Abstract
Reprogramming factors have been used to induce pluripotent stem cells as an alternative to somatic cell nuclear transfer technology in studies targeting disease models and regenerative medicine. The neuronal repressor REST maintains self renewal and pluripotency in mouse embryonic stem cells by maintaining the expression Oct3/4, Nanog, and cMyc. We report that primary hepatocytes express REST and most of the reprogramming factors in culture. Their expression is upregulated by hepatocyte growth factor (HGF) and epidermal growth factor (EGF). REST inhibition results in downregulation of reprogramming factor expression, increased apoptosis, decreased proliferation, and cell death. The reprogramming factors are also upregulated after 70% partial hepatectomy in vivo. These findings show that genes inducing iPS phenotype, even though expressed at lower levels than embryonic stem cells, they nonetheless are associated with control of apoptosis and cell proliferation in hepatocytes in culture and may play a role for such processes during liver regeneration.
Keywords: Reprogramming factors, REST, Oct4, Klf4, Growth factors
In recent years many efforts have been aimed at generating pluripotent stem cells from somatic cells by inducing high levels of expression of a combination of transcription factors including Sox2, Oct3/4 (henceforth referred to as Oct4), Nanog, Klf4, and cMyc (1–5). Initially, retroviral transduction of four reprogramming factors, namely Oct4, Sox2, cMyc, and Klf4, was shown to be sufficient to convert mouse fibroblasts to embryonic stem cell like phenotype (6). Later, pluripotent stem cells were generated from adult mouse liver and stomach cells (7), human somatic cells (8), and human primary hepatocytes (9). Recently generation of pluripotent stem cells without viral integration by repeated transfection of expression plasmids (10–11) and by protein transfer (12–13) have overcome the risks of tumorigenicity associated with viral integration. While these reprogramming factors are expressed in stem cells, their expression in adult somatic cells with high potential for clonal expansion such as hepatocytes has been less explored. Primary hepatocytes show limited ability to proliferate in culture except under the influence of chemically defined HGM medium containing the primary mitogens hepatocyte growth factor (HGF), and epidermal growth factor (EGF) (henceforth referred to as growth factors(GF)) (14). Under these conditions, hepatocytes undergo multiple proliferative cycles, express altered levels of hepatocyte-associated transcription factors and lose characteristic gene expression markers such as albumin. In the presence of specific matrix components such as Matrigel, they redifferentiate and revert to a mature hepatocyte phenotype (15). In the present study, we examined whether primary hepatocytes express any of the iPS-reprogramming factors in culture and if their expression changes as a result of growth factor-induced proliferation. Transcription factor REST (RE-1 silencing transcription factor; also called NRSF) has been shown to regulate the expression of these self-renewal and reprogramming factors in mouse embryonic stem cells (16). Thus we looked at REST expression to see if it was expressed in our model and if it regulates the expression of any of the reprogramming factors. Primary rat hepatocytes were plated on collagen coated plates and incubated in the presence of HGM medium with (+GF) or without growth factors (−GF) over a period of 10 days. Plates were harvested at day 0 (2h plated), 2, 4, 6, 8, and 10 after plating for analysis of message and protein by real time RTPCR (qRT-PCR) and Western blot analysis, respectively.
Experimental Procedures
Male Fisher 344 rats (150–200 g) were purchased from Charles River Laboratories (Fredrick, MD). Animals were allowed food and water ad libitum. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences.
Isolation and culture of heaptocytes
Hepatocytes were isolated by adaptation of the calcium two step collagenase perfusion technique as previously described (15, 17). Hepatocytes were plated on collagen coated six-well plates (BD Biosciences, CA) at 250,000 cells/well. After the initial 2h attachment period, plating media was changed to either HGM complete with growth factors (+GF) HGF (supplemented at 40ng/ml) and EGF (supplemented at 20ng/ml) or without growth factors (−GF) and every 48 h thereafter. Cells were harvested on day 0 (2h plated), 2, 4, 6, 8, and 10 for RNA and protein.
Total RNA extraction
Total RNA was extracted from plated cells using the RNABee reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s protocol. The isolated RNA was treated with Turbo DNA-free (Ambion, Austin, TX) according to the manufacturer’s instructions. RNA was quantified by spectrophotometry at 260 nm, and purity was assessed by optical density 260/280 ratio. The RNA was stored at −80°C. The experiment was repeated in three rats and their mRNA was pooled for further processing in primary hepatocytes as well as PHx experiments.
cDNA synthesis
Four micrograms RNA per sample was reverse transcribed using random hexamer to cDNA by using SuperScriptIII reverse transcriptase (Invitrogen, CA) according to the manufacture’s protocol. A no reverse transcriptase (RT) control was also included.
Real time reverse transcriptase polymerase chain reaction
The gene-specific primers used for rat were as follows. REST, cMyc, Klf4, Nanog (SuperArray Bioscience Corporation Cat. # PPR45101A, PPR45580A, PPR43919A, and PPR59663A, respectively).
Oct4 forward: 5′- GGC GTT CTC TTT GCA AAG GTG TTC -3′
Oct4 reverse: 5′- CTC GAA CCA CAT CCT TCT CT -3′
Expression levels of REST, Oct4, cMyc, and Klf4 were determined by quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) using SYBR green, and levels were normalized relative to expression of cyclophillin in each sample. Fold change in gene expression was calculated by using the 2(−ΔΔCt) method (18). Reverse transcribed samples were amplified in parallel on an ABI prism 7000 SDS instrument (Applied Biosystems, Foster city, CA). qRT-PCR for each sample was performed in triplicate in a 20 μl reaction with 50ng of cDNA, 5 picomoles of each primer, and 1X SYBR green PCR mix (Applied Biosystems). The standard conditions for real time PCR were as follows: 2 minutes at 50°C, 10 minutes at 95°C followed by 40 cycles of 15 seconds denaturation at 95°C, and elongation at 60°C for 45 seconds. A dissociation curve analysis was performed at the end of every run. A no RT and a no template control were also included in every run.
A different set of primers designed in a common region with same sequence for rats and mice for comparison with MESC were as follows:
REST forward: 5′ – AACCAT TTTCCCAGGAAAGTCTACAC
REST Reverse: 5′ – AGTTCACATTTATACGGGCGTTCTC
Oct4 forward: 5′- GAGGGCCAGGCAGGAGCACGAGTGG-3′
Oct4 reverse: 5′- TTCTGCAGGGCTTTCATGTCCTGG -3′
Klf4 forward: 5′- CTCAAGGCACACCTGCGAACTCACA-3′
Klf4 reverse: 5′- TGTGTTTGCGGTAGTGCCTGGTCAGT -3′
cMyc forward: 5′- AGTGTTCTCTGCCTCTGCCCGCGA-3′
cMyc reverse: 5′- GTCGTAGTCGAGGTCATAGTTCCT -3′
Effect of Matrigel on expression of reprogramming factors
Primary hepatocytes were plated in 100 mm collagen coated plates at 2.4 million cells/plate. Matrigel (Cat. # 354234) was obtained from BD Biosciences, San Jose, CA. After the 2h attachment period hepatocytes were treated as follows: 1) HGM medium with growth factors and 0.233 mg/ml Matrigel (+MTG+GF), 2) HGM without growth factors and 0.233 mg/ml Matrigel (+MTG−GF), 3) HGM with growth factors but no Matrigel (−MTG+GF), and 4) HGM without growth factors and without Matrigel (−MTG−GF). Plates were harvested on days 2, 6, and 10 for RNA.
Semi-quantitative PCR for Matrigel experiment
Standard PCR was performed using 50ng cDNA and Amplitaq DNA polymerase (Applied Biosystems). PCR products were resolved on 2% agarose gels and visualized with ethidium bromide staining. The bands on gel were scanned for optical density using Image J software for quantitation purposes.
REST inhibition studies
shRNA for REST: we used a commercially available kit from Invivogen (catalog # ksirn4-gz21) to generate the plasmid containing shRNA targeted against REST. The shRNA vector employed also encodes a red-shifted variant of the jellyfish GFP. This plasmid is specifically designed for the cloning of small synthetic oligonucleotides that encode 2 complementary sequence of 21 nt, homologous to a segment of REST. The insert is cloned downstream of a human 7SK promoter. It is transcribed into a short dsRNA with a hairpin structure (shRNA) consisting of a 21 bp double stranded region corresponding to REST and a small loop formed by the spacer region. Sequences for REST shRNA insert:
Forward: 5′- ACC TCTTGGTGAAGAGAGACAGATTCAAGAGATCTGTCTCTCTTCACCAAT T -3′
Reverse: 5′ - CAAAAATTGGTGAAGAGAGACAGATCTCTTGAATCTGTCTCTCTTCACCAA G -3′
Primary hepatocytes were plated at a density of 1×106 cells per 100 mm dish or 0.25×106 cells per well (6-well plate) on day 1. After the 2 h attachment period, plating media was replaced with HGM complete without growth factors. On the 2nd day, hepatocytes were either transfected with shRNA for luciferase (C), or shRNA for REST (R). The transfection media was replaced with fresh HGM without growth factors after 6h. On the next day (day 3) media was changed to HGM with growth factors and thereafter replaced every 48h throughout the time course. Cells were harvested at day 0, 1, 2, 3, 4, and 5 after transfections for RNA and protein. MTT assay was done on days 2, 3, 4, and 5 as a marker of live cells. Tritiated Thymidine incorporation was measured on day 1–2 after transfections to assess proliferation of hepatocytes. TUNEL assay was done on 3 days after transfections to assess apoptosis in these cells.
MTT assay
Cell death was assessed by measuring the live mitochondrial activity using the TOX-1 in vitro toxicology assay kit (Sigma Aldrich corp., St. Louis, MO) according to the manufacturer’s protocol.
Proliferation assay
[3H]Thymidine was added to the medium on day 3 (24 h after transfections) at a concentration of 2.5 μCi/ml. The medium was removed after 24 h, and hepatocytes were fixed with ice-cold 5% trichloroacetic acid. Trichloroacetic acid was removed and the plates were washed in running tap water and air dried completely. 750 μl 0.33 M NaOH was added to each well for 30 minutes to solubilize the cells. The solution was transferred to a new tube, and 250 μl of 40% TCA/1.2 M HCl was added for precipitation. The tubes were centrifuged at 12000 g for 10 minutes, and the pellets were redissolved in 500 μl 0.33 M NaOH. A 200 μl aliquot was used to measure cpm/dpm in a Beckman LS6000IC scintillation counter (Beckman Coulter, CA), and 100 μl was used to determine optical density value of total DNA. Data are plotted as CPM/μg DNA.
Apoptosis assay
The extent of apoptosis in hepatocytes was measured 3 days after transfection using TUNEL assay according to manufacturer’s protocol (ApopTag Peroxidase In Situ Apoptosis Detection Kit, S7100, Chemicon International). Brown stained apoptotic nuclei were counted in 5 different fields along with the total number of cells in the field for each group. Percent apoptotic nuclei were calculated and plotted.
Protein analysis by Western Blot
Protein levels in nuclear extracts were assessed by Western blot analysis by harvesting cells at different time points. Nuclear extracts pooled from 3 rats were prepared using NE-PER Nuclear and cytoplasmic extraction kit according to manufacturer’s protocol (Pierce Biotechnology Cat. # 78833, Rockford, IL). Nuclear extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis in 4% to 12% NuPage Bis-Tris gels with 1× MOPS buffer (Invitrogen, Carlsbad, CA), then transferred to Immobilon-P membranes (Millipore, Bedford, MA) in NuPAGE transfer buffer containing 10% methanol. Membranes were stained with Ponceau S to verify equal loading of total protein and transfer efficiency. Membranes were probed with primary and secondary antibodies in Tris-buffered saline with Tween 20 containing 1.5% nonfat milk, then processed with SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, IL) and exposed to a X-ray film (Pierce, Rockford, IL). Horseradish peroxidase-conjugated secondary antibodies were used at a 1:50,000 dilution (Chemicon, Temecula, CA). Primary antibodies used were as follows: REST (07-579, Millipore, Temecula, CA), Oct4 (ab52014, Abcam Cambridge, MA), cMyc, Nanog, and Klf4 (sc-764, sc-33760, and sc-20691, respectively, Santa Cruz Biotechnology, Santa Cruz, CA). Since our model involves proliferation, Tata binding protein used as a loading control for nuclear extracts changed because of the treatment. Hence ponceau stain was used to verify equal loading of samples.
70% Partial Hepatectomy (PHx) on rats
Isoflurane inhalation (Baxter, IL) was used to anesthetize animals. Partial hepatectomy was performed as described previously (19).
Immunohistochemisrty on rat tissues
Paraffin-embedded liver sections (4 μm thick) were used for immunohistochemical staining. Antigen retrieval was achieved by heating the slides in the microwave at high power in 1× citrate buffer for 10 minutes. The tissue sections were blocked in blue blocker for 20 minutes followed by incubation with primary antibody overnight at 4°C. The primary antibody was then linked to biotinylated secondary antibody followed by routine avidin-biotin complex method. Diaminobenzidine was used as the chromogen, which resulted in a brown reaction product. Primary antibodies used were as follows: NRSF (Millipore 07-579; 1:1000), Oct4 (ab27985; 1:250), KLF4 (SC-20691; 1:500), Nanog (ab80892; 1:700), and Myc (ab32072; 1:100).
Statistical analysis
Data are expressed as means ± S.E. Statistical differences were determined by one way analysis of variance (ANOVA) followed by Tukey’s honest significant difference test to determine which means were significantly different from each other or from controls using the JMP software (SAS Institute INC., Cary, NC). The criterion for significance was p < 0.05.
Results
Expression of reprogramming factors in primary hepatocytes
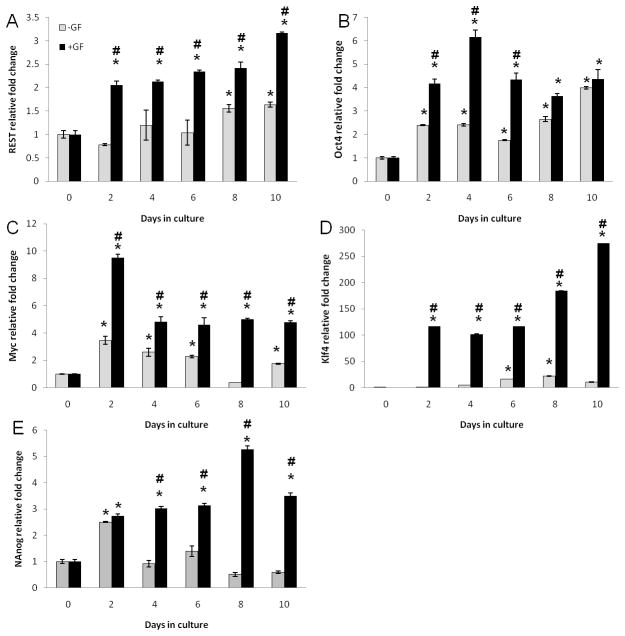
We report that primary hepatocytes do express REST, Oct4, cMyc, Klf4, and Nanog in culture (Fig. 1 and 2). Their expression is upregulated with time in culture. However, this upregulation is further enhanced in cultures incubated with growth factors (Fig. 1 and 2). REST and Klf4 message showed a steady increase with time in both the groups peaking at day 10 in culture (Fig. 1A and D). Oct4, cMyc, and Nanog message showed earlier peaks at 4d, 2d, and 8d, respectively (Fig. 1B, C, E, and Fig. 2). Previous studies have shown maximum proliferation of dedifferentiated hepatocytes when plated in culture with GF between days 4 and10 by BRDU labeling and tritiated thymidine incorporation (15). Expression of REST, Oct4, Nanog, Myc, and Klf4 protein was maximally induced when there was maximum proliferation in culture (days 4 to10) suggesting their role in GF-mediated proliferation of hepatocytes in culture.
Figure 1.
mRNA levels assessed by qRT-PCR over a time course in the presence/absence of growth factors. (A) REST; (B) Oct4; (C) cMyc; (D) Klf4; (E) Nanog expressed as fold change relative to cyclophillin. *Significantly different from the corresponding 0h time point. #Significantly different form −GF group at the same time point.
Figure 2.
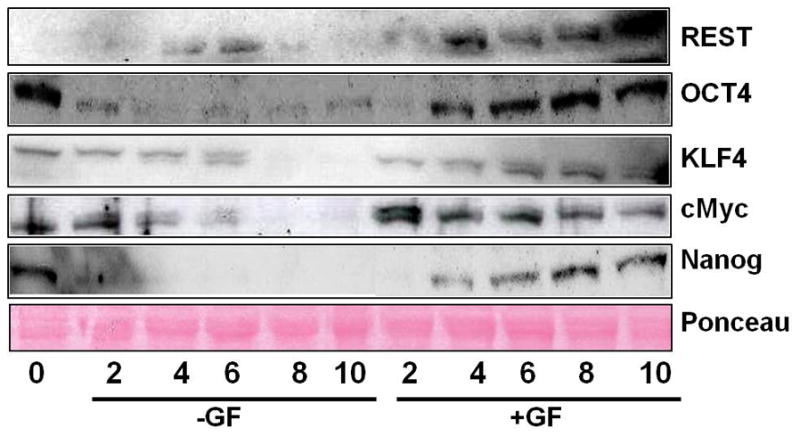
Western Blot analysis of protein in nuclear extracts of hepatocytes incubated with (+GF) or without (−GF) growth factors in culture over a time course of 0 to 10 days.
Effect of Matrigel on reprogramming factors’ expression
To test if expression of REST and reprogramming factors is dependent on the differentiation status of cells, Matrigel was used to induce hepatocyte differentiation as previously described. Hepatocytes were incubated as follows: with Matrigel with GF (+MTG+GF), with Matrigel without GF (+MTG−GF), without Matrigel with GF (−MTG+GF), or without Matrigel without GF (−MTG−GF). Matrigel-induced differentiation downregulated the growth factor induced expression of REST, Klf4, cMyc, and Oct4 message (Fig. 3A, B, C, and D). The group without Matrigel and with growth factors (−MTG+GF) showed maximum expression of REST, Klf4, and cMyc, whereas the group with Matrigel and without growth factors (+MTG−GF) showed the least expression. The other two groups lay in between (Fig. 3). Albumin (Fig. 3E) was used as a marker of differentiation and its expression was found to be higher in groups treated with Matrigel as compared to groups without Matrigel, suggesting that Matrigel did in fact induce differentiation in these groups. These data suggest that Matrigel-induced hepatocyte differentiation downregulates the expression of transcription factor REST as well as reprogramming factors Klf4, cMyc, and Oct4.
Figure 3.
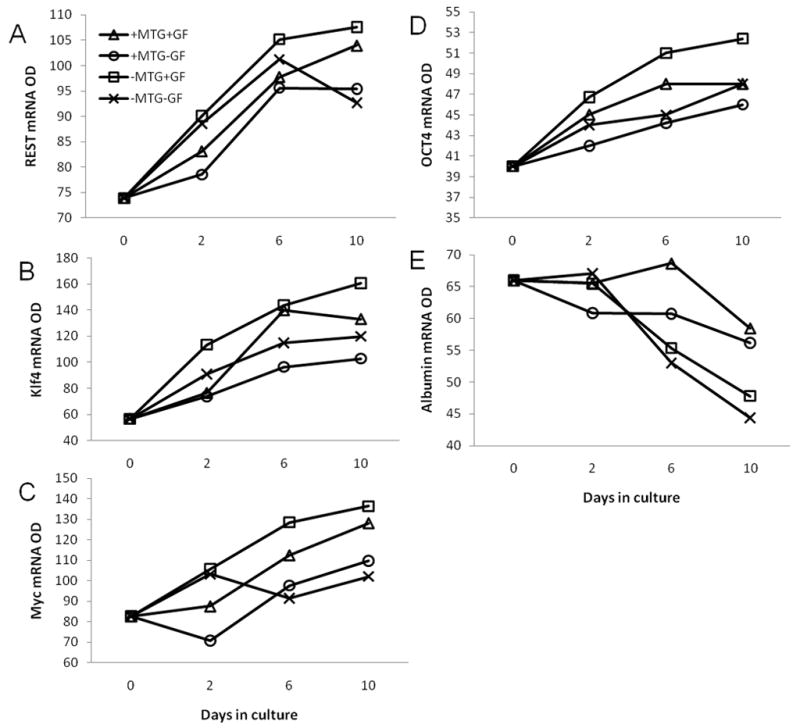
Effect of Matrigel-induced differentiation on expression of REST, cMyc, and Klf4. Optical densities of semi-quantitative PCR product of: (A) REST, (B) Klf4, (C) cMyc, (D) Oct4, and (E) Albumin.
REST inhibition studies
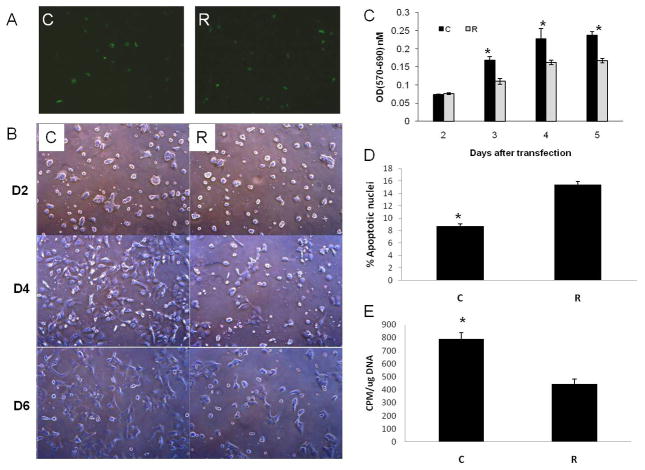
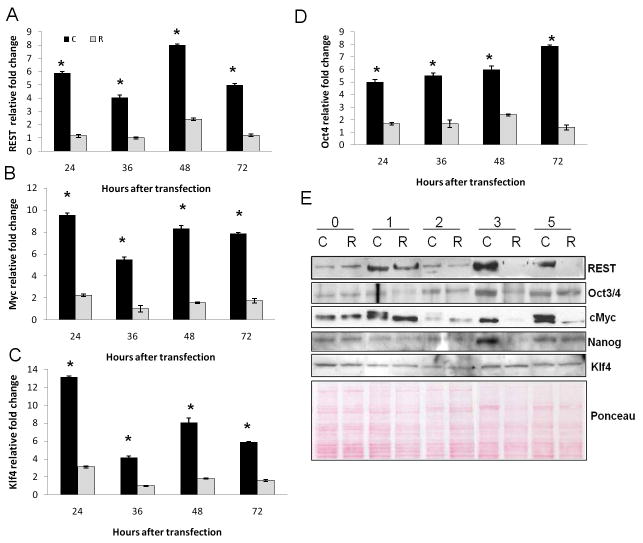
To find out if the expression of these reprogramming factors in primary hepatocyte culture is regulated by REST, we transiently inhibited REST in these hepatocytes using a short hairpin RNA (shRNA) for REST. There was 50% transfection of hepatocytes as assessed by green fluorescence protein (GFP) in REST-inhibited (R) and luciferase control (C) groups (Fig. 5A). REST mRNA and protein levels were inhibited as compared to luciferase control (Fig. 4A and E) suggesting efficient transfection and REST inhibition in hepatocytes. This was also accompanied by downregulated expression of Oct4 (Fig. 4D and E), cMyc (Fig. 4B and E), and Nanog protein (Fig. 4E) in these cells suggesting that REST might be regulating these self-renewal factors. Klf4 protein levels did not change suggesting possible post-transcriptional changes (Fig. 4C and E).
Figure 5.
Effect of REST-inhibition on hepatocytes. (A) Comparable transfection in REST-inhibited (R) and luciferase control (C) groups as assessed by GFP. (B) Photomicrographs of REST-inhibited(R) or control (C) hepatocytes over a time course. Original magnification 5x. (C) Cell survival as assessed by live mitochondrial (MTT) assay between control (C) and REST-inhibited (R) hepatocytes over a time course. (D) % apoptotic nuclei measured 3 days after transfection by TUNEL assay. (E) Rate of proliferation as assessed by tritiated thymidine incorporation 24h after transfections. *Significantly different from R group.
Figure 4.
Effect of REST inhibition on expression of reprogramming factors. C represents luciferase control, R represents REST-inhibited group. qRT-PCR for message of (A) REST; (B) cMyc; (C) Klf4; and (D) Oct4 expressed as fold change relative to cyclophilin. *Significantly different from REST-inhibited (R) group. (E) Western blot analysis of reprogramming factors 0–5 days after transfection.
There was significant cell death in the REST-inhibited (R) group compared to control (C) (Fig. 5B), as assessed by MTT assay (Fig. 5C). TUNEL assay performed 3 days after transfections showed increased apoptosis in REST-inhibited cells as compared to luciferase controls (Fig. 5D). Rate of proliferation was assessed by measuring tritiated thymidine incorporation in the REST-inhibited and control groups on day 3 (24h after transfection). The REST-inhibited group showed significant decrease in the rate of proliferation as compared to the controls suggesting that REST-inhibition was affecting proliferation of hepatocytes (Fig. 5E). The increased cell death observed by MTT assay could be a combination of direct effect of REST inhibition on hepatocyte survival by upregulating apoptotic pathways and decreased proliferation of hepatocytes.
Reprogramming factors after PHx
To test if these self-renewal factors are expressed in vivo and to further assess their role in liver regeneration, we studied their expression after 70% partial hepatectomy (PHx) in rats. REST, Oct4, cMyc, Klf4, and Nanog message was induced as early as 3h after PHx (Fig. 6). These data were corroborated by Western Blot analysis of their protein levels (Fig. 7B &D) as well as immunohistochemical staining of these reprogramming factors after PHx (Fig. 8), both of which indicated upregulation of reprogramming factors after PHx. Peak proliferation after PHx is observed at day 1 in rats (20). Immunohistochemical staining showed that both hepatocytes and billiary cells express these factors in their nuclei. Their expression by IHC was significantly upregulated one day following PHx (Fig. 8a, b, c, and e), except for Klf4 (Fig. 8d), which is consistent with Western Blot data (Fig 7D & E). These data suggest that expression of these factors may play a role in hepatocyte proliferation and survival during liver regeneration in vivo as well.
Figure 6.
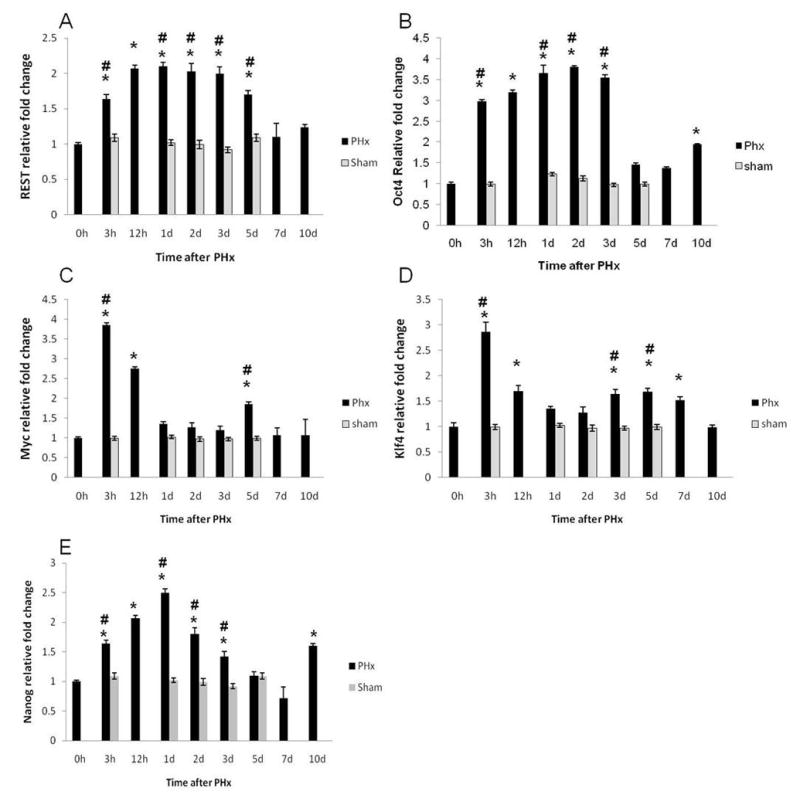
Expression of reprogramming factors in regenerating livers after PHx. qRT-PCR analysis of mRNA levels of (A) REST, (B) Oct4, (C) cMyc, (D) Klf4, and (E) Nanog over a time course after PHx expressed as fold change relative to cyclophilin. *Significantly different from the corresponding 0h time point. #Significantly different from shams at the same time point.
Figure 7.
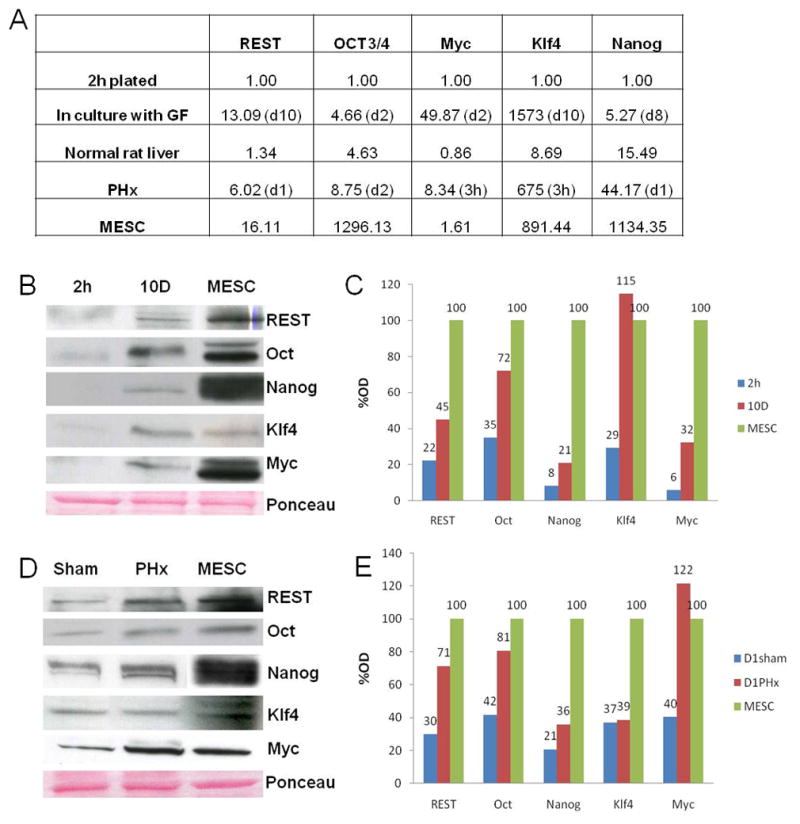
Expression of reprogramming factors after 70% PHx compared to MESC. (A) Fold induction of reprogramming factors as assessed by qRT-PCR in culture and after PHx relative to MESC. (B) Western blot of reprogramming factors in culture at 2h after plating, 10 days incubated with GF, and MESC. (C) Densitometric analysis of protein in culture expressed as a percentage of MESC. (D) Western blot of reprogramming factors in sham controls, one day after PHx, and MESC. (E) Densitometric analysis of protein after PHx expressed as a percentage of MESC.
Figure 8.
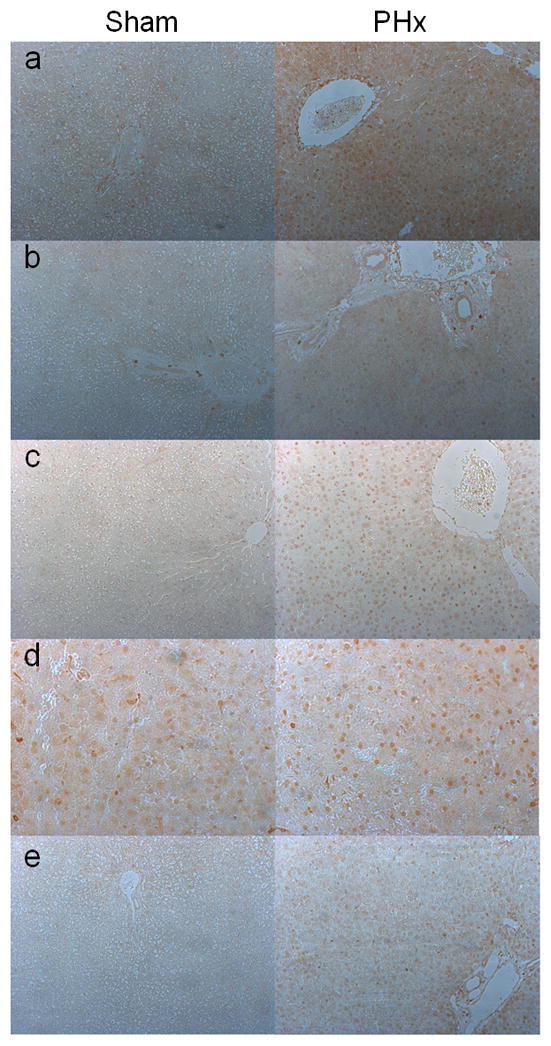
Immunohistochemical staining of reprogramming factors in shams operated and one day after PHx livers. Representative photomicrographs of (a) REST, (b) Oct4, (c) Nanog, (d) Klf4, and (e) Myc. (Original magnification 100X for a–c, and e. d:200X). All proteins show an increase in nuclear presence after PHx, except for KLF4.
Relative expression of reprogramming factors
To estimate the induction of these reprogramming factors in relation to their expression in mouse embryonic stem cells (MESC), we looked at their message by qRT-PCR under various conditions (Fig. 7A). Time points that showed peak expression in culture and after PHx from previous experiments were chosen for comparison. To make the results more comparable, primers were designed in a common region with same sequence for rats and mice. Considering the specific gene expression for hepatocytes plated for 2 hours as one-fold, we found that the expression of cMyc and Klf4 at mRNA level was more in culture (49 and 1573-fold, respectively) than in MESC (1.63 and 891-fold, respectively). Oct4 and Nanog expression was more in MESC, and REST expression in culture (13 fold) was close to that in MESC (16-fold). Oct4 and Nanog induction was more after PHx than in culture whereas that of cMyc, Klf4, and REST was less than that in culture. Oct4 induction in culture was 4-fold, which was close to the levels in normal rat liver.
Protein expression of reprogramming factors one day after PHx was compared to the protein expression of MESC (Fig. 7B, C, D, and E). While expression of REST, Oct4, Myc, and Nanog were less than that expressed in MESC, KLF4 expression was in fact more in cultured hepatocytes with growth factors as compared to MESC (Fig. 7B and C). On the other hand KLF4 expression did not seem to change much after PHx (Fig. 7D and E). At the same time Myc protein expression after PHx was more than in MESC (Fig. 7D and E).
Discussion
Our study suggests that the expression of transcription factor REST and the downstream reprogramming factors Oct4, cMyc, Nanog is crucial for the survival of normal hepatocytes in culture and that their expression might have an anti-apoptotic effect on hepatocytes. The fact that inhibition of REST leads to cell death suggests that REST, Oct4, cMyc, and Nanog act as survival factors for hepatocytes in culture. The fact that Klf4 is upregulated during hepatocyte proliferation (Fig. 1 and 2) but its unchanged protein levels after REST-inhibition are not sufficient to save the hepatocytes from apoptotic death (Fig. 4), suggests that Klf4 may have a role in proliferation but not in survival of hepatocytes.
We saw high levels of Oct4, Nanog, and Klf4 protein at 0d (2h after plating, Fig. 2). While the mRNA for these reprogramming factors seems to increase with time in culture (Fig. 1), their protein levels seem to decrease in culture without growth factors and the levels are simply maintained in culture with growth factors. This can be explained based on our data from Fig. 7A where we compare the mRNA for reprogramming factors under varied experimental conditions. Considering the specific mRNA levels for hepatocytes plated for 2 hours as one-fold, Oct4 mRNA expression in normal rat liver was 4-fold. Considering that the protein levels seen at 2h after plating are from a perfused normal rat liver, the higher protein seen at 0d (2h after plating) is not surprising. Also, the data show that there is a decrease in Oct4 mRNA after plating. Similarly, Nanog’s mRNA was 15-fold in normal rat liver, again indicating a drop in Nanog levels after plating. These data for Oct4 and Nanog mRNA in Fig. 7A support their high protein levels seen at day 0 (2h after plating) in the initial experiment (Fig. 2). Hepatocytes cultured with growth factors brought back these levels in contrast to hepatocytes cultured without the growth factors. The present data signify the fact that even though the expression of these reprogramming factors in cultures with GF is not comparable to MESC, the level of expression is nevertheless important for the proliferation and normal survival of these hepatocytes in culture.
We also found that these reprogramming factors are upregulated after PHx as seen by qRTPCR, Western Blot, and IHC. In fact, the expression of REST, and Oct4 is close to that of MESC, whereas that of Myc is more than that in MESC (Fig. 7D and E). In view of our results with hepatocytes in culture, it is reasonable to speculate that they may play a role in liver regeneration in vivo as well. The fact that primary hepatocytes express these reprogramming factors but are not acting as stem cells is intriguing. The mechanism(s) that inhibit hepatocytes from behaving like stem cells in spite of expressing reprogramming factors is unknown and it probably relates to relative levels of expression. On the other hand, previous studies have shown that hepatocytes can transdifferentiate to biliary epithelial cells in vivo (21). Other studies have also shown that mouse (22) and rat hepatocytes have a high capacity of clonal growth in recolonization of liver of mice with FAH deficiency (23–25). In these studies it was estimated that one mouse hepatocyte was capable of generating 50 mouse livers. It is conceivable that the coordinated and growth factor induced expression of REST and the reprogramming factors underlies the capacity of hepatocytes for such high clonal growth, documented in several models of liver recolonization (26–28). It is also possible, however, that the expression of reprogramming factors may occur in other cell types under normal growth conditions. Our studies should prompt further investigation of both of these possibilities.
Supplementary Material
Acknowledgments
Financial Support
This work is supported by NIH grants 5R01CA103958-05 and 5R01CA035373-27(GKM).
We thank John Stoops for his assistance with the partial hepatectomy experiments.
List of Abbreviations
- HGF
Hepatocyte growth factor
- EGF
epidermal growth factor
- GF
growth factors
- REST
RE-1 silencing transcription factor
- MTG
Matrigel
- PHX
70% Partial Hepatectomy
- and MESC
mouse embryonic stem cells
Footnotes
Potential conflict of interest: Nothing to report
References
- 1.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 3.Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, et al. Generation of functional human hepatic endoderm from human inducedpluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stemcells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggenschwiler R, Cantz T. Induced pluripotent stem cells generated without viral integration. Hepatology. 2009;49:1048–1049. doi: 10.1002/hep.22827. [DOI] [PubMed] [Google Scholar]
- 11.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebhardt R, Cruise J, Houck KA, Luetteke NC, Novotny A, Thaler F, Michalopoulos GK. Differential effect of growth factors on growth stimulation and phenotypic stability of glutamine-synthetase-positive and -negative hepatocytes in primary culture. Differentiation. 1986;33:45–55. doi: 10.1111/j.1432-0436.1986.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 15.Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, et al. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Stolz DB, Mars WM, Petersen BE, Kim TH, Michalopoulos GK. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 20.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 21.Limaye PB, Bowen WC, Orr AV, Luo J, Tseng GC, Michalopoulos GK. Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium. Hepatology. 2008;47:1702–1713. doi: 10.1002/hep.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 23.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 24.Grompe M. Liver repopulation for the treatment of metabolic diseases. J Inherit Metab Dis. 2001;24:231–244. doi: 10.1023/a:1010375203539. [DOI] [PubMed] [Google Scholar]
- 25.Overturf K, Al-Dhalimy M, Finegold M, Grompe M. The repopulation potential of hepatocyte populations differing in size and prior mitotic expansion. Am J Pathol. 1999;155:2135–2143. doi: 10.1016/S0002-9440(10)65531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laconi E, Oren R, Mukhopadhyay DK, Hurston E, Laconi S, Pani P, Dabeva MD, et al. Long-term, near-total liver replacement by transplantation of isolated hepatocytes in rats treated with retrorsine. Am J Pathol. 1998;153:319–329. doi: 10.1016/S0002-9440(10)65574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braun KM, Thompson AW, Sandgren EP. Hepatic microenvironment affects oval cell localization in albumin-urokinase-type plasminogen activator transgenic mice. Am J Pathol. 2003;162:195–202. doi: 10.1016/S0002-9440(10)63810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/−mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.