Abstract
Inflammasomes are a family of cytosolic multiprotein complexes that initiate innate immune responses to pathogenic microbes by activating the CASPASE1 (CASP1) protease1,2. Although genetic data support a critical role for inflammasomes in immune defense and inflammatory diseases3, the molecular basis by which individual inflammasomes respond to specific stimuli remains poorly understood. The inflammasome that contains the NLRC4 (NLR family, CARD domain containing C4) protein was previously shown to be activated in response to two distinct bacterial proteins, flagellin4,5 and PrgJ6, a conserved component of pathogen-associated type III secretion systems. However, direct binding between NLRC4 and flagellin or PrgJ has never been demonstrated. A homolog of NLRC4, NAIP5 (NLR family, Apoptosis Inhibitory Protein 5), has been implicated in activation of NLRC47–11, but is widely assumed to play only an auxiliary role1,2, since NAIP5 is often dispensable for NLRC4 activation7,8. However, Naip5 is a member of a small multigene family12, raising the possibility of redundancy and functional specialization among Naip genes. Indeed, we show here that different NAIP paralogs dictate the specificity of the NLRC4 inflammasome for distinct bacterial ligands. In particular, we found that activation of endogenous NLRC4 by bacterial PrgJ requires NAIP2, a previously uncharacterized member of the NAIP gene family, whereas NAIP5 and NAIP6 activate NLRC4 specifically in response to bacterial flagellin. We dissected the biochemical mechanism underlying the requirement for NAIP proteins by use of a reconstituted NLRC4 inflammasome system. We found that NAIP proteins control ligand-dependent oligomerization of NLRC4 and that NAIP2/NLRC4 physically associates with PrgJ but not flagellin, whereas NAIP5/NLRC4 associates with flagellin but not PrgJ. Taken together, our results identify NAIPs as immune sensor proteins and provide biochemical evidence for a simple receptor-ligand model for activation of the NAIP/NLRC4 inflammasomes.
A fundamental question in immunology is how host defense is initiated in response to specific microbial ligands. The inflammasome containing the NLRC4 protein activates CASP1 in response to the C-terminus of bacterial flagellin6,7, as well as in response to the inner rod protein of the type III secretion systems of diverse bacterial species (e.g., PrgJ of Salmonella Typhimurium)6. Activated CASP1 processes interleukin-1β and −18 inflammatory cytokines and induces a rapid and inflammatory host cell death called pyroptosis13. In certain cases, NLRC4 activation requires NAIP5, as Naip5−/− mice fail to activate NLRC4 or CASP1 in response to infection with Legionella pneumophila or in response to the C-terminus of flagellin7,8. Interestingly, however, NAIP5 is not essential for NLRC4 activation in response to Salmonella Typhimurium or PrgJ7,8.
In addition to Naip5, C57BL/6 mice express three additional Naip genes (Naip1, Naip2, and Naip6), the functions of which remain unknown12. We hypothesized that each NAIP paralog may have evolved to be specific for a unique bacterial ligand. We first focused on NAIP2, as it appeared to be highly expressed in C57BL/6 mice14. We used specific shRNAs to knock down Naip2 expression in primary bone marrow-derived macrophages. ShRNA#1 and #2 specifically reduced NAIP2 protein levels without targeting other NAIP paralogs, whereas empty vector, shRNA#3 or a scrambled control shRNA had little effect on NAIP2 protein levels (Supplementary Fig. 1a, b). Macrophages expressing these shRNAs were then infected with flagellin-deficient Listeria strains that inducibly express PrgJ (Listeria-PrgJ) or flagellin (Listeria-FlaA)8. A Listeria-based system was chosen because it is an efficient means for delivering PrgJ to macrophages8, and because it allows for controlled comparisons of PrgJ and FlaA within a single experimental system. Remarkably, knockdown of Naip2 prevented pyroptosis and CASP1 activation by Listeria-PrgJ (Fig. 1a–c). By contrast, Naip2 knockdown did not affect inflammasome activation by Listeria-FlaA (Fig. 1b, c) or L. pneumophila, which expresses flagellin but not PrgJ (Supplementary Fig. 1c). Instead flagellin-dependent inflammasome activation depended on Naip5, as previously shown7–11. Inflammasome activation by wild-type Salmonella, which encodes both flagellin and PrgJ, was not significantly affected by Naip2 knockdown (Fig. 1d, e). However, knockdown of Naip2 in Naip5−/− macrophages significantly reduced or abolished inflammasome activation by wild-type Salmonella (Fig. 1d, e), indicating that both NAIP2 and NAIP5 recognize Salmonella. Interestingly, inflammasome activation by flagellin-deficient (FliC/FljB−) Salmonella, which still express PrgJ, depended entirely on Naip2 (Fig. 1d, e). Taken together, these data indicate that Naip2 is specifically required for activation of the NLRC4 inflammasome by PrgJ, in contrast to Naip5, which appears to be specifically required for NLRC4 activation by flagellin.
Figure 1. NAIP2 is required in macrophages for inflammasome activation in response to PrgJ.
(a–c) Primary bone-marrow derived macrophages expressing shRNAs targeting NAIP2 (or controls) were infected with flagellin-deficient Listeria monocytogenes (MOI = 5) expressing a secreted ActA100-PrgJ (pPrgJ) or ActA100-FlaA (pFlaA) fusion protein under IPTG-inducible control. (a, b) Cell death (± s.d.) was measured in triplicate by LDH release 6 hours after infection, or (c) Active CASP1 (p10) was measured by western blotting of cell supernatants. (d, e) NAIP2 knockdown cells were infected with wildtype or flagellin-deficient (FliC/FljB−) Salmonella Typhimurium and inflammasome activation was measured by (d) LDH release (± s.d.) at 3h after infection or (e) CASP1 processing. Data shown are representative of two (c, e) or three (a, b, d) independent experiments. *, p<0.02 as compared to scramble (Student’s t-test, two-tailed).
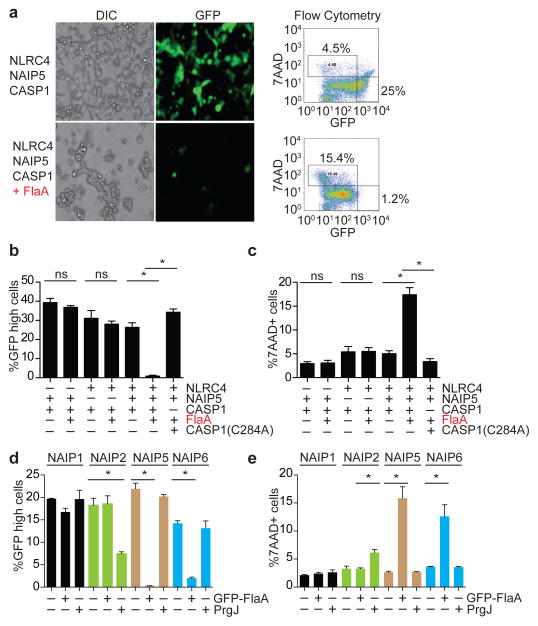
Biochemical analysis of the inflammasome in macrophages is complicated by the expression of multiple NAIP proteins and by their low expression levels. We therefore decided to reconstitute the NLRC4 inflammasome in non-immune 293T cells, which do not express NLRC4 or NAIPs, so that the functions of individual NAIP proteins could be analyzed. 293T cells transiently transfected with green fluorescent protein (GFP)-marked vectors encoding wild-type NLRC4, NAIP5 and CASP1 did not exhibit significant spontaneous inflammasome activation, and instead, a majority of cells expressed GFP (Fig. 2a). However, when flagellin (FlaA) from L. pneumophila was co-expressed with NLRC4/NAIP5/CASP1, we observed a significant loss of GFPhigh cells and an increase in the number of dead (7AAD+) cells (Fig. 2a). This result was highly reminiscent of flagellin-dependent activation of the endogenous NAIP5/NLRC4 inflammasome in macrophages, which also results in a rapid CASP1-dependent cell death, loss of membrane integrity, and release of cytosolic contents and GFP7. Similar to the genetic requirement for Nlrc4, Naip5 and Casp1 in macrophages4,5,7–11,15, we found that NAIP5, NLRC4, catalytically active CASP1, and FlaA are all required to trigger cell death and loss of membrane integrity/GFP in reconstituted 293T cells (Fig. 2b, c). The reconstituted NAIP5/NLRC4 inflammasome also recapitulated the ability of native inflammasomes to process CASP1 and IL-1β in response to cytosolic flagellin (Supplementary Fig. 2). Consistent with a lack of a role for NAIP5 in recognition of PrgJ by macrophages8, the reconstituted NAIP5/NLRC4 inflammasome did not respond to PrgJ (Fig. 2d, e). By contrast, a reconstituted NAIP2/NLRC4 inflammasome responded specifically to PrgJ but not flagellin (Fig. 2d, e). Taken together, we conclude that we have successfully reconstituted NAIP2/NLRC4 and NAIP5/NLRC4 inflammasomes that exhibit all the known requirements and specificities of the native inflammasomes.
Figure 2. Reconstitution of the NAIP5/NLRC4 inflammasome in 293T cells.
(a) GFP-marked expression vectors encoding NLRC4, NAIP5, CASP1 and/or flagellin (FlaA) were transiently transfected into 293T cells. Cells were imaged for differential interference contrast (DIC) and GFP fluorescence 48 hours later. Dead cells were stained with 7AAD. (b) GFP-high cells and (c) 7AAD positive cells were quantified (± s.d.) as in a, but with specific expression vectors omitted from the transfection as indicated. CASP1(C284A) is a catalytically dead mutant. (d, e) 293T cells were transfected as indicated and analyzed as above. Data shown (± s.d.) are representative of at least three independent experiments. *, p<0.02 (Student’s t test, two tailed); ns, not significant.
It is believed that activated inflammasomes assemble into high molecular-weight multiprotein complexes16, but this has not been demonstrated for the NLRC4 inflammasome. To visualize inflammasome assembly, 293T cells were transfected with NAIP5, NLRC4, and FlaA in various combinations, but CASP1 was omitted so that cell death and loss of cellular contents (and assembled inflammasomes) would not occur. Digitonin-solubilized cell lysates were resolved on Blue Native (BN) PAGE gels17. A dramatic shift of NLRC4 from a monomer (~120kDa) to an oligomeric complex (~1000kDa) was seen in the presence of NAIP5 and FlaA. NAIP5 was also contained within the high molecular weight oligomeric complex (Fig. 3a). The association of NAIP5 and NLRC4 in the same complex was validated by co-immunoprecipitation (Supplementary Fig. 3)11,18. NLRC4 oligomerization was induced by either untagged FlaA or a GFP-FlaA fusion protein (Fig. 3a), both of which activate NLRC4/CASP1. Importantly, assembly of the NLRC4 inflammasome required FlaA (Supplementary Fig. 4a) and was not observed in the absence of NAIP5 (Fig. 3a), indicating that a biochemical function of NAIP5 is to promote NLRC4 oligomerization.
Figure 3. NAIP5 is required for formation of a hetero-oligomeric complex that contains NLRC4, NAIP5 and flagellin.
(a) 293T cells were transfected as indicated, followed by analysis by Blue Native-PAGE or SDS-PAGE, and western blotting. *NS, non-specific band. (b) 293T cells were transfected as indicated and lysates were separated by a first dimension of Blue Native-PAGE followed by a second dimension of SDS-PAGE. (c, d) 293T cells were transfected as indicated and samples were processed and analyzed as in a. Data shown are representative of at least three independent experiments.
Despite strong genetic evidence that NLR proteins, such as NAIP5 and NLRC4, function as microbial ‘sensors’, there is no biochemical evidence that NLRs interact directly with microbial ligands. In fact, some studies of the NLRP3 inflammasome19–21, as well as analyses of analogous proteins from plants22, suggest that at least some NLRs recognize pathogens indirectly. In order to determine if the oligomerized NAIP5/NLRC4 complex also contains flagellin, we subjected samples separated in the first dimension by native PAGE to a second dimension of SDS-PAGE. To facilitate detection of flagellin, we used a 6x-Myc-tagged flagellin, which activates the inflammasome identically to native flagellin (data not shown). This approach revealed that FlaA was indeed present in a high-molecular weight complex, along with NAIP5 and NLRC4 (Fig. 3b). NAIP5 exhibited a weak flagellin-dependent mobility shift in the absence of NLRC4 (Supplementary Fig. 4b) suggesting that NLRC4 is not essential for flagellin recognition, though formation/stabilization of the oligomerized complex appears to be significantly enhanced by NLRC4. FlaA expressed alone was present in cell extracts only as a monomer (Supplementary Fig. 4c). Taken together, these observations provide evidence for a simple receptor-ligand model of NAIP5/NLRC4 activation by flagellin.
Consistent with the autoinhibitory function of the leucine rich repeats (LRRs) in other NLRs, we found that NAIP5ΔLRR and NLRC4ΔLRR constitutively activated CASP1-dependent cell death, independent of the presence of flagellin (Supplementary Fig. 5). Interestingly, NLRC4ΔLRR was able to activate CASP1 in the absence of NAIP5, whereas constitutively active NAIP5ΔLRR required wild-type NLRC4 in order to activate CASP1. This result suggests that NAIP5 functions upstream of NLRC4. Indeed, NAIP5ΔLRR was able to induce the oligomerization of wild-type NLRC4 (Fig. 3c), whereas the spontaneous oligomerization of NLRC4ΔLRR did not require NAIP5 (Fig. 3d). Spontaneous oligomerization of NLRC4ΔLRR did require the nucleotide binding domain (NBD) of NLRC4, as a K175R mutation previously shown to disrupt NBD function23 abolished NLRC4ΔLRR auto-oligomerization (Fig. 3d). The ability of NAIP5 to induce oligomerization of NLRC4 in response to flagellin required both the NBD and N-terminal BIRs of NAIP5, but did not require the N-terminal CARD of NLRC4 (Supplementary Fig. 4d, e), whereas functional CASP1 activation required all these domains (Supplementary Fig. 5, 6). Taken together, these data suggest a working model (Supplementary Fig. 7) in which NAIP5 is activated by flagellin and induces downstream NLRC4 oligomerization and CASP1 activation.
Consistent with a specific role for NAIP2 in recognition of PrgJ, we found that PrgJ did not induce the oligomerization of NAIP5/NLRC4 (Fig. 4a), but did induce oligomerization of NAIP2/NLRC4. Oligomerization of NLRC4 did not occur when co-expressed with NAIP2 alone or with NAIP2/FlaA (Fig. 4b). Interestingly, NAIP6 resembled NAIP5 and supported NLRC4 oligomerization in response to FlaA but not PrgJ (Fig. 4c), perhaps providing an explanation for the previously puzzling observation that Naip5−/− cells can respond to high levels of flagellin7. In contrast, NAIP1 is an ‘orphan’ NAIP since it responded neither to PrgJ or flagellin (Fig. 2d, e; Supplementary Fig. 8).
Figure 4. NAIP Paralogs Confer Specificity to the NLRC4 Inflammasome.
(a) 293T cells were co-transfected with wild-type NAIP5 and NLRC4, alone or in combination with 6x-Myc-FlaA or 6x-Myc-PrgJ followed by Blue Native PAGE 48 hours later. *NS, non-specific band. Whole cell lysates were also separated by conventional 4–12% SDS-PAGE to control for expression of each transfected gene construct (left panel). (b) 293T cells were transfected with wild-type NAIP2 and NLRC4 and analyzed as in a. (c) 293T cells were transfected with wild-type NAIP6 and NLRC4, and analyzed as in a. Data shown are representative of at least three independent experiments.
Our results demonstrate that the ability of the NLRC4 inflammasome to assemble and functionally activate CASP1 in response to specific bacterial ligands is dictated by NAIP family members. The most parsimonious model to account for our results is that NAIP proteins function as direct receptors for bacterial ligands (Supplementary Fig. 7). Although NLRC4 was previously suspected to be the cytosolic flagellin sensor1,2, we hypothesize that a main function of NLRC4 may instead be to serve as an adaptor, downstream of NAIP proteins, to recruit CASP1 via a CARD-CARD interaction. NLRC4 may also play an important role in ligand binding or in stabilizing NAIP/NLRC4/ligand complexes, but the specificity of the complexes for particular ligands appears to be controlled by NAIP proteins.
The number and sequence of Naip paralogs varies significantly among inbred mouse strains, and has been suggested to be evolving rapidly24. Indeed, the murine Naip locus was originally identified by a forward genetic approach which took advantage of the widely varying susceptibility of inbred mouse strains to L. pneumophila infection14,25. The single known human NAIP ortholog may also exist within a rapidly evolving locus24; our results suggest that it will be of great interest to establish the specificity of the human NAIP protein. We propose that Naip gene evolution represents a fascinating example of the molecular arms race between bacteria and their hosts.
Methods Summary
Naip2 knockdown
Primary C57BL/6 bone marrow cells were transduced with pLKO.1-based lentivirus encoding shRNAs that specifically target Naip2 or controls. Bone marrow cells were differentiated into macrophages by culture in media containing MCSF. On day 4 of culture, transduced macrophages were selected by addition of puromycin (5μg/ml). On day 8 of culture, macrophages were replated and infected the next day with Listeria monocytogenes or Salmonella Typhimurium expressing flagellin or PrgJ8 and inflammasome activation was measured by assaying release of the cytosolic enzyme lactate dehydrogenase (LDH)7 or by western blotting for processed (p10) CASP1.
Reconstituted inflammasome
The inflammasome was reconstituted by transfection of 293T cells with MSCV2.2-IRES-GFP-based expression vectors encoding various mouse (C57BL/6-derived) Naip genes, Nlrc4 and Caspase-1. Inflammasome oligomerization was assessed in digitonin (1%) lysates using a Bis-Tris NativePAGE system (Invitrogen) followed by western blotting.
Statistical Analysis
Statistical differences were calculated with an unpaired two-tailed Student’s t-test using GraphPad Prism 5.0b.
Supplementary Material
Acknowledgments
Work in R.E.V.’s laboratory is supported by Investigator Awards from the Burroughs Wellcome Fund and the Cancer Research Institute and by NIH grants AI075039, AI080749, and AI063302. We thank Jakob von Moltke, Alex Kintzer and Bryan Krantz for provision of LFn-FlaA and PA, S. Mariathasan and V. Dixit for the gift of anti-NLRC4 antibodies and Nlrc4−/− mice, J.D. Sauer and D. Portnoy for development of Listeria strains to deliver PrgJ and FlaA, and E. Michelle Long and W. Dietrich for pCDNA3-NAIP constructs and anti-NAIP antibodies. We thank A. Roberts for her initial efforts to knock down NAIP2, M. Fontana for validating NAIP knockdowns, and Jakob von Moltke and members of the Barton and Vance Labs for helpful discussions.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions: E.M.K. and R.E.V. conceived experiments and wrote the paper. E.M.K. performed the experiments.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Latz E. The inflammasomes: mechanisms of activation and function. Curr Opin Immunol. 2010;22:28–33. doi: 10.1016/j.coi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Ting JP, Kastner DL, Hoffman HM. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol. 2006;6:183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- 4.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 5.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 6.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lightfield KL, et al. Differential requirements for NAIP5 in activation of the NLRC4 (IPAF) inflammasome. Infect Immun. 2011;79:1606–1614. doi: 10.1128/IAI.01187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 12.Growney JD, Dietrich WF. High-resolution genetic and physical map of the Lgn1 interval in C57BL/6J implicates Naip2 or Naip5 in Legionella pneumophila pathogenesis. Genome Res. 2000;10:1158–1171. doi: 10.1101/gr.10.8.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright EK, et al. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 15.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 17.Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 18.Damiano JS, Oliveira V, Welsh K, Reed JC. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem J. 2004;381:213–219. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008 doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 21.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 22.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, et al. Nucleotide binding to CARD12 and its role in CARD12-mediated caspase-1 activation. Biochem Biophys Res Commun. 2005;331:1114–1119. doi: 10.1016/j.bbrc.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Romanish MT, Lock WM, de Lagemaat LN, Dunn CA, Mager DL. Repeated Recruitment of LTR Retrotransposons as Promoters by the Anti-Apoptotic Locus NAIP during Mammalian Evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diez E, et al. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet. 2003;33:55–60. doi: 10.1038/ng1065. [DOI] [PubMed] [Google Scholar]
- 26.Krantz BA, et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.