Abstract
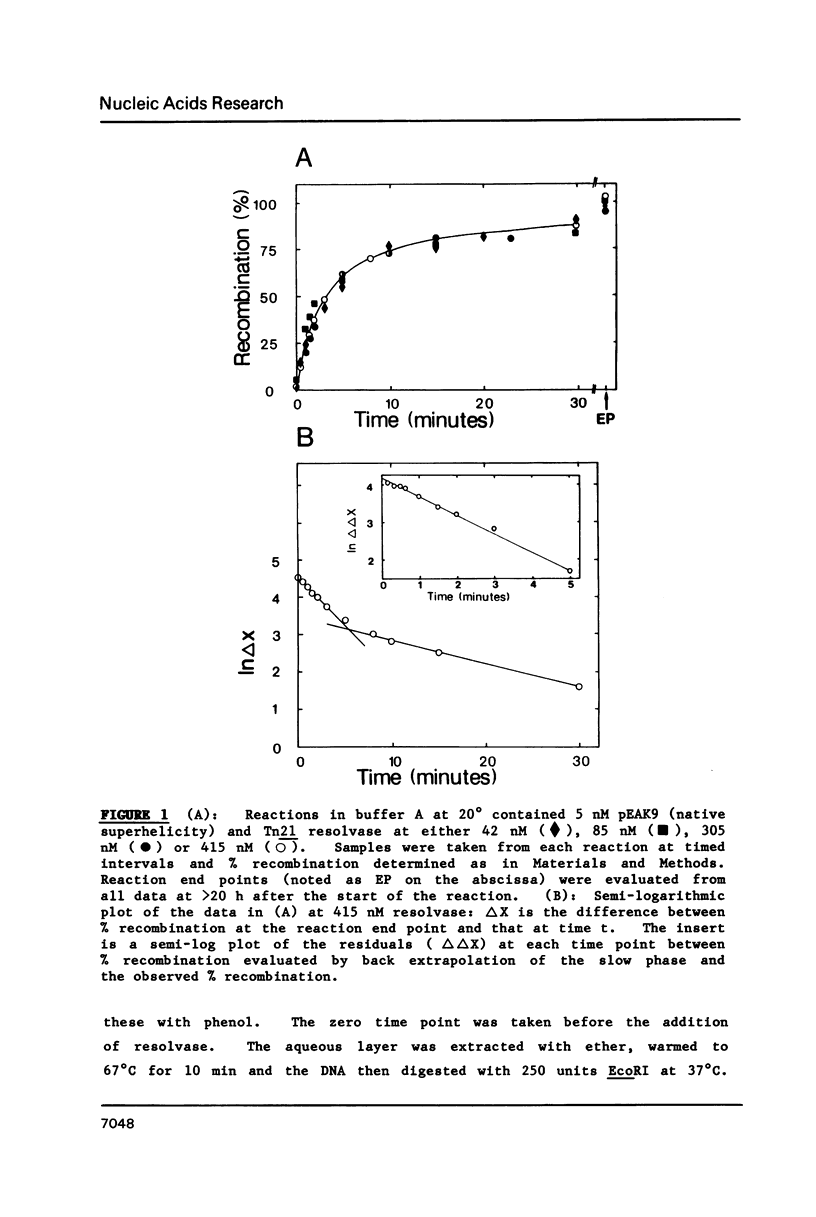
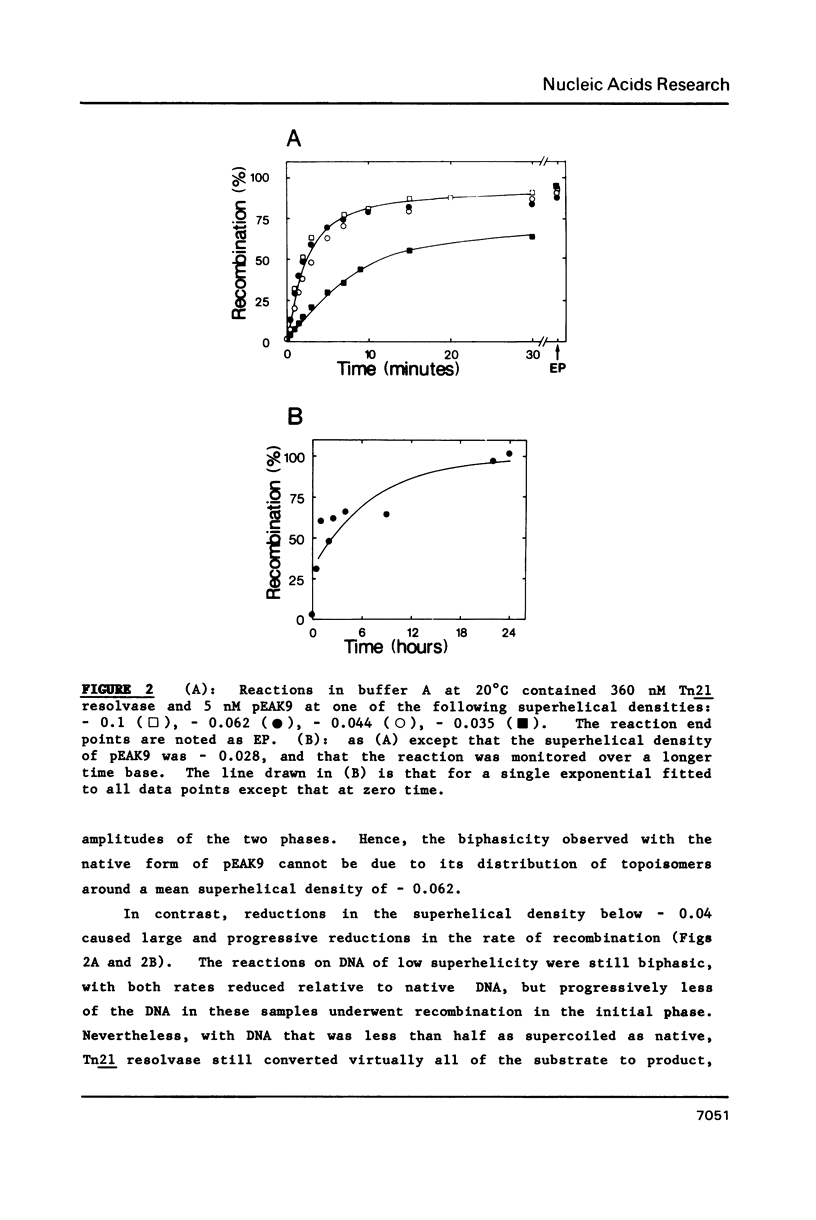
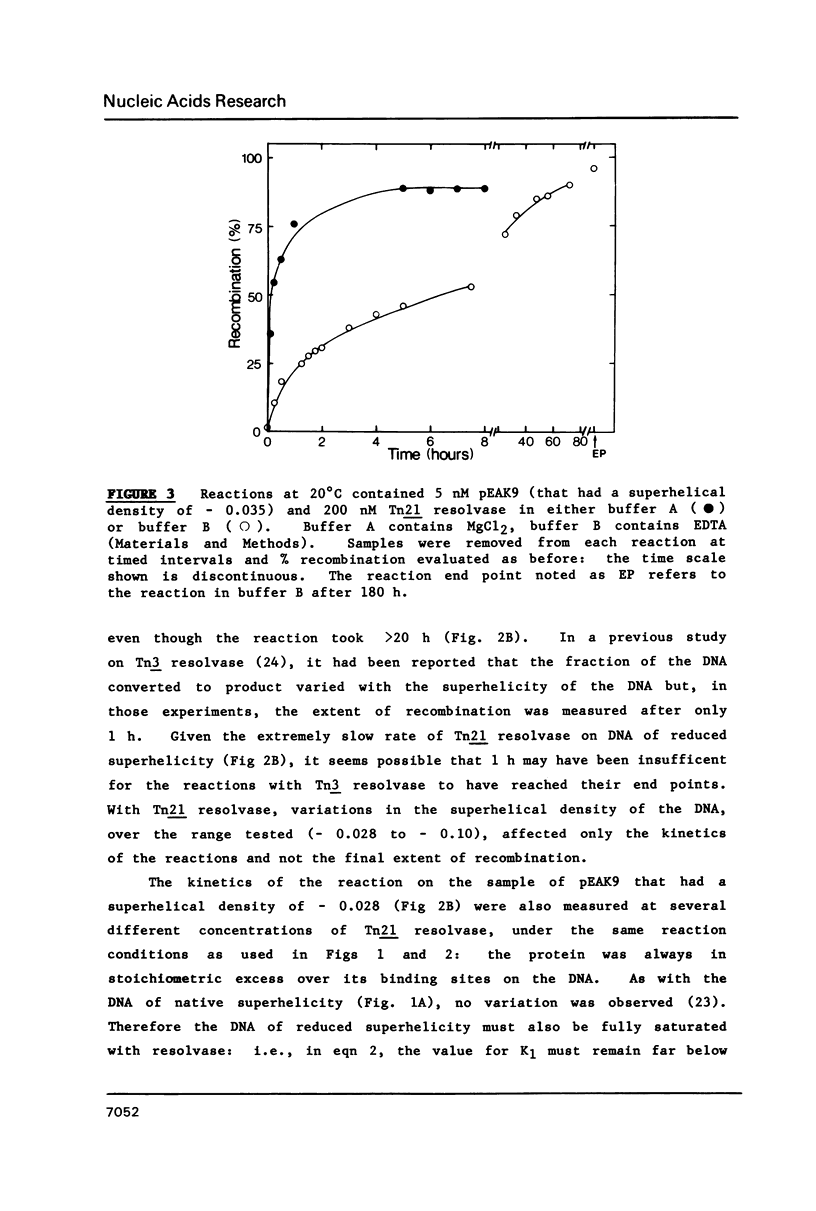
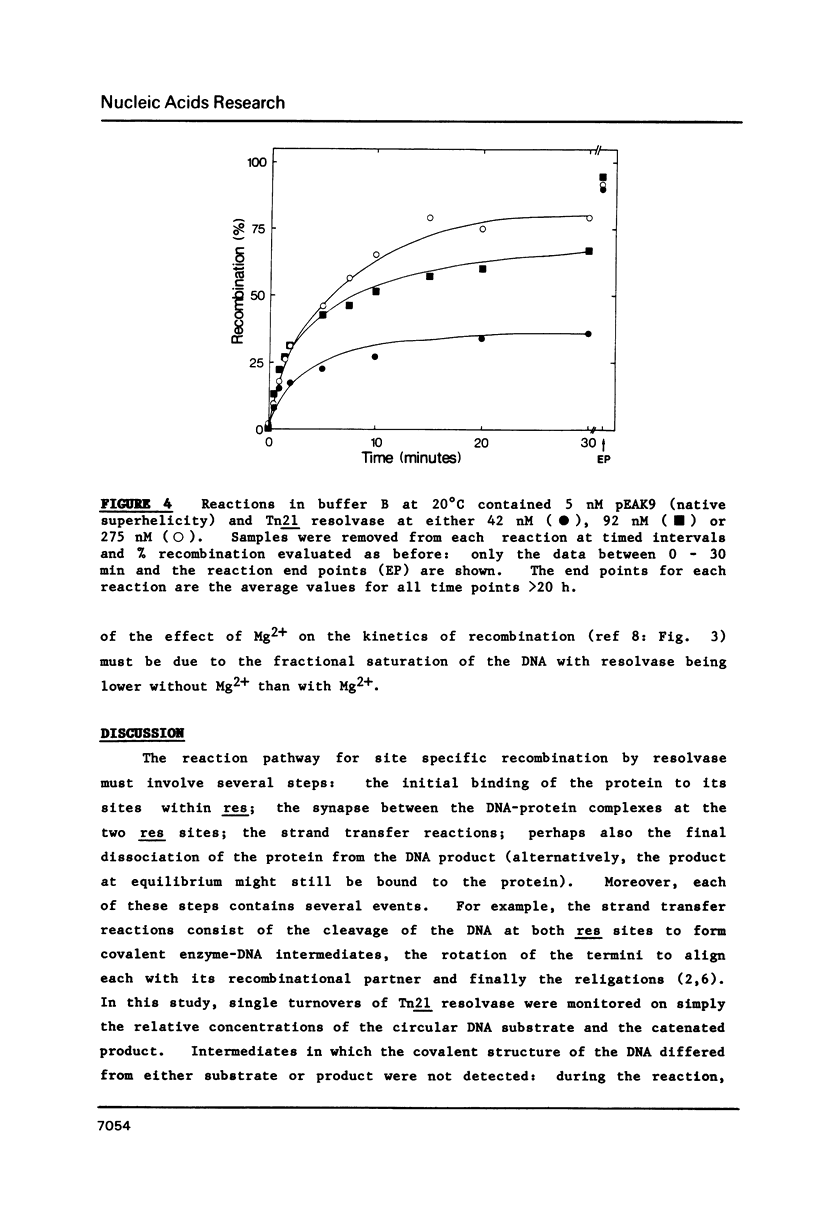
A kinetic analysis of site specific recombination by Tn21 resolvase has been carried out using DNA substrates of varying superhelicities. The rates for the formation of the recombinant product increased with increasing superhelicity up to a maximum value, after which further increases in superhelicity caused no further increase in rate. The reactions with DNA of reduced superhelicity were extremely slow, yet they eventually led to virtually all of the substrate being converted to product. Hence, the level of DNA superhelicity must determine the activation energy barrier for at least one of the steps within the reaction pathway that can be rate-limiting. In the presence (but not in the absence) of Mg2+ ions, the DNA was fully saturated with resolvase whenever the protein was in stoichiometric excess over resolvase binding sites on the DNA. Thus the process affected by DNA supercoiling cannot be coupled to the binding of resolvase. Instead, the step whose rate is determined by supercoiling seems to be located within the reaction pathway after the synapse. However, these reactions may involve two forms of the synaptic complex that are converted to the recombinant product at different rates.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P., Bauer W. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 1978 Feb 21;17(4):594–601. doi: 10.1021/bi00597a006. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Isolation and characterization of the Tn3 resolvase synaptic intermediate. EMBO J. 1988 Jun;7(6):1897–1905. doi: 10.1002/j.1460-2075.1988.tb03023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Matzuk M. M., Krasnow M. A., Cozzarelli N. R. Recombination site selection by Tn3 resolvase: topological tests of a tracking mechanism. Cell. 1985 Jan;40(1):147–158. doi: 10.1016/0092-8674(85)90318-6. [DOI] [PubMed] [Google Scholar]
- Bennett S. P., Halford S. E. Recognition of DNA by type II restriction enzymes. Curr Top Cell Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- Castell S. E., Jordan S. L., Halford S. E. Site-specific recombination and topoisomerization by Tn21 resolvase: role of metal ions. Nucleic Acids Res. 1986 Sep 25;14(18):7213–7226. doi: 10.1093/nar/14.18.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Hatfull G. F., Grindley N. D. Uncoupling of the recombination and topoisomerase activities of the gamma delta resolvase by a mutation at the crossover point. Nature. 1988 Apr 28;332(6167):861–863. doi: 10.1038/332861a0. [DOI] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P. Single turnovers of the EcoRI restriction endonuclease. Biochem J. 1983 May 1;211(2):405–415. doi: 10.1042/bj2110405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S. E., Jordan S. L., Kirkbride E. A. The resolvase protein from the transposon Tn21. Mol Gen Genet. 1985;200(1):169–175. doi: 10.1007/BF00383331. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. B., Glaccum M. B., Simon M. I. In vitro analysis of Hin-mediated site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:751–760. doi: 10.1101/sqb.1984.049.01.085. [DOI] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Luke P. A., Halford S. E. Solubility of the EcoRI restriction endonuclease and its purification from an over-producing strain. Gene. 1985;37(1-3):241–246. doi: 10.1016/0378-1119(85)90278-1. [DOI] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M. Integrative recombination of bacteriophage lambda: in vitro study of the intermolecular reaction. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1111–1114. doi: 10.1101/sqb.1979.043.01.123. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Kanaar R., van de Putte P. A genetic switch in vitro: DNA inversion by Gin protein of phage Mu. Proc Natl Acad Sci U S A. 1984 May;81(9):2689–2692. doi: 10.1073/pnas.81.9.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. The interaction of recombination proteins with supercoiled DNA: defining the role of supercoiling in lambda integrative recombination. Cell. 1986 Sep 26;46(7):1011–1021. doi: 10.1016/0092-8674(86)90700-2. [DOI] [PubMed] [Google Scholar]
- Rogowsky P., Halford S. E., Schmitt R. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 1985 Aug;4(8):2135–2141. doi: 10.1002/j.1460-2075.1985.tb03904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Trentham D. R., Eccleston J. F., Bagshaw C. R. Kinetic analysis of ATPase mechanisms. Q Rev Biophys. 1976 May;9(2):217–281. doi: 10.1017/s0033583500002419. [DOI] [PubMed] [Google Scholar]



