Abstract
Spatial normalization and segmentation of pediatric brain MRI data with adult templates may impose biases and limitations in pediatric neuroimaging work. To remedy this deficiency, we created a single database made up of a series of pediatric, age-specific MRI average brain templates. These average, age-specific templates were constructed from brain scans of individual children obtained from two sources: (1) the NIH MRI Study of Normal Brain Development and (2) MRIs from University of South Carolina’s McCausland Brain Imaging Center. Participants included young children enrolled at ages ranging from 8 days through 4.3 years of age. A total of 13 age group cohorts spanning the developmental progression from birth through 4.3 years of age were used to construct age-specific MRI brain templates (2 weeks, 3, 4.5, 6, 7.5, 9, 12, 15, 18 months, 2, 2.5, 3, 4 years). Widely-used processing programs (FSL, SPM, ANTS) extracted the brain and constructed average templates separately for 1.5T and 3T MRI volumes. The resulting age-specific, average templates showed clear changes in head and brain size across ages and between males and females, as well as changes in regional brain structural characteristics (e.g., myelin development). This average brain template database is available via our website (http://jerlab.psych.sc.edu/neurodevelopmentalmridatabase) for use by other researchers. Use of these age-specific, average pediatric brain templates by the research community will enhance our ability to gain a clearer understanding of the early postnatal development of the human brain in health and in disease.
Introduction
The study of brain magnetic resonance images (MRI) in young children allows for a quantitative and qualitative assessment of neurodevelopment that can enhance our understanding of early brain growth patterns and morphological changes during both normal and abnormal brain development. However, MRI research with normal, healthy human infants, toddlers and preschoolers has been hindered by a number of problematic factors, including (1) a reliance on the use of sedated clinical populations for MRI, especially for children ranging from infancy through preschool ages, and (2) a reliance on the use of adult brain templates and atlases for analyses (e.g., normalization and segmentation processing) of MRI scans obtained at infant through preschool ages. Sedation in neuroimaging research is not an ideal procedure and the use of clinical pediatric populations and adult brain data as the standard for neuroimaging analysis in young children is a questionable practice. The first problematic factor has been resolved somewhat by the website availability of a large MRI brain scan data repository from the NIH MRI Study of Normal Brain Development (NIHPD): Objective 1 for ages 4.5 to 18+ years, and Objective 2 for ages birth to 4.3 years (Almli et al., 2007; Brain Development Cooperative Group, 2006, 2011; Waber et al., 2007; Leppert et al., 2009; Lange et al., 2010). This repository includes MR brain scans of normal, healthy neonates, infants, children, adolescents, and young adults that were acquired during natural sleep (i.e., without sedation) or while awake. The second problematic factor regarding the use of adult brain templates with very young children is the target of this report, as we have constructed average MR brain scan templates of normal, healthy infants, toddlers and preschoolers that will be made available via website to developmental neural and behavioral researchers and clinicians.
It is important to emphasize that MR image analysis methods with adults requires adequate normalizations procedures which serve to align an individual subject’s MR image with a common reference space/template for inter-subject, inter-sample, or inter-population types of comparisons (Evans, 2005). Although some studies have reported that children as young as 7 years of age can be adequately normalized with adult templates (Burgund et al., 2002; Kang et al., 2003), MRI templates for older children between 4.5 and 18.5 years of age have recently become available (Fonov et al., 2011; Sanchez et al., 2010; Wilke et al., 2008) thereby diminishing dependence on adult templates for pediatric research.
Research using the Objective 1 (4.5 to 18+ years of age) NIHPD database has implemented different methods to produce MRI templates for school-age children. These methods utilized various (1) normalization and registration procedures (i.e., linear vs. nonlinear, iterative vs. noniterative), (2) age grouping sizes, and (3) NIH database releases. For example, Wilke et al. (2008) created a template-building platform (Template-O-Matic) through which researchers could specify the age range and sex of the resulting templates which were based on linear registration techniques. Fonov et al. (2011) constructed age-appropriate atlases that provided templates with significant anatomical detail for six age ranges with a width of 4 to 6 years each that were grouped according to estimated pubertal status: 4.5–8.5 years, pre-puberty; 7.0–11.0 years, pre- to early- puberty; 7.5–13.5 years, pre- to mid-puberty; 10.0–14.0 years, early to advanced puberty; and 13.0–18.5 years, mid- to post- puberty. Also, Sanchez et al. (2010) built age-specific templates grouped in 6 month increments from 4.5 years through young adulthood based primarily on the Objective 1 data. Both Fonov et al. (2011) and Sanchez et al (2010) utilized nonlinear registration and iterative techniques to build the templates. Linear registration has been described to blur anatomical details and decrease overlap between subjects when compared to nonlinear registration (Ashburner & Friston, 1999). Iterative techniques, wherein subsequent iteration processing is based on the previous average, avoids biasing the templates to adult reference data. Whereas Fonov (2011) grouped ages in ≥4 year increments, both Wilke et al. (2008) and Sanchez et al. (2010) allowed for more discrete age ranges. Finally, Sanchez and Fonov created the templates from the later releases of the NIHPD database, which allowed averaging from a larger participant pool. Nevertheless, all of the aforementioned MRI templates for school ages are useful to researchers for different purposes and represent the first pediatric atlases/templates based on an epidemiological sample of children.
The rapid growth occurring during birth through preschool years creates large variability within and between children for brain size, shape and tissue classes (Muzik et al., 2000; Wilke et al., 2002; Joshi et al., 2004; Prastawa et al., 2005). The use of the adult template for the spatial normalization process for MRI of infants and young children poses significant biases given the great differences between the infant brain image and an adult reference image (Muzik et al., 2000; Wilke et al., 2002). As an example, Altaye and colleagues (2008) report that the use of adult templates in normalization and segmentation of infant MR images results in misclassifications of infant tissue, and that the misclassifications are reduced when using an infant template, even though the infant template was constructed from scans that were averaged over the first 12 months postnatal (i.e., from birth through one-year of age). It thus appears that establishment of a comprehensive set of age-specific MRI brain templates would greatly facilitate our understanding of human brain development between the ages of birth and four years ( Altaye et al., 2008; Gaillard et al., 2001; Xue et al., 2007).
Although some MRI templates for children ranging between birth and four years of age have been published (e.g., Aubert-Broche et al., 2008; Altaye et al., 2008; Joshi et al., 2004; Kazemi et al., 2007; Prastawa et al., 2005; Shi et al., 2010; Shi et al., 2011; Srinivasan et al., 2007), many of those reports have included clinical populations of sedated infants and young children of questionable health status. In addition, those published templates have primarily focused on either the newborn brain only (Prastawa et al., 2005), or have studied only a small age range (Altaye et al., 2008), or have averaged scans across relatively wide / broad age-ranges within an individual template (e.g., an individual template constructed by collapsing and averaging scans of children from birth through one year of age [Altaye et al., 2008; Shi et al., 2011]). Thus, most of the currently published MRI templates for early human brain development between the ages of birth and four years have used clinical populations as subjects, and the resulting templates for the birth through four year age range are mostly incomplete or overly-averaged. The lack of a comprehensive and fine-grained set of published MRI templates for children ages birth through four years may account for the current reliance on adult templates or incomplete pediatric templates for use in MRI research with young children.
In order to fully understand the dynamic early postnatal development of the human brain through four years of age, an average brain template specific to each of multiple age time-points (i.e., a relatively fine grain) would serve as a beginning for the accurate representation of important developmental brain changes. This report provides a starting point as we have created 13 age-specific, average templates to cover the age span from birth through four years of age. The age choice for the age-specific templates was determined by the NIHPD Objective-2 sampling plan which included 11 age cohorts (Almli et al., 2007; Brain Development Cooperative Group, 2006). Two additional age cohorts from MRI conducted at the University of South Carolina’s McCausland Brain Imaging Center (i.e., USC-MCBI) were added to the NIHPD sequence, yielding a total of 13 age group cohorts.
This report describes application of the methodology for the template averaging processes (Evans et al., 1993) and how state-of-the-art averaging programs (Avants et al., 2008, 2011; Klein et al., 2009) can be used to create templates of averaged MRI volumes with manual and automated procedures based on widely available software. Automatic averaging procedures are important for dealing with large numbers of brain scans (e.g., the NIHPD and USC-MCBI data sets of this report) and for the usefulness of such templates in pediatric research and clinical applications. For this report on infants and preschoolers, we followed the same methodology as in Sanchez et al. (2010) who created average age-specific templates for children from 4.5 years of age through young adulthood.
Methods
Participants
The MRI images for this study came from the NIH MRI Study of Normal Brain Development (NIHPD) and the University of South Carolina McCausland Center for Brain Imaging (USC-MCBI). The NIHPD MRI images came from infants and young children ranging in age from 8 days to 4.3 years. The participants were screened for the presence of factors that might adversely affect brain development (e.g., preterm birth, physical growth delay, perinatal complications, learning disabilities, psychiatric disorders of first order family members). The infants were scanned with 1.5T MRI, while awake, or during natural sleep without sedation. The Objective-2 cohort ages (EDC adjusted) included (see Table 1 for ages and number of participants): 2 weeks (8 to 29 days); 3, 6, 9, 12, and 15 months (each ± 2-weeks); 1.5, 2, 2.5, 3 years (each ± 4-weeks); and ages 4 years to 4 years 5 months. The NIHPD had a combined cross-sectional and longitudinal design, gathered from 105 participants for a total of 294 MRI scans. Detailed methodology for the NIHPD Objective-2 can be found in recent publications (Brain Development Cooperative Group, 2006; Almli et al., 2007; Leppert et al., 2009), as well as from the Objective-2 Procedure Manuals available at the NIHPD Database Repository website (https://nihpd.crbs.ucsd.edu/nihpd/info/index.html).
Table 1.
Age at brain scan, number of participants (by gender) providing scans for each age group, and total number of scans completed for each age group for NIHPD and MCBI.
| Age | Days, Range | Days, Mean | Number of participants* Female / Male | Total number of brain scans** |
|---|---|---|---|---|
| 2 weeks NIHPD | 8–29 | 19.7 | 12 / 11 | 23 |
|
| ||||
| 3 Months NIHPD | 75–105 | 90.3 | 10 / 11 | 22 |
| 3 Months MCBI | 95–110 | 103.3 | 6 / 4 | 10 |
|
| ||||
| 4.5 Months MCBI | 142–145 | 143.2 | 4 / 6 | 10 |
|
| ||||
| 6 Months NIHPD | 167–197 | 182.1 | 15 / 17 | 32 |
| 6 Months MCBI | 179–203 | 194.8 | 5 / 5 | 10 |
|
| ||||
| 7.5 Months MCBI | 223–245 | 234.5 | 6 / 4 | 10 |
|
| ||||
| 9 Months NIHPD | 257–287 | 273.8 | 16 / 13 | 29 |
| 9 Months MCBI | 274–288 | 281.6 | 2 / 2 | 4 |
|
| ||||
| 12 Months NIHPD | 352–376 | 366.2 | 11 / 14 | 25 |
| 12 Months MCBI | 365–383 | 372.7 | 3 / 3 | 6 |
|
| ||||
| 15 Months NIHPD | 445–475 | 461.4 | 14 / 18 | 32 |
|
| ||||
| 18 Months NIHPD | 526–571 | 550.9 | 14 / 18 | 32 |
|
| ||||
| 2.0 Years NIHPD | 722–767 | 740.2 | 9 / 18 | 27 |
|
| ||||
| 2.5 Years NIHPD | 886–958 | 927.0 | 13 / 18 | 31 |
|
| ||||
| 3.0 Years NIHPD | 1080–1131 | 1107.5 | 13 / 9 | 22 |
|
| ||||
| 4.0 Years NIHPD | 1468–1552 | 1508.0 | 9 / 10 | 19 |
Number of participants (female / male) providing brain scans to the specific age-groups from NIHPD and MCBI. Note that an individual participant from the NIHPD could contribute a scan to multiple age groups based on the longitudinal design, not so for the MCBI [Participant pool: NIHPD = 105 (46 females/59 males), MCBI = 49 (25 females/24 males)].
Total number of brain scans completed for each specific NIHPD and MCBI age group.
The USC-MCBI sample consisted of smaller sample of children (N = 49, 25 F / 24 M). The USC-MCBI sample were healthy participants who had been screened for the presence of preterm birth, perinatal complications, and any other significant health problems. The infants were scanned with 3.0T MRI during natural sleep (without sedation). The MCBI ages included (see Table 1): 3, 4.5, 6, 7.5, 9, or 12 months of age, and were scanned within 30 days after the nominal age (e.g., the 3 month age-group participants were scanned during the interval of 3 months, 5 days through 3 months, 20 days), and the USC-MCBI design was cross-sectional only. Thus, the USC-MCBI sampling strategy created four age groups that overlapped with the NIHPD cohorts (i.e., 3, 6, 9, 12 months), as well as adding two new age cohorts (i.e., 4.5 and 7.5 months). The USC-MCBI used only a cross-sectional design, i.e., without a longitudinal component.
Institutional review board approval and informed consent were obtained for all NIHPD and USC-MCBI participants.
MRI Data Acquisition
The MRI procedures for the NIHPD Objective-2 are described in detail by others (Brain Development Cooperative Group, 2006; Almli et al., 2007; Leppert et al., 2009), as well as in the Objective-2 Procedure Manuals available at the NIHPD Database Repository website (https://nihpd.crbs.ucsd.edu/nihpd/info/index.html). Briefly, the Objective 2 MRI acquisition generally lasted 30–45 minutes on a 1.5T scanner with a 2D sequence that minimized scan duration for the birth to 4 year age-groups. The axial scans consisted of a 2D T1-weighted spin echo and a T2-weighted 2D Fast Turbo spin echo sequence. The T1 and T2 scans were nominally 1×1×3 resolution (1×1×3 or .97×.97×3), with a FoV of 192 X 256 or 256 × 256 mm, and ranged from 46 to 66 slices in the axial plane. This FoV and resolution was enough to cover from the top of the head to at least below the bottom of the braincase. The NIHPD Objective-2 scans were conducted at two different sites/scanners. The site using a Siemens Medical Systems (Sonata, Magnetom) scanner provided the majority of the scans, while the other site used a GE (Signa Excite) scanner and provided less than half of the scans. The scans that we used were obtained from the NIHPD website in compressed NIFTI format.
The MRI data from the USC-MCBI were collected on a Siemens Medical Systems 3T Trio with an overall duration of about 15 min. A 3D T1-weighted “MPRAGE” RF-spoiled rapid flash scan in the sagittal plane and a T2/PD-weighted multi-slice axial 2D dual Fast Turbo spin-echo scan in the axial plane were used. The USC-MCBI T1 scans had 1 mm3 resolution and sufficient FoV to cover from the top of the head down to the neck, whereas the T2 scans were 1×1 mm2 in the axial plane, but varied from 1 to 2.5 mm in the axial slices, with enough FoV and resolution to cover the entire brain and surrounding CSF. The USC-MCBI (3T) files were read from DICOM files to compressed NIFTI format (http://nifti.nimh.nih.gov/). All MRI volumes, whether from the NIHPD database or from USC-MCBI, were processed in the same manner using NIFTI compressed format and 32 bit floating point resolution.
File Preparation
The MRI images were prepared in three steps.
Step-1. The brains were extracted from the whole-head MRI volume using the FSL and SPM computer programs. The extraction was done with procedures recommended by the FMRIB group (Jenkinson et al., 2005, and Smith, 2002). It includes registering the head to an average template that already has a brain mask, using the registered brain mask on the child head volume to extract an initial brain; use the FSL procedures betsurf and bet2 to extract the brain; visually inspect the brain for accuracy and adjust parameters to get a well-formed brain.
The MNI head template (MNI-152 or ICBM-152 defined in Mazziotta et al, 2001; Joshi et al., 2004) cannot be automatically registered to the infant heads because of size and structural differences between the adult template and infant heads. The initial step of our procedure involved extensive manually guided brain extraction from a subset of the 3T USC-MCBI 6-month-old MRIs. We constructed a preliminary template of the 6-month-old head and brain from four participants using the FSL FLIRT, “FMRIB’s Linear Image Registration Tool” (Jenkinson & Smith, 2001). This preliminary template for the head and brain then guided the automated brain extraction procedure. Next, the preliminary template was then applied to all participants of the same age (6 months) and a complete template was constructed for the 3T volumes. The average template for the 3T volumes at 6 months of age (N = 10) successfully worked as the initial brain mask for all 6-month NIHPD 1.5T volumes in the current study. The same 6-month-old average template was then used to guide the brain extraction for the other ages and preliminary templates were constructed. The age-specific preliminary template then was used to extract the brain for the infants at that age. This iterative procedure resulted in average templates that could be applied to any new brain at that age to extract the brain using the automated extraction procedure (e.g., the average head and brain template for the 3T MRI volumes worked for the brain extraction of all 1.5T volumes at the same age).
Step-2. The cerebrospinal fluid was identified in the head volumes. Cerebrospinal fluid is easily identified in all ages as voxels with the brightest intensities in the T2W MRI volume. Our procedure first computed preliminary voxel probability for T2W-CSF in the T2W volume by thresholding the voxel intensity of the T2W voxels with the intensity values found in the lateral ventricles. The average voxel intensity of the lateral ventricles in the T2W scans was used to threshold the T2W voxel intensities, which gives an approximate map of cerebrospinal fluid in the entire T2W brain. This map was transformed by relative voxel intensity into a probability map representing the CSF. The T2W-CSF probability maps for individual participants may be averaged to form an average, age-specific template for each age group. We also have found that the average T2W-CSF template, inverse transformed to the individual MRI volume, and used as a prior template in the SPM8b segment method accurately identifies individual cerebrospinal fluid distributions in MRI volumes of individual participants.
Step-3. Some intensity variations occur in the MRI scans. First, bias field inhomogeneity was corrected with a N4 bias field correction procedure (Avants et al., 2011; Tustison et al., 2010). Second, intensity variations were corrected in the T1W scans by segmenting the scan into GM, WM, and CSF using the FSL FAST procedure. The identification of GM, WM, and CSF was not intended as a final tissue segmentation step, but the process resulted in a putative GM intensity that closely matched the histogram peak for GM in the brain volume. The scan was renormed by masking putative GM in the T1W, finding the MRI voxels with partial volume estimates of 1.0 in the GM segments, and norming the T1W (head or brain) so that the GM intensity had an average value of ~100 for all MRIs. The T2W scans were normed to find the cerebrospinal fluid in the lateral ventricles with an average intensity of 100.
Iterative Average Procedure
Iterative routines constructed the average templates (Guimond, 2000; Sanchez et al., 2010; Yoon et al, 2009). The whole-head and brain-extracted MRI volumes were performed separately to provide both head and brain templates. Figure 1 is a schematic representation of the steps used in the construction of the average template for a specific age group for both whole-head and brain-extracted templates. Before the iterative non-linear procedure, a preliminary template was made by averaging a rigid rotation (FLIRT 6 parameter linear registration and transformation; Jenkinson & Smith, 2001) of each participant image to a preliminary brain template that was in the same orientation as the MNI-152 adult template (ICBM-152 defined in Mazziotta et al, 2001; Joshi et al., 2004). The original MRI volumes were then registered to this template and transformed in size and orientation with non-linear registration (using ANTS, “Advanced Normalization Tools”; Avants et al., 2008, 2011) into this template space (Figure 1B, Vn), and averaged (Figure 1B, An-1). This then became the next reference template for the registrations (An). This procedure was iteratively applied separately for each of the study ages, and separately for the head and brain volumes. A gross resolution (50 steps maximum at 8 mm resolution) began the first template normalization step followed by the second step with finer resolution (4 mm resolution), and the final steps used 1 mm voxels. The first non-linear registration with 8 mm resolution retained the approximate size and shape of the head / brain for a specific age; subsequent steps used a size adjustment to retain the initial size and orientation for the entire sequence (affine 9-dof registration). The RMS (root mean square) difference between intensity values of successive reference templates was calculated and the iterative procedure was performed until the successive RMS values reached a minimum. The resulting average, age-specific template served as the final reference template.
Figure 1.
The processing steps for the age-specific template creation. (A). In Step 0, the rigid registration occurred using FLIRT to a average template with 6 DOF rotation, with the resulting MRI volume the same size as the original to keep pediatric sizes. The rigidly registered brains (V0) were averaged to create a rough template (A0). This template was used as the first tentative template in Step 1. (B) With each iteration of step N, the rigidly registered brains were nonlinearly registered to an iterative average (An-1), and transformed and then averaged to create a new average (An) for the next iteration.
A similar iterative procedure constructed the T2W image templates. The T2W construction contained an additional step where the current reference template was registered with affine parameters (FLIRT) to the relevant T1W volume on each step to retain the T2W in the same size and orientation as the T1W volume (e.g., Figure 1B, “Next Iteration” includes FLIRT of T2W average to the T1W average template).
The procedures constructed templates for T1W head, T1W brain, T2W head, and T2W brain separately at each age. The T2W head construction resulted in a MRI volume that was co-registered (ANTS) to the corresponding T1W head template. Likewise, the T1W and T2W brains are co-registered. The average brain template(s) are not precisely registered to the corresponding head template(s) since the whole head and extracted-brain constructions were separate. We therefore extracted the brain from the completed head template to provide a brain model corresponding to the space of the average head MRI template.
Results
Developmental brain and behavioral researchers realize that gaining understanding of early neurobehavioral development (i.e., birth through school ages) will be facilitated by the establishment of average, age-specific MRI brain templates throughout early developmental periods. Such brain templates are necessary for at least two major reasons: (1) adult average MRI brain templates hardly resemble the MR images of the young infant brain (i.e., newborn to three months), and (2) even within a narrowly defined developmental age, such as “three months” of age (defined here as three months +/− two weeks of age), individual differences in whole brain and regional brain sizes, shapes, and tissue classes are readily apparent with MRI.
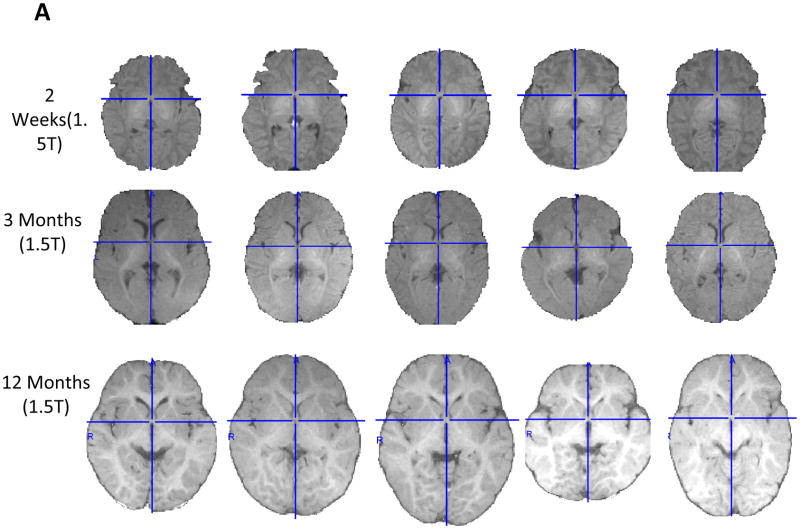
Examples of individual differences in brain size and structural characteristics are shown in Figure 2(A and B) for infants and young children of the same age for easy comparison of the images. Figure 2A shows an axial slice of a 1.5T MRI (T1w) for five participants each at ages 2 weeks, 3, 12, and 18 months, and 4 years. Figure 2B shows 3T MRI (T1w) for five participants each at ages 3 and 6 months. For the brain slices in Figures 2A and 2B, the left side of brain is on the reader’s right, and the crosshairs on the individual axial slices are placed on the anterior commissure (AC) and run through the posterior commissure (PC). These axial slices are oriented to the Talairach stereotaxic space which is the approximate alignment of the adult MNI-152 average template (Joshi et al., 2004; Talairach & Tournoux, 1988). The individual MRI slices shown preserve the relative size of the brain for children within a single age-group, and the differences across age retain the approximate relative size of the head.
Figure 2.
Axial slice of extracted brain for individual infants at 2 weeks, 3, 12, or 18 months, or 4 years of age for 1.5T (2A), or 3 and 6 months of age for 3T (2B). The crosshairs are placed on the anterior commissure (AC) and run through the posterior commissure (PC). The individual figures preserve the relative size of the brain for children within one age-group, and the differences across age retain the approximate relative size of the head. Note: all axial slices shown in figures are oriented to the Talairach stereotaxic space, which places a line drawn from the AC to the PC orthogonal to the sides of the MRI volume, and is the approximate alignment of the MNI-152 average template (. All axial MRI volumes in all figures are shown with the left side of the brain on the right side of the figure.
For all age groups under study with either 1.5T or 3T scanners, there were clear individual variations in structure, size, and topology of brains within specific individual age groups. Examples for individual differences within single age groups are presented below for size, shape, and myelination of the brain during early development. Figure 2A shows clear individual differences in the specific size and shape of the axial slices from 5 participants at birth (2 weeks), and also shows that individual differences for size and shape are present through the 3, 12, and 18 month ages, as well as at 4 years of age. Figure 2A and 2B show individual differences in the timing and pattern of myelination development across axial slices of different participants. For newborns (2 weeks) in Figure 2A, there is a dearth of myelin present in the axial slices of the 5 participants except for some early (immature) myelin seen in the midline diencephalic (i.e., thalamic) region, and the degree of this early myelination differs across participant slices. Figure 2A and 2B show that the posterior limb of the internal capsule is becoming myelinated by 3 months of age, while the anterior limb of the internal capsule does not have significant myelin at 3 months. Different participants at 3 months of age (Figure 2A and 2B) show individual differences for myelin development at this age. At 6 months of age (Figure 2B), the poster regions of the hemispheres (e.g., occipital lobes) are displaying clear myelin increases with individual differences, while the myelination within the frontal lobes lags significantly with individual differences. Despite individual differences (Figure 2A), the amount of myelin in the frontal lobes has clearly increased by 12 months of age, and the extent of myelin development is still increasing in both hemispheres and the entire brain through 4 years of age. Thus, although there is a relatively consistent pattern for brain myelin development between age groups, there are obvious individual differences in the timing and extent myelin development within each of the specific age groups. The sample of MR images shown in Figure 2A and 2B are representative of those used to create the age-specific, average brain templates described below.
Template Database
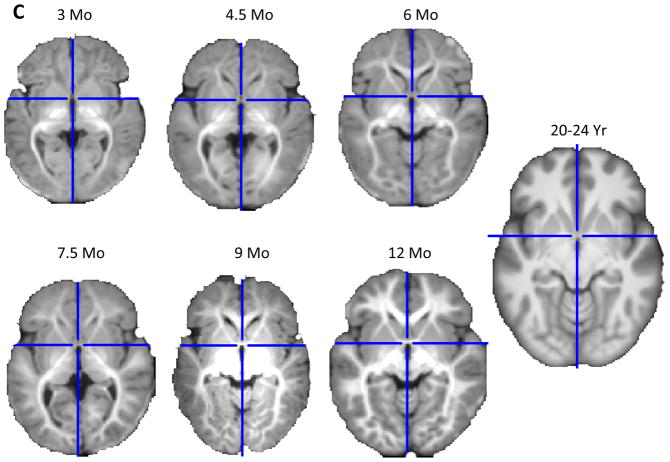
The template database consists of average templates of 1.5T MRI scans at the nominal ages defined by the NIHPD (i.e., all NIHPD Objective 2 age groups; 2 weeks; 3, 6, 9, 12, 15, 18 months; 2, 2.5, 3, 4 years), and average templates consisting only of 3T scans at 3, 4.5, 6, 7.5, 9, and 12 months1. Figure 3(A and B) show mid-sagittal (A) and axial (B) slices for each of the 11 age-specific, average head templates based on the NIHPD 1.5T MRIs. Figure 3C shows an axial slice for the 6 age-specific, average brain templates at 3, 4.5, 6, 7.5, 9, and 12 months for the MCBI 3T MRIs. Figure 3(A, B, and C) also shows an average MRI (3T) template made up of 20–24 year olds (i.e., adults) that were created with the same methods as used here in this report. The adult template can be used for comparisons with the birth through 4.3 year old templates (from Sanchez et al., 2010).
Figure 3.
Age-specific templates for 1.5T scans showing mid-sagittal slice (A) and axial slice at AC-PC commissure (B) of whole head average template T1W MRI volumes across ages of study. The individual figures preserve the relative size of the head for children at that age. Age-specific templates for 3T scans showing axial slice (C) of brain average template T1W MRI volumes. Note: all axial slices shown in figures are at the origin location of the Talairach stereotaxic space, which is a line drawn from the anterior commissure to the posterior commissure. The scans come from 1.5T scans for 2 weeks, 3, 6, 9, 12 months, 1.5, 2, 2.5, 3, and 4 yrs; 3T scans for 3, 4.5, 6, 7.5, 9, and 12 months. The 20–24 year scans come from participants aged 20 through 24.9 years and are combined 1.5T and 3T scans (3A, 3B) or only 3T scans (3C).
Figure 3A (mid-sagittal) and 3B (axial) show that average template head size increases rapidly through about 15–18 months of age, and continues at a slower pace through 4 years of age. The average brain templates preserve the overall pattern of brain development results that were presented above regarding individual participant differences. For example, beginning with an absence of myelination at birth (2 weeks of age), the average templates reveal regional patterns of myelin development that show myelination of (1) the posterior limb of the internal capsule at 3 months; (2) the anterior limb of internal capsule at 4.5 months; (3) the posterior regions of the hemispheres (e.g., occipital and posterior temporal lobes, subcortical) at 6 to 7.5 months; and (4) the frontal lobe at 9 to 12 months (note the lag between 4.5 to 6 months). In comparison to the average adult template shown, it is clear that brain development is not complete by 4 years of age.
Figure 4 shows the change in age-specific template fit with successive iterations. Each iteration represents the registration of the individual participant MRIs to the tentative average template, and the averaging of the transformed participant MRIs. The RMS (root mean square) difference measures the intensity difference between successive iterations at each voxel. Figure 4 compares the iteration sequence convergence for scans consisting of only 1.5T scans (3, 6, and 9 months) or only 3T scans (3, 4.5, 6, and 7.5 months) for T1 Head (Left), and T1 Brain (Right). The iterations appeared to converge at a minimum level after approximately 6 or 7 iterations for both scanner strength types. Two of the 3T average brain templates took slightly longer to begin to converge, but all templates showed convergence by about the same iteration level. The convergence patterns for the T2W head and brain and data collapsed across scanner strength (data not shown) were similar to those of the T1W MRI volumes.
Figure 4.
Degree of fit between successive iterations for T1W head and T1W brain. Each line represents an age group template undergoing iterations. The figures do not preserve the relative size for the ages. For the bottom figures, the red lines are for 3T MRI volumes and the black lines are for 1.5T MRI volumes at comparable ages. The dependent variable (RMS) represents the root-mean-squared difference between the intensity of voxels at the same location in the successive iterations.
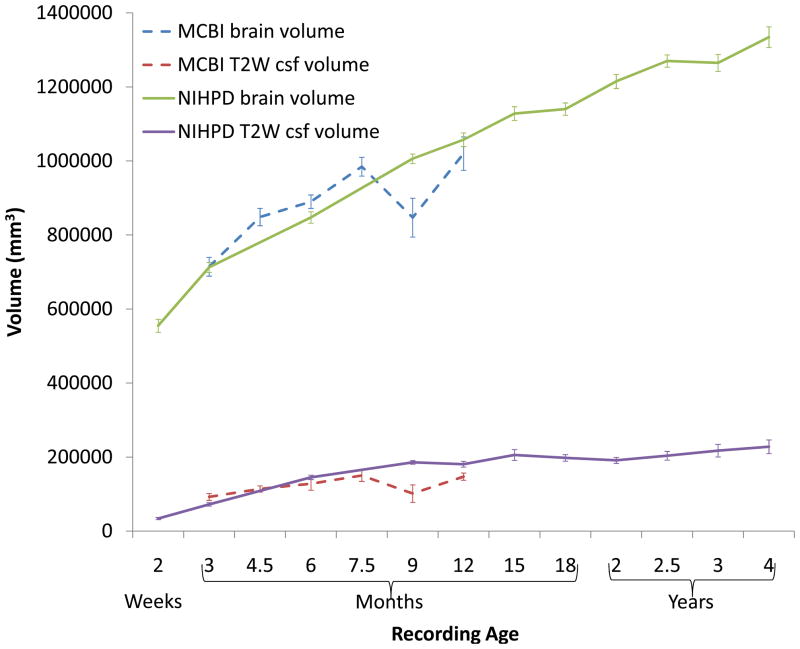
Average Brain Volume across Ages
This neurodevelopmental database of MRI volumes can be used to quantitatively measure developmental changes in total brain volume and volumes of specific brain structures. For example, we used our final age-specific, average brain templates (see Figure 3A, B, and C) and an automatic brain extraction procedure on individual participants to calculate total brain volume. The total brain volume was quantified from the extracted brain and consists of cortex, ventricles, brainstem, and cerebellum. The total brain volume was calculated for each individual, and the mean and standard error were computed for each age group. Figure 5 shows the total brain volume as a function of age, separated by the NIHPD (1.5T) and the MCBI (3T) volumes. There was a stable increase in brain volume across the full NIHPD age range (240% brain volume increase from birth to 4 years of age); and the values from the MCBI and NIHPD datasets were similar for ages 3 to 12 months. There was a small dip in the 9-month-old 3T total brain volume, which may be due to small numbers of participants at that age. The CSF was also identified in the T2W MRI volumes as voxels with the brightest intensities (see Methods). The volume change across age of the T2W-derived CSF is shown in Figure 5. There was a slight increase in T2W-CSF through the first year, followed by a smaller and more gradual increase across ages to 4 years.
Figure 5.
Total brain volume as a function of scan age. The solid lines are from NIHPD MRI volumes (1.5T), and the dashed lines are from the USC-MCBI MRI volumes (3T). The error bars represent the standard error of the mean (SE).
The total brain volume was examined for its relation to gender. Figure 6 shows the total brain volume as a function of age, separated for male and female participants, for the NIHPD (1.5T) volumes. The average brain volume of the males was larger than that of the females (F (1, 265) = 83.04, p < .001). There was no interaction for age and gender, implying that the difference between male and female brain size held steady over this age range.
Figure 6.
Total brain volume as a function of scan age and gender. The data are from the NIHPD MRI volumes (1.5T). The error bars represent the standard error of the mean (SE).
Discussion
The current work contributes to the developmental research and clinical community by providing (1) a method for creating pediatric MRI average brain templates and (2) T1W and T2W average templates for head and brain for infants and young children ages birth through 4 years. The average templates produced are unique as they used MRIs from normal, healthy children representative of the U.S. census for gender (approximately half males, half females), race/ethnicity, and family income levels (Almli et al., 2007; Brain Development Cooperative Group, 2006), and the age-specific average templates provided were fine grained with 3-month increments from birth through 18 months of age, 6-month increments from 18-months through 36-months, and a final 12-month increment to 4 years of age (Almli et al., 2007; Brain Development Cooperative Group, 2006). These average MRI templates are useful for normalizing MRIs for most neuroimaging work; including, for example, voxel-based morphometry, functional task-based MRI, and functional-connectivity (BOLD) neuroimaging, as well as providing realistic head models for EEG/MEG source analysis.
Procedures and methods for creating templates
The existence of the Objective 2 database of the NIHPD dataset provided a normal, healthy, and age-appropriate database of MRIs from 2 weeks to 4 years of age (Brain Development Cooperative Group, 2006; Almli et al., 2007). The procedures used here yield a uniform methodology that can be used for creating age-specific average templates for infancy and early childhood, as well as adolescence and adulthood, and advanced aging. Similar procedures have been used by Fonov et al., (2011) and Sanchez et al. (2010) for NIHPD Objective -1 templates constructed with participants 4.5 years of age and older.
The present research aimed to provide a method for doing MRI structural work on very young children with publicly available software and a relatively automated post-processing pipeline. Template averaging processes (Guimond et al., 2000), iterative routines based on Sanchez et al. (2010; cf. Yoon et al, 2009), and publicly available software (ANTS, FSL, SPM8b) contributed to the construction of the average templates. The average templates produced retain approximate brain volume size of the age of the participants and work well with children from the same ages. Multiple iterations allowed for enhanced non-linear registration, such that imperfections in registration are eventually minimized through the use of successive templates as registration targets. The non-linear registration and transformation procedure, ANTS (Avants et al., 2008, 2011), is an open-source and accessible computer program that allows diffeomorphic registration between individual MRI volumes and the average template. The ANTS procedure has performed well in tests of registration to a “gold standard” atlas-based segmentation (Klein et al., 2009). The procedure for template construction developed in Sanchez et al. (2010) and used in this paper will serve as a useful tool for template construction with pediatric MRI data.
The template building strategy employed in this study favorably compares to other pediatric average atlases (Fonov et al. 2011). As mentioned previously, Fonov et al. created pediatric templates with the Objective 1 data from the NIH Study of Normal Brain Development. Similar to our strategy, they utilized non-linear iterative registration techniques to average groups of subjects with intensity-matching algorithms. While the our study averaged fewer subjects to create the age-specific templates than Fonov et al. (2011), our study represents a departure from the use of broad-aged templates to describe a period of extremely rapid brain development. Instead, the current study developed templates based on narrow age ranges over the age period spanning infancy through early childhood. The methods of Fonov et al. (2011), Sanchez et al. (2010) and the current study strived to create age-specific templates with high anatomical detail to capture the subtleties of brain development and reduce reliance on adult-based templates.
The averaging process for the current templates involved considerable manually guided application of the programs and some manual editing. Several differences in the MRIs of infant brains and adult brains likely contributed to this difficulty, including: contrast-to-noise ratio (Mewes et al., 2006), poor spatial resolution due to small head size or the lower resolution for the 2D, 3 mm3 voxels, and 1.5T scans of the NIHPD data (Xue et al., 2007). Having overcome these difficulties with the successful creation of age-relevant average MRI templates, the templates can be applied to participants at very young ages with automatic procedures similar to those used in adult MRI analysis pipelines.
Average age-specific templates
The templates created for this report uniquely fill a major void for a comprehensive series of MRI templates covering the ages from birth through 4.3 years, as the understanding of brain changes during early development will be greatly enhanced with the use of a comprehensive set of age-specific templates (Gaillard et al., 2001; Xue et al., 2007). Similar to the templates constructed by Fonov et al., (2011), Sanchez et al. (2010), and Wilke et al., (2008) from the NIHPD Objective 1 (4.5 years of age and older), the current report begins the process of filling the template void for ages 2 weeks (birth) through 4.3 years.
Although other datasets (i.e., non-NIHPD datasets) have been used to produce infant and/or early childhood MRI templates (i.e., birth through 4.3 years of age), most have studied clinical participants, and most templates are incomplete by only constructing templates for a single subject, a single age group, or only a narrow age range (Joshi et al., 2004; Kazemi et al., 2007; Prastawa et al., 2005; Shi et al., 2011; Srinivasan et al., 2007). Notably, Shi et al. (2011) created infant brain atlases from neonates to 2 years of age using longitudinal data. The atlases created by Shi et al. were centered at three, non-overlapping age time points (i.e., neonate, 1-year-old and 2-year-old atlases) from 3T scanners. In comparison, we constructed averaged brain templates centered at ten (non-overlapping) age time points (e.g., 2 weeks, 3, 4.5, 6, 7.5, 9, 12 months in the first year, and 15, 18 and 24 months in the second year) from 1.5T and 3T scanners. Thus, our finer-grain age time point templates overlap the age ranges of Shi et al. and they continue through 4 years of age. A strength of the Shi et al. longitudinal study is the relatively large number of participants (n=95) contributing to each of the three age-specific templates. However, their sample included at risk children born at less than 37 weeks gestational age (i.e., premature delivery), and their report did not include any assessment of neuropsychological or behavioral status of their sample. Such was not the case for our normal, healthy NIHPD sample. Nevertheless, the results from our study and Shi et al. (2011) accentuate the differences in templates between one year olds and neonates; thus underscoring the necessity of fine grained age-appropriate templates when working with brain imaging data from children under 1 and 2 years of age. Both of these sets of atlases/templates will serve to enhance understanding of brain development at younger ages so that unified procedures can be established for constructing future templates and atlases of the developing brain.
The NIHPD database, with a fine-grained cohort sampling plan design (i.e., 11 individual, non-overlapping, age cohorts across ages birth through 4 years) allows for the creation of age-appropriate templates with good age specificity over the entire age range from birth through young adulthood (Brain Development Cooperative Group, 2006; Almli et al., 2007). Aubert-Broche et al. (2008) also used the NIHPD database to characterized white matter (WM) development from birth to 4.5 years of age. T2 relaxation times were used provide an assessment of brain myelination and were used to create age-specific segmention priors. Specifically, the T2 relaxation values were used to discriminate between myelinated and unmyelinated WM. Aubert-Broche et al. (2008) helped to characterize WM development in infancy and early childhood, however, average age specific templates are still needed for accurate normalization before the segmentation process occurs.
The existence of the MRI templates in the current study may be used for other neuroimaging methods. For example, segmented tissue maps may be created from the averages, or from individuals at each age, to show the distribution of GM, WM, CSF, and other head media. With older children (e.g., 4.5 year olds in Sanchez et al., 2010; or Fonov et al., 2011), adult-based segmentation procedures are relatively easily applied to MRI volumes either with simple application of voxel-intensity procedures or segmentation priors. These typical segmentation procedures were designed for MRI volumes with distinct and ordered peaks in histogram intensity. Infant brains have reversed GM/WM contrast levels (Barkovich, 2005; Xue, et al., 2007) and large amounts of nonmyelinated axons (Weisenfeld & Warfield, 2009), properties which make segmentation of infant brains particularly difficult (Gilmore et al., 2007; Prastawa et al., 2005; Weisenfeld & Warfield, 2009; Xue et al., 2007). Importantly, we plan to create such segmentation templates with GM, WM, CSF, and with nonmyelinated axons (NMA) in the MRI volumes across different infant age groups. Our segmentation technique will involve both T1W and T2W classification (Shi et al. 2010) to aid in tissue discrimination. In addition to segmentation templates, other types of templates can be developed. This might include stereotaxic atlases that identify major anatomical locations, Talairach stereotaxic space (Taliarich & Tournoux, 1988) or Brodmann locations, or specialized axonal or white matter tracts.
Summary
Normative imaging data of healthy infant populations has historically been scarce due to previous limitations imposed by the data collection of this population. The incorporation of methods that bypass the use of sedation has greatly increased the number of healthy infants scanned (Almli et al., 2007). Objective 2 of the NIH MRI Study of Normal Brain Development (NIHPD) compiled a normal, healthy, age-appropriate pediatric brain image reference database of children from two weeks to 4 years to characterize healthy brain maturation (Almli et al., 2007; Evans, 2006). The NIHPD database allowed the opportunity to create age-specific templates with a large sample of unified brain scan protocols across ages. We (present study and Sanchez et al., 2010) and others (Fonov et al. 2011; Shi et al., 2011) created average templates with a specificity that was developmentally appropriate (e.g., 3 month range for birth through 18 months; 6 months or 1 year range through 4 years [Almli et al., 2007]). The methods applied in this study followed an automated pipeline procedure. These age templates allow for accurate spatial normalization with infants and children, which is a key procedure implemented in neuroimaging studies involving volumetric analysis, voxel-based morphometry, functional and connectivity MR neuroimaging. While the infant templates characterize discrete periods of early childhood development, and combined templates corresponding to the ages under study may be necessary to create a common atlas space for conducting inter-age group comparisons. We anticipate that other researchers will use similar methods to create templates comprised of wider age ranges.
These age-specific templates may be used for a wide range of neuroimaging studies in developmental psychobiology and developmental neuroscience. The creation of pediatric and infant templates for normalization represents the first step in improving accuracy in pediatric neuroimaging work. Segmentation maps are particularly useful for these age groups. Axonal myelination is often used as an explanatory mechanism for cognitive development (e.g., Klingsberg, 2008). The volume mean and standard errors for each age group may be used to determine the relative fit of an individual volume to white matter age-based norms. Additionally, near infrared spectroscopy (NIRS) is becoming a popular technique for infant and child neuroimaging (Aslin & Mehler, 2005; Mehler et al., 2008). Future work might use these templates to create age-specific atlases. For example, the age templates could provide an approximate atlas for the depth of cortical tissue under the scalp and skull, or specify the anatomical areas under the infrared emitters and collectors. Neuroimaging with electroencephalographic or magnetoencephalographic measures use quantitative cortical source analysis to infer the cortical sources of the scalp-recorded electrical or magnetic signals. Such techniques work best with realistic models of head tissue, e.g., finite element models, and realistic head models are particularly important for infants and young children (Reynolds & Richards, 2009; Richards, 2010). Realistic head models for age groups may be derived from the average templates provided in this study (e.g., Reynolds et al., 2010). These tools will be useful for clinicians and for researchers interested in neural and behavioral development.
Template Database Availability
The present brain templates, and those of Sanchez et al (2010), are publicly available to researchers for clinical and experimental studies of normal and pathological brain development. In addition, we are currently working on an adult version of this template construction with comparable techniques for adults from 20 through 90 years of age. The template database is available for use by other investigators and clinicians for their developmental studies of behavior, neuroimaging, electrophysiology, and so on. Data access is limited to scientific professionals for research purposes. The data that are available are average templates for T1W head, T2W head, T1W brain and T2W brain, and brain extracted from the average template of the T1W head. The template volumes are in compressed NIFTI format (http://nifti.nimh.nih.gov/). The data are on a file server that may be accessed with the Secure Shell (SSH) file transfer protocols (SCP or SFTP). Instructions for access are given online (http://jerlab.psych.sc.edu/neurodevelopmentalmridatabase). Interested users should contact John E. Richards (richards-john@sc.edu). The original, individual MR brain scans and behavioral data from the NIHPD can be obtained from their website (https://nihpd.crbs.ucsd.edu/nihpd/info/index.html).
Acknowledgments
This work was supported by the NIH grant to JER, R37 HD18942, and by grants from the National Institute of Child Health and Human Development, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke (Contract #s N01-HD02-3343, N01-MH9-0002, and N01-NS-9-2314, -2315, -2316, -2317, -2319 and -2320) to CRA.
We would like to acknowledge Ben Davis for his help with ANTS programming, Michael Stevens who collected the MRI volumes at the USC-MCBI, and Allison Connington for data processing work. We thank the Brain Development Cooperative Group of the NIHPD for the Objective 2 MRI data.
Footnotes
Most of the data used in the preparation of this article were obtained from the NIH MRI Study of Normal Brain Development, Pediatric MRI Data Repository (referred to as the “NIHPD in this article). The NIHPD is a multisite, combined cross-sectional and longitudinal study of normal, healthy developing children (representative of U.S. Census 2000 statistics for gender, family income, race/ethnicity) from ages newborn through young adulthood. A listing of the participating sites and a complete listing of the study investigators can be found at http://www.bic.mni.mcgill.ca/nihpd/info/participating_centers.html.
This manuscript reflects the views of the authors and may not reflect the opinions or views of the Brain Development Cooperative Group Investigators or the NIH.
We also constructed average templates consisting of both 1.5 and 3T scans combined together, but which are not presented in this paper. They are available from the online database.
References
- Almli CR, Rivkin MJ, McKinstry RC. The NIH mri study of normal brain development (objective-2): Newborns, infants, toddlers, and preschoolers. Neuroimage. 2007;35:308–325. doi: 10.1016/j.neuroimage.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Altaye M, Holland SK, Wilke M, Gaser C. Infant brain probability templates for mri segmentation and normalization. Neuroimage. 2008;43:721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison N, Song G. Release 1.5, Penn Image Computing and Science Laboratory. 2011. Advanced Normalization Tools (ANTS) [Google Scholar]
- Aubert-Broche B, Fonov V, Leppert I, Pike GG, Collins DL. Medical Image Computing and Computer-Assisted Intervention, Prt II. Vol. 5242. LNCS; 2008. Human brain myelination from birth to 4.5 years; pp. 180–187. [DOI] [PubMed] [Google Scholar]
- Brain Development Cooperative Group. The nih mri study of normal brain development. Neuroimage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Brain Development Cooperative Group. Cerebral Cortex. 2011. Total and regional brain volumes in a population-based normative sample from 4 to 18 years: The NIH study of normal brain development. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgund ED, Kang HC, Kelly JE, Buckner RL, Snyder AZ, Petersen SE, et al. The feasibility of a common stereotactic space for children and adults in fmri studies of development. Neuroimage. 2002;17:184–200. doi: 10.1006/nimg.2002.1174. [DOI] [PubMed] [Google Scholar]
- Evans AC. Large-scale morphometric analysis of neuroanatomy and neuropathology. Anatomy And Embryology. 2005;210:439–446. doi: 10.1007/s00429-005-0045-1. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3d statistical neuroanatomical models from 305 mri volumes. Paper presented at the Proc IEEE Nucl Science Symp Medl Imaging Conf..1993. [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54:313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard WD, Grandin CB, Xu B. Developmental aspects of pediatric fmri: Considerations for image acquisition, analysis, and interpretation. Neuroimage. 2001;13:239–249. doi: 10.1006/nimg.2000.0681. [DOI] [PubMed] [Google Scholar]
- Guimond A, Meunier J, et al. Average brain models: a convergence study. Computer Vision and Image Understanding. 2000;77:192–210. [Google Scholar]
- Jenkinson M, Smith S. A global optimization method for robust affine registration of brain images. Medical Image Analysis. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23:S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Kang HC, Burgund ED, Lugar HM, Petersen SE, Schlaggar BL. Comparison of functional activation foci in children and adults using a common stereotactic space. Neuroimage. 2003;19:16–28. doi: 10.1016/s1053-8119(03)00038-7. [DOI] [PubMed] [Google Scholar]
- Kazemi K, Moghaddam HA, Grebe R, Gondry-Jouet C, Wallois F. A neonatal atlas template for spatial normalization of whole-brain magnetic resonance images of newborns: Preliminary results. Neuroimage. 2007;37:463–473. doi: 10.1016/j.neuroimage.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain mri registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N, Froimowitz MP, Bigler ED, Lainhart JE. Associations between IQ, regional brain volumes and demography in a large normative sample of healthy children and adolescent. Developmental Neuropsychology. 2010;3:296–317. doi: 10.1080/87565641003696833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Simpson G, Woods R, Paus T, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Schormann T, Amunts K, Palomero-Gallagher N, Parsons L, Narr K, Kabani N, LeGoualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain. Philosophical Transactions of the Royal Society of London: B, Biological Sciences. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzik O, Chugani DC, Juhasz C, Shen CG, Chugani HT. Statistical parametric mapping: Assessment of application in children. Neuroimage. 2000;12:538–549. doi: 10.1006/nimg.2000.0651. [DOI] [PubMed] [Google Scholar]
- Prastawa M, Gilmore JH, Lin WL, Gerig G. Automatic segmentation of mr images of the developing newborn brain. Medical Image Analysis. 2005;9:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Rutherford MA. MRI of the neonatal brain. W.B Saunders; 2002. [Google Scholar]
- Sanchez CE, Richards JE, Almli CR. Age-specific MRI brain templates for healthy brain development from 4 to 24 years of age. Unpublished ms. 2010 ( http://jerlab.psych.sc.edu/pdf/expt77.pdf)
- Shi F, Fan Y, Tang SY, Gilmore JH, Lin WL, Shen DG. Neonatal brain image segmentation in longitudinal MRI studies. Neuroimage. 2011;49:391–400. doi: 10.1016/j.neuroimage.2009.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Dutta R, Counsell SJ, Allsop JM, Boardman JP, Rutherford MA, et al. Quantification of deep gray matter in preterm infants at term-equivalent age using manual volumetry of 3-tesla magnetic resonance images. Pediatrics. 2007;119:759–765. doi: 10.1542/peds.2006-2508. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers; 1988. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Schmithorst VJ, Holland SK. Assessment of spatial normalization of whole-brain magnetic resonance images in children. Human Brain Mapping. 2002;17:48–60. doi: 10.1002/hbm.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Warfield SK. Automatic segmentation of newborn brain mri. Neuroimage. 2009;47:564–572. doi: 10.1016/j.neuroimage.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Srinivasan L, Jiang SZ, Rutherford M, Edwards AD, Rueckert D, et al. Automatic segmentation and reconstruction of the cortex from neonatal mri. Neuroimage. 2007;38:461–477. doi: 10.1016/j.neuroimage.2007.07.030. [DOI] [PubMed] [Google Scholar]