA-binding cleft have far greater effects in the context of the 12mer binding mode (Table 4; Figure 6). Five of the six alanine mutations tested result in little to no change in Pot1-DBD binding to 15mer, with only the F88A mutation having a major impact on binding. Each alanine substitution has a larger impact on 12mer binding as compared to 15mer, with the most deleterious being the L122, D64, and F88 mutations. The effects of nucleotide and amino acid substitutions on 12mer binding parallel those observed for Pot1pN binding to 6mer (24, 29) and indicate that the Pot1pN/6mer interface is likely conserved in the context of Pot1-DBD. Taken together, the data define alternate hot-spots of recognition (Figure 7) and suggest a model in which H-bonding and other specificity-defining contacts are individually nonessential in the optimal 15mer binding mode, but are required to maintain the same binding affinity in complex with a smaller ligand.
Finally, another difference between the binding modes may be the salt dependence of the specificity profile. We observe that the 12mer pattern is different from the profile we previously determined for Pot1-DBD under high-salt binding conditions (34). Binding conducted in buffer containing 400 mM NaCl yielded a perfectly symmetric specificity requirement for only the two GTT repeats (GGTTACGGTTAC) versus the skewed profile observed at low salt GGTTACGGTTAC (Figure 4A). In contrast, we see no evidence for a salt-dependent alteration in 15mer recognition (data not shown), nor is there a change in specificity of binding for the S. cerevisiae TEP protein Cdc13 between 50 and 750 mM NaCl ((32, 33) (J. Earley, D.S.W unpublished results). Salt-dependent changes in binding mode are observed for the non-specific ssDNA-binding proteins RPA and SSB (44–46); however, the changes in these systems are characterized by drastic alterations in length preference resulting from a change in the number of OB-folds participating in binding. Thus, the SpPot1 differences in 12mer specificity indicate an unexpected alteration in binding mode between 50 and 400 mM salt not predicted by the log-log linearity of the measured salt-dependence for 12mer binding (34).
Proposed Basis for Binding Mode Discrimination
While full assessment of the binding mode will require structural data of the Pot1-DBD complex, the 12mer binding mode exhibits sensitivity to protein and ssDNA alterations similar to that of the Pot1pN/6mer complex (24, 29). Thus, using the 6mer-bound Pot1pN structure, we can speculate on the origin of some of the substitution effects and differences between 12mer and 15mer binding. As T4 and F88 make the largest contributions to the binding affinity of SpPot1 for 15mer, this supports the idea that the T3-T4-F88 aromatic stack observed in the Pot1pN/6mer structure is maintained in both binding modes and contributes critically to binding. The importance of this aromatic stack is emphasized by the observation that the F88A mutation is lethal in vivo (24). In addition to participating in this stacking interaction, T3 and T4 participate in several specific H-bonds with both amino acids and other nucleotides. The phosphate backbones of both T3 and T4 are coordinated by the A5 base and specific H-bonding is observed between the 4-carbonyl group of T4 and the 2-amino group of G2. While the G2I, A5T and D64A mutations, which presumably disrupt these interactions, impact binding to 12mer, they have little to no effect on 15mer binding (Figures 5 and 6; Tables 2 and 4). Our biochemical data suggest a model in which the aromatic stack and G2-T3-T4 interactions contribute stability and specific affinity to the 15mer binding mode and the combination of the other 5'-recognition elements adds specificity and increased affinity. Consistent with this model, SpPot1 exhibits no binding to the 15mer complement sequence; however, the affinity increases to ~10 nM with the inclusion of the specified GTT motif (CGTTTGCCAATGCCA), and wild-type affinity is restored with addition of all six bases of the first repeat (GGTTACCCAATGCCA) (Table 3).
High Affinity Recognition of the 5'-end by Telomere-end Protection Proteins
SpPot1 displays a strong preference for nucleotides within the first telomeric repeat of the 12mer and 15mer sequences. The human and S. cerevisiae TEP proteins also display preference for the 5'-end of their cognate substrates (25, 33). The human POT1 specificity profile for binding two telomere repeats (GGTTAGGGTTAG) reveals that hPOT1 strongly specifies positions T3 and T4. Similarly to Pot1pN/6mer, these positions stack against a phenylalanine in the hPOT1 crystal structures (25, 26). However, unlike hPOT1, SpPot1 displays no preference for the final 3'-oligonucleotide position in either 12mer or 15mer. As S. pombe telomeres do not consist of simple direct repeats, SpPot1 may maintain a malleable interaction with the 3'-end of the sequence in order to accommodate variability resulting from the heterogeneous spacer elements. 5'-specificity may also play a role in defining the binding register of the 3'-ssDNA overhang. Human POT1 establishes the ssDNA binding register by specifying the final base pair of the double-stranded portion of the telomere (8). Thus, the telomere overhang always begins with GGTTAG and presents the optimal binding register for hPOT1. While the binding register of the S. pombe telomeric overhang is not known, a similar system may exist in S. pombe, whereby SpPot1 sets the overhang register such that it begins with the specifically recognized sequence.
The SpPot1 characteristics of extremely tight binding and accommodation of heterogeneous sequence are reminiscent of S. cerevisiae Cdc13-DBD, which binds its cognate telomeric DNA sequence with 3 pM affinity (47–49). Similarly to SpPot1 binding to 15mer, Cdc13-DBD specifically recognizes only three nucleotides in the 5'-end of the telomere sequence (GTGTGGGTGTG) (33). As S. cerevisiae telomeric DNA is composed of degenerate sequence, Cdc13 is capable of binding myriad GT-rich sequences (31, 50, 51). However, the extent of sequence accommodation by SpPot1 is far greater than that of Cdc13. We see that complementary change of the entire 3'-end has little to no effect on either SpPot1 binding mode (Table 3), whereas Cdc13 binding is drastically compromised by fewer nucleotide alterations (33, 51).
The strikingly high affinities maintained by SpPot1 in spite of drastic changes in sequence seem at odds with the role of specific telomere end protection. As SpPot1 is targeted to the telomere through interaction with the telomere-associated protein Tpz1 (17), it may be that SpPot1 coordinates maintenance of the 3'-telomere overhang via discrimination against the C-strand, rather than G-strand specificity per se. These data indicate that specific targeting of SpPot1 to the telomere is essential in order to prevent non-specific interaction with other ssDNA sequences. However, this remarkable accommodation of non-telomeric sequence may be relevant to a recently described mechanism of linear chromosome maintenance in S. pombe in the absence of telomeric sequence (52). Termed HAATI survivors, some cells retain a “telomere” composed of ribosomal DNA, complete with a 3'-overhang and co-localization of SpPot1. Thus, SpPot1 binding of nontelomeric ssDNA may play unforeseen roles in genome maintenance.
Supplementary Material
Acknowledgment
We gratefully acknowledge Dr. Jayakrishnan Nandakumar and Professor Thomas Cech (University of Colorado, Boulder) for thoughtful discussion and the plasmid encoding FL-Pot1. We thank Lisa Warner for critical review of this manuscript.
We gratefully acknowledge the National Institutes of Health Training Appointment (NIH) GM065103 (to S.E.A. and T.H.D.), NIH GM-059414 (to D.S.W.), University of Colorado Cancer Center (to D.S.W.), and National Science Foundation (NSF) MCB-0617956 (to D.S.W.) for funding this research.
Abbreviations
- βME
β-mercaptoethanol
- BSA
bovine serum albumin
- DBD
ssDNA-binding domain
- CTD
C-terminal domain
- DTT
dithiothreitol
- EMSA
electrophoretic mobility shift assay
- FL-Pot1
full-length S. pombe Pot1
- H-bond
hydrogen bond
- hOB1
N-terminal OB-fold of human POT1
- hOB2
second OB-fold of human POT1
- hPOT1
human Pot1
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- KD
apparent equilibrium binding constant
- OB-fold
oligonucleotide/oligosaccharide-binding fold
- Pot1-DBD
residues 1–389 of full length S. pombe Pot1
- Pot1pC
residues 178–389 of full length S. pombe Pot1
- Pot1pN
residues 1–185 of full length S. pombe Pot1
- ssDNA
single-stranded deoxyribonucleic acid
- SpPot1
Schizosaccharyomyces pombe protection of telomeres 1
- ssDNA
single-stranded deoxyribonucleic acid
- TEP
telomere-end protection
Footnotes
Supporting Information Available A comprehensive table containing apparent KD values for FL-Pot1 and Pot1-DBD specificity profiles and FL-Pot1/15mer and Pot1-DBD/15mer multiangle light scattering data are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Croy J, Wuttke D. Themes in ssDNA recognition by telomereend protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flynn RL, Zou L. Oligonucleotide/oligosaccharide-binding fold proteins: a growing family of genome guardians. Crit. Rev. Biochem. Mol. Biol. 2010;45:266–275. doi: 10.3109/10409238.2010.488216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 7.Veldman T, Etheridge K, Counter C. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr. Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, De Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Q, Zheng YL, Harris CC. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 2005;25:1070–1080. doi: 10.1128/MCB.25.3.1070-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat. Struct. Mol. Biol. 2008;15:79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 12.Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bunch JT, Bae NS, Leonardi J, Baumann P. Distinct requirements for Pot1 in limiting telomere length and maintaining chromosome stability. Mol. Cell. Biol. 2005;25:5567–5578. doi: 10.1128/MCB.25.13.5567-5578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei M, Zaug A, Podell E, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 15.Palm W, De Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 16.Dehé P-M, Cooper JP. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 2010;584:3725–3733. doi: 10.1016/j.febslet.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 18.Jain D, Cooper JP. Telomeric strategies: means to an end. Annu. Rev. Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- 19.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, De Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Mitton-Fry R, Anderson E, Hughes TR, Lundblad V, Wuttke D. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 23.Mitton-Fry R, Anderson E, Theobald D, Glustrom L, Wuttke D. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 24.Lei M, Podell E, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 25.Lei M, Podell E, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 26.Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc. Natl. Acad. Sci. USA. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiraoka Y, Henderson E, Blackburn EH. Not so peculiar: fission yeast telomere repeats. Trends Biochem. Sci. 1998;23:126. doi: 10.1016/s0968-0004(98)01176-1. [DOI] [PubMed] [Google Scholar]
- 29.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 30.Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- 31.Förstemann K, Höss M, Lingner J. Telomerase-dependent repeat divergence at the 3' ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson E, Halsey WA, Wuttke D. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 33.Eldridge AM, Halsey WA, Wuttke D. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croy J, Podell E, Wuttke D. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Croy J, Altschuler SE, Grimm NE, Wuttke D. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–6875. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trujillo KM, Bunch J, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J. Biol. Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 37.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 38.Theobald D, Cervantes RB, Lundblad V, Wuttke D. Homology among telomeric end-protection proteins. Structure. 2003;11:1049–1050. doi: 10.1016/s0969-2126(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 39.Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell. Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 40.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 41.Theobald D, Wuttke D. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Croy J, Fast JL, Grimm NE, Wuttke D. Deciphering the mechanism of thermodynamic accommodation of telomeric oligonucleotide sequences by the Schizosaccharomyces pombe protection of telomeres 1 (Pot1pN) protein. Biochemistry. 2008;47:4345–4358. doi: 10.1021/bi701778x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Croy J, Wuttke D. Insights into the dynamics of specific telomeric single-stranded DNA recognition by Pot1pN. J. Mol. Biol. 2009;387:935–948. doi: 10.1016/j.jmb.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 45.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozlov AG, Eggington JM, Cox MM, Lohman TM. Binding of the dimeric Deinococcus radiodurans single-stranded DNA binding protein to single-stranded DNA. Biochemistry. 2010;49:8266–8275. doi: 10.1021/bi100920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 48.Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson E, Halsey WA, Wuttke D. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Förstemann K, Lingner J. Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 2001;21:7277–7286. doi: 10.1128/MCB.21.21.7277-7286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zappulla DC, Roberts JN, Goodrich KJ, Cech TR, Wuttke D. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. Nucleic Acids Res. 2009;37:354–367. doi: 10.1093/nar/gkn830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010;467:223–227. doi: 10.1038/nature09374. [DOI] [PubMed] [Google Scholar]
- 53.Schrodinger LLC. The PyMOL Molecular Graphics System. Version 1.3r1 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.