Abstract
Magnetic resonance imaging (MRI) is now a leading diagnostic technique. As technology has improved, so has the spatial resolution achievable. In 1986 MR microscopy (MRM) was demonstrated with resolutions in the tens of microns, and is now an established subset of MRI with broad utility in biological and non-biological applications. To date, only large cells from plants or aquatic animals have been imaged with MRM limiting its applicability. Using newly developed microsurface coils and an improved slice preparation technique for correlative histology, we report here for the first time direct visualization of single neurons in the mammalian central nervous system (CNS) using native MR signal at a resolution of 4–8µm. Thus MRM has matured into a viable complementary cellular imaging technique in mammalian tissues.
Keywords: Microscopy, magnetic resonance, MRM, neurons, brain tissue
Introduction
Since its inception in the 1970’s, MRI has grown dramatically and has arguably transformed clinical imaging and the practice of medicine with broad applications in a wide range of biological and non-biological systems. Foremost in the evolution of MRI has been a drive to improve the signal-to-noise ratio (SNR) through a combination of hardware and techniques, lead primarily by the transition to ever higher magnetic fields. As the SNR is improved, it can be traded usually for improved spatial and/or temporal resolution. Sub-millimeter spatial resolutions were achievable in the 1970s and early 1980s. However, the pioneers of MRI quickly realized that, with sufficient SNR, microscopic resolutions may be achievable (Mansfield and Morris, 1982). Practically, this was first published in 1986 through MR microimages of single cells (frog ovum) (Aguayo et al, 1986) at in-plane resolutions on the order of tens of microns. Since that time MRM has been applied in a variety of applications, including studies of biological tissues, plants and materials (Callaghan, 1994, Aiken et al, 1995, Ciobanu et al, 2003). It is generally accepted that MR imaging transitions to MR microscopy at resolutions below 100 microns (Benveniste and Blackband, 2005). To date, isotropic resolutions of 3 to 4 microns have been reported (Ciobanu et al, 2002, Weiger et al, 2008), with the highest reported in-plane resolution being 1×1 microns (with a 75 micron slice thickness) (Lee et al, 2001). It is important to remember that the resolution alone is only one consideration in MR studies. The contrast mechanism employed is also important for differentiating structure in different samples with different MR properties, which may, in turn, require resolution compromises to maintain adequate SNR.
MRM studies of single biological cells have been limited to plant cells and two animal cells (see Aiken et al, 1995, and Ciobanu et al, 2003 for reviews). Although the work of Aguayo et al, 1986, featured the first MR images of animal cells, the cell they studied was very large: i.e. frog ova up to 1mm in diameter. To this point, the only other animal cell imaged via MR is the L7 neuron of the gastropod Aplysia californica (Schoeniger et al, 1994). On its own, this represented a significant technical achievement in that the L7 neuron, at approximately 300 µm in diameter, has a 30-fold smaller volume than the fog ovum. Resolutions of 20 to30 microns are achieved through the use of small so-called microcoils (usually solenoids 0.5–1mm in diameter) and high magnetic fields. The large size of the frog ovum and its robustness facilitates studies of live cells with physiological perturbation (Hsu et al, 1996) or proton spectroscopy localized to the cell cytoplasm and nucleus (Grant et al, 2000). Still, these resolutions are barely acceptable for imaging even the largest cell bodies in mammalian neural tissue that are approximately 5 to 100 microns in diameter. Since then, the drive has been to further improve MRM so that cellular resolutions could be obtained in mammalian tissues: particularly the brain. This would expand the utility of MRM for animal and human studies on excised tissues which could act as a complement to optical techniques and which could lead to physiological MRM studies on live tissues at the cellular level.
Using newly developed microsurface coils at high magnetic fields, we recently reported the observation of small dark regions in rat neural tissue that corresponded broadly with the expected dimension of neural cells (Flint et al, 2008). However, without direct correlative histology, image interpretation was problematic. In the present study, using these new microsurface coils and corresponding histological analysis, we demonstrate for the first time the ability of MRM to visualize single mammalian neurons using indigenous MR signal.
Methods
All imaging was performed on a 600MHz Bruker imaging spectrometer with a microimaging accessory providing gradient strengths of 3000mT/m. The console was interfaced with microsurface coils developed by Bruker Instruments Inc. (Massin et al, 2002, Weiger et al, 2003) shown in Figure 1.
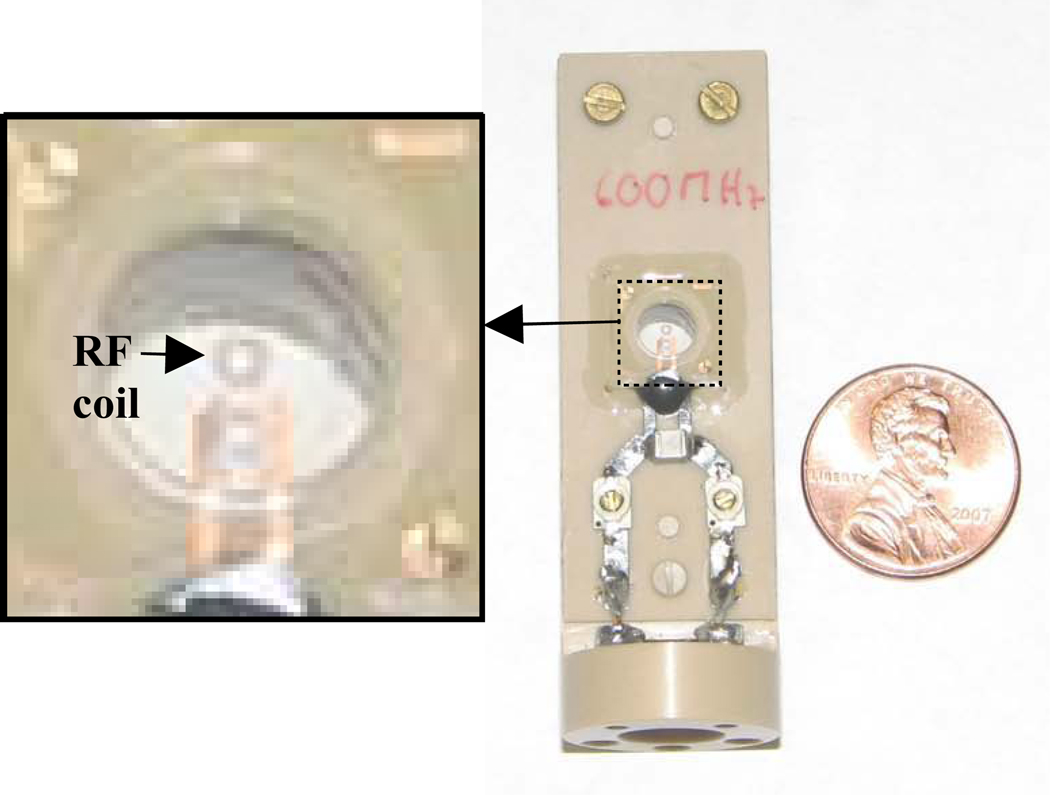
Figure 1.
Photograph of the 500µm surface microcoil developed by Bruker, Switzerland (Z76409). The 500µm four-turn surface microcoil sits inside a 5mm diameter, 500µm deep tissue well better visualized in the expanded section on the left. A 200µm coil was also employed.
The key to successful MR microimaging and acquiring the corresponding histology of mammalian neural tissue at the cellular level lies in the sample preparation. For the spinal cord tissue preparation, Male Sprague Dawley rats (150g) were anesthetized with isoflurane and exsanguinated by perfusion of phosphor-buffered saline (300ml; 137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, and 1.8mM KH2PO4: pH 7.4 at 300mOsm) through an intracardial catheter secured in the aortic arch. Normal saline perfusion was followed by an equivalent volume of saline containing 4% formaldehyde as a fixing agent. Following perfusion fixation, spinal cords were removed by gross dissection and the cervical and lumbar enlargements stored in saline solution containing 4% formaldehyde until use. Transverse sections (25µm) were cut from within the cervical enlargement using a Lancer vibratome (Ted Pella, series 1000) and samples were washed in PBS overnight as a means of removing formaldehyde prior to imaging experiments. Spinal cord sections (25µm) were bisected along the median fissure to permit placement of the sample into the tissue well of the 500µm surface microcoil. The tissue section was placed such that the ventral horn of the spinal cord near the boundary of white and gray matter was contacting the coil face. The tissue was secured in place using a retention system developed in-house consisting of a circular, nylon mesh screen with a viewing window cut into its center held in place by a nylon ring. Once secured, PBS was placed into the tissue well to prevent drying of the sample and the well was sealed using adhesive PCR film (ABgene, AB-0558). Conventional spin echo diffusion-weighted images (b = 4025 s/mm2, Δ = 17ms, δ = 6ms, slice thickness = 80µm, 256×256 matrix, spatial resolution = 7.8µm in-plane, total acquisition time = 7h7min, bandwith = 50kHz, read and phase gradient amplitudes = 591 and 650 mT/m, respectively) were acquired. Although the nominal slice thickness for the excitation pulse was set to the minimum value possible within the boundaries of our scan parameters (80µm), heavy diffusion weighting resulted in total abolition of signal from the PBS surrounding the sample yielding an effective slice thickness to be that of the tissue sample itself: i.e. 25µm. Following the collection of MR data, tissue slices were removed from the imaging coil and placed in a Nissl stain (0.5% Cresyl Violet, 0.3% glacial acetic acid in ddH2O) for one minute. Slices were then placed temporarily in a destaining bath (0.3% glacial acetic acid in ddH2O) to remove excess Nissl stain before being wet mounted to pre-cleaned, optical microscopy slides (Fisher, 12-550-13) using Histomount mounting medium (National Diagnostics, HS-103). False-color histological images were taken using a digital camera (QImaging, Retiga 4000R Fast 1394 Color) attached to a Zeiss microscope (Axioplan 2, Zeiss) and processed with software (QCapture Pro 6.0) available from QImaging. Images were acquired at 100X magnification and employed an emission filter in the Texas Red range.
For the preparation of striatal brain samples, Male, Sprague Dawley rats (150g) were anesthetized with isoflurane and euthanized by decapitation. The striatum was excised in 500µm thick slices and immersion fixed in PBS containing 4% formaldehyde for no less than 24h. Following fixation, samples were washed in PBS overnight. Washed tissue was placed inside the sample well of a 200µm diameter microsurface coil (Bruker, Z76412) and MR data were acquired. Three dimensional image data sets were collected using a conventional gradient echo sequence (TE=10ms, TR=150ms, matrix=1283, FOV = 0.6mm, total acquisition time=22h, bandwidth = 25kHz, read and phase gradient amplitudes = 1980 and 1830 mT/m, respectively) acquired at 4.7µm isotropic resolution. Following imaging, samples were snap-frozen in cryosectioning medium (Richard-Allan, 6502), sliced in 25µm increments (Microm, HM 505 E Cryostat), and affixed to poly-L-coated microscope slides. After allowing tissue slices to adhere overnight, slides were Nissl stained and photographed as described above. Segmentation of the MR images taken in the striatum was performed using AMIRA visualization software (Visage Imaging, AMIRA 3.1.1).
Note that in MR microscopy, molecular diffusion may lead to image blurring if the gradients are not strong enough to dominate these effects (Callaghan 1994). In all our imaging studies, the gradients strengths are sufficient for the intrinsic resolution to be real for the diffusion coefficients observed in these samples of approximately 1×10−9 m2/s or less (Shepherd et al, 2009) using the criteria provided by Ciobanu et al, 2003. Note also that no filtering or line broadening is applied to the image data sets, so the spatial resolution reported is the intrinsic resolution.
Results
Tissue samples were isolated from rats using modified procedures designed to minimize sample distortion and tissue water loss (see methods). In preliminary studies, relatively thick sections of tissue that fit within the microcoil sample chamber were imaged. Figure 2 shows a representative MR image taken of the striatum using a 200µm microsurface coil with representative histology from a similar piece of striatum. Although direct correlative histology is not possible with this approach (it proves impractical to section the sample for subsequent optical studies and unambiguously match the data), a descending white mater tract is visible and manifests as a dark band running vertically through the coil’s field of view. In the striatal gray matter, dark circular structures are visible throughout. Some of the more distinct structures were segmented in order to attain an estimate of their size and shape (Figure 3). Although the segmentation analysis showed the structures to be approximately the same size (approximately 15µm diameter (12)) as cell bodies of medium spiny neurons which are ubiquitous in the basal ganglia, a direct histological correlation was not achieved.
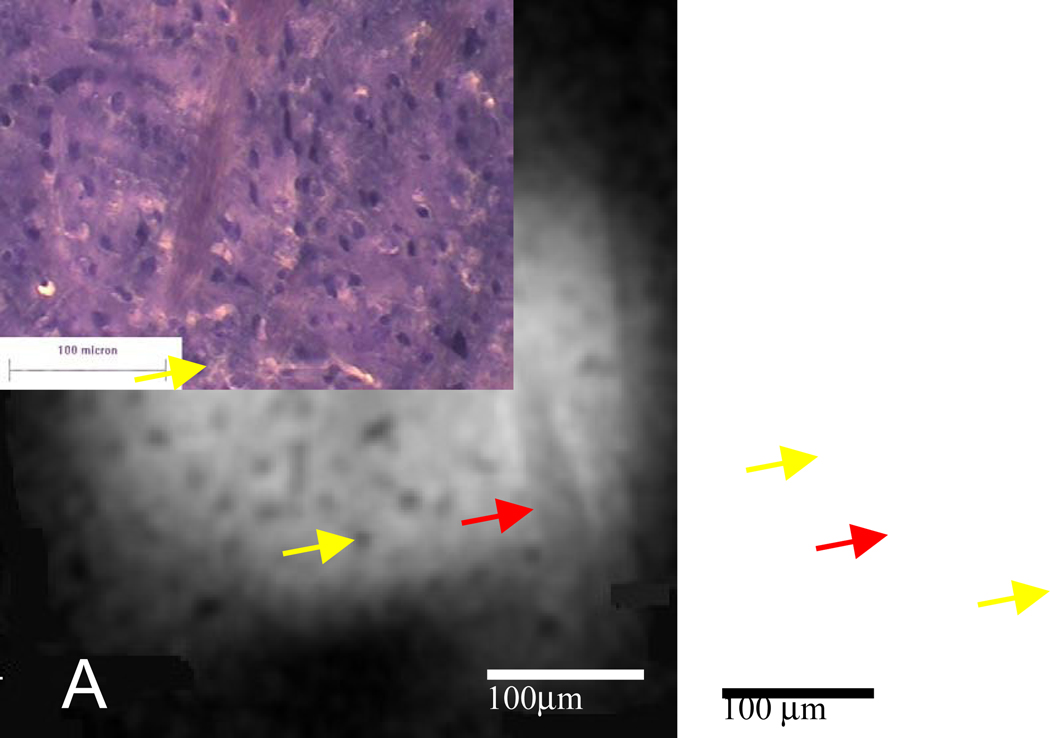
Figure 2.
MR microscopy and histology of rat striatal tissue. (A) Representative MR image (4.7µm isotropic) from a three-dimensional gradient echo data set acquired at 4.7µm isotropic resolution (TE=10ms, TR=150ms, matrix=1283, 0.6mm FOV, acquisition time=22h) with (B) histology from a similar piece of tissue. The image and histology show descending white matter tracts (red arrows). Structures which resemble cell bodies of medium spiny neurons in size, shape, and distribution are present throughout (yellow arrows show two examples).
Figure 3.
3D segmentation reconstruction (70µm depth) of 15 image frames from a 3D MRM data set. A descending white matter tract (pink) and numerous structures described in 2A (red) are visible.
By preparing spinal tissue containing larger cell bodies and tissue slices 25µm in thickness for MRM (see methods), and then performing histology on the very same slice (no additional sectioning), direct comparison of MRM and histology becomes possible. Figure 4 shows what we believe to be the first MRM image in which mammalian cells are resolved using native tissue contrast with direct histological confirmation. The hypointense, circular structures present in the MR image are approximately 50µm in diameter which further supports our conclusion that the signal we see from these regions originates from the cell bodies of alpha motor neurons. These data provide further support to the conclusion that the dark regions in the striatum in Figure 2 are also cell bodies.
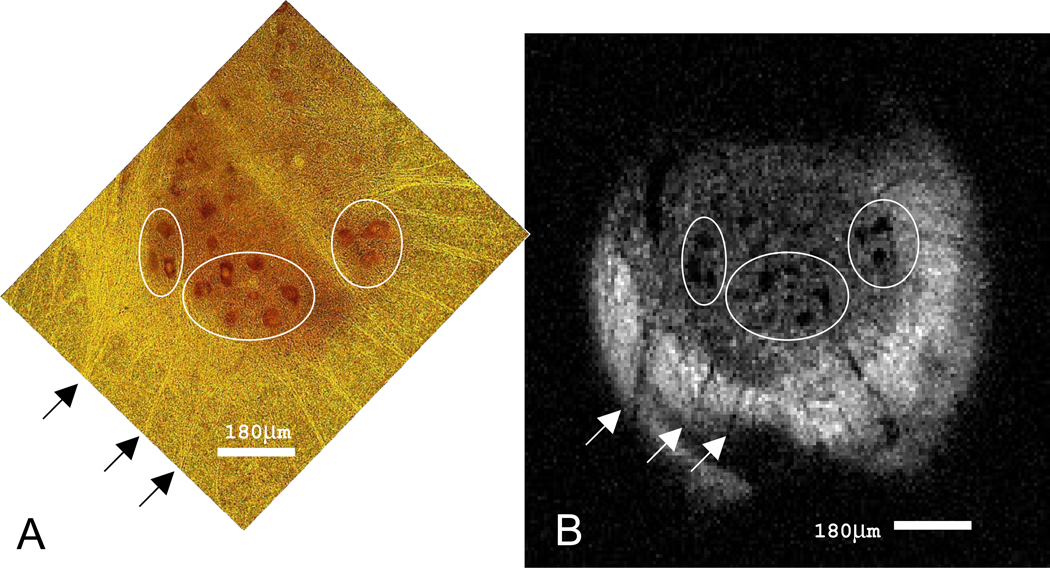
Figure 4.
MR microscopy and histology of rat spinal cord tissue. (A) Nissl stained 25µm section of the ventral horn in the rat spinal cord. Alpha motor neurons (dark red, ~50µm diameter) and their nuclei (yellow) are clearly seen. The outermost border of the ventral horn and its projections are visible (bright yellow) against the backdrop of the spinal cord white matter (dark yellow). (B) MR image (7.8 µm in-plane, acquisition time = 7h 7minutes) of the tissue slice exhibited in (A). The boundary between gray and white matter of the spinal cord is clearly visible as the signal intensity in this diffusion-weighted image is greater in the surrounding white matter. Individual cell bodies of alpha motor neurons appear as dark spots in the image and correlate spatially and morphologically with the cells in (A).
Discussion
Cellular level MRI in animal and human brain has been a long term goal of MRI. Although large cells have been imaged (frog ova and Aplysia neurons), direct imaging of single cells in mammalian neural tissue has so far been elusive. The presence of cells has been detected using exogeneous contrast agents, in which the local MR signal is distorted and amplified, such as those generated by the introduction of iron oxide particles (Shapiro et al, 2004) in order to generate contrast between cells and their surrounding tissues. However, these techniques require contrast agents and report only cell position: they do not provide information on the cell itself, nor offer the possibility of quantitating the indigenous water signal characteristics used to generate MR contrast in tissues. Although we have previously observed hypointense areas on microimages of neural tissue (Flint et al, 2007), direct histological correlation was not feasible, and thus it could not be ascertained unambiguously if these areas were cells or other features (such as blood vessels or artifacts). In the present work, this challenge is solved by sectioning the tissue before MRM and using diffusion weighting to eliminate the perfusate signal. In this way, the exact same tissue is visualized in the MR and optical images and the correlation then becomes unambiguous. A similar method was employed to image histological samples of animal tissues on conventional histology slides (Meadowcroft et al, 2007), but not at the resolutions reported here. Similarly studies of small sheets of onion skin cells were also performed on histological slides with the coil mounted on the slide, facilitating accurate optical correlation (Glover et al, 1994).
This paper represents the first MR imaging of individual neural cells in mammalian tissue achieved using a combination of high magnetic fields and microsurface coils. Corresponding optical histology clearly indicates the cell bodies that appear as hypointense areas using diffusion weighted MRI. The apparently increased water diffusion within the cell body (cell cytoplasm) compared with surrounding tissue is not consistent with reduced diffusion observed in both frog ova and Aplysia neurons (Aguayo et al, 1986, Scheniger et al, 1995), but is consistent with the hypointense pyramidal cell layer observed in rat hippocampal slices (Shepherd et al, 2003). These observations will be validated and explored in future quantitative studies, and also addressed in studies on live tissues.
Future studies include quantitation of the MR signals and the development of a microperfusion chamber. This will facilitate the investigation of cells undergoing physiological perturbations (such as ischemia) and therapeutic interventions so that the microstructural and physiological origins of MR signals in tissues may be understood. Our long term goal is to use the data to develop meaningful and realistic working mathematical models of tissues that can be used to interpret lower spatial resolution MRI, including human imaging, and then use these models to improve the sensitivity and specificity of clinical MRI.
Acknowledgements
Funding provided by the NIH (P41 RR16105 and RO1 NS36992), the NSF through the National High Magnetic Field Laboratory, the KTI (Switzerland, CTI project 6364.1 KTS-NM), the Danish National Research Foundation (95093538- 2458, project 100297), the Denmark-America Foundation, Dagmar Marshall's Foundation, Julie von Mullen's Foundation and the Oticon Foundation. Thanks to Charles Massin for preliminary work with microcoils, and for optical support from the McKnight Brain Institute Cell and Tissue Analysis Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguayo JB, Blackband SJ, Schoeniger J, Mattingly MA, Hintermann M. Nuclear magnetic resonance imaging of a single cell: the NMR microscope. Nature. 1986;322:190–191. doi: 10.1038/322190a0. [DOI] [PubMed] [Google Scholar]
- Aiken NR, Hsu EW, Blackband SJ. A review of NMR microimaging studies of single cells. J Magn Reson Anal. 1995;1:41–48. [Google Scholar]
- Benveniste H, Blackband SJ. Translational neuroscience and MR microscopy. Lancet Neurology. 2005;5(6):536–544. doi: 10.1016/S1474-4422(06)70472-0. [DOI] [PubMed] [Google Scholar]
- Callaghan PT. Principles of Nuclear Magnetic Resonance Microscopy. Oxford University Press; 1994. [Google Scholar]
- Ciobanu L, Seeber DA, Pennington CH. 3D MR microscopy with resolution 3.7 micron by 3.3 micron by 3.3 micron. J Magn Reson. 2002;158(1–2):178–182. doi: 10.1016/s1090-7807(02)00071-x. [DOI] [PubMed] [Google Scholar]
- Ciobanu L, Webb AG, Pennington CH. Magnetic resonance imaging of biological cells. Progress in Nuclear Magnetic Resonance Spectroscopy. 2003;42:69–93. [Google Scholar]
- Flint JJ, Lee CH, Hansen B, Fey M, Schmidig D, Bui JD, King MA, Vestergaard-Poulsen P, Blackband SJ. Magnetic resonance microscopy of discreet microstructures in 5µm-isotropic images of the excised rat striatum. Proceedings of the ISMRM; 19–25th May; Berlin. 2007. [Google Scholar]
- Glover PM, Bowtell RW, Brown GD, Mansfield P. A microscope slide probe for high resolution imaging at 11.7 Tesla. Magn Reson Med. 1994;31(4):423–428. doi: 10.1002/mrm.1910310411. [DOI] [PubMed] [Google Scholar]
- Grant SC, Aiken NR, Plant HD, Gibbs S, Mareci TH, Webb AG, Blackband SJ. NMR spectroscopy of single neurons. Magn Reson Med. 2000;44:19–22. doi: 10.1002/1522-2594(200007)44:1<19::aid-mrm4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Hsu EW, Aiken NR, Blackband SJ. NMR microscopy of single neurons under hypotonic perturbation. Am J Physiol. 1996;271:C1895–C1900. doi: 10.1152/ajpcell.1996.271.6.C1895. [DOI] [PubMed] [Google Scholar]
- Lee SC, Kim K, Kim J, Lee S, Yi J, Kim S, Ha K, Cheong C. One micrometer resolution NMR microscopy. J. Magn. Reson. 2001;150:207. doi: 10.1006/jmre.2001.2319. [DOI] [PubMed] [Google Scholar]
- Mansfield P, Morris PG. In: NMR Imaging in Biomedicine. Advances in Magnetic Resonance. Waugh JS, editor. New York: Academic Press; 1982. [Google Scholar]
- Massin C, Boero G, Vincent F, Abenhaim J, Besse PA, Popovic RS. High Q-factor RF planar microcoils for micro scale NME spectroscopy. Sensors and Actuators A: Physical. 2002;97:280–288. [Google Scholar]
- Meadowcroft MD, Zhang S, Liu W, Park BS, Connor JR, Collins CM, Smith MB, Yang QX. Direct magnetic resonance imaging of histological tissue samples at 3.0T. Magn Reson Med. 2007;57(5):835–841. doi: 10.1002/mrm.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeniger JS, Aiken N, Hsu EW, Blackband SJ. Relaxation-time and diffusion NMR microscopy of single neurons. J Magn Reson B. 1995;103(3):261–273. doi: 10.1006/jmrb.1994.1039. [DOI] [PubMed] [Google Scholar]
- Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP. MRI detection of single particles for cellular imaging. Proc Natl Acad Sci. 2004;101(30):10901–10906. doi: 10.1073/pnas.0403918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Wirth ED, Thelwall PE, Chen HX, Roper SN, Blackband SJ. Water diffusion measurements in perfused human hippocampal slices undergoing tonicity changes. Magn Reson Med. 2003;49(5):856–863. doi: 10.1002/mrm.10456. [DOI] [PubMed] [Google Scholar]
- Shepherd TM, Flint JJ, Thelwall PE, Stanisz GJ, Mareci TH, Yachnis AT, Blackband SJ. Postmortem interval alters the water relaxation and diffusion properties of rat nervous tissue--implications for MRI studies of human autopsy samples. Neuroimage. 1. 2009;44(3):820–826. doi: 10.1016/j.neuroimage.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiger M, Schmidig D, Denoth S, Massin C, Vincent F, Schenkel M, Fey M. NMR-microscopy with isotropic resolution of 3 µm using dedicated hardware and optimized methods. Concepts in Magnetic Resonance Part B. 2008;33B(2):84–93. [Google Scholar]