Abstract
Protein ubiquitination plays critical roles in the regulation of multiple cellular processes including cell proliferation, signal transduction, oncogenesis and hypoxic response. TS20 is a Balb3T3-derived cell line in which ubiquitination is inhibited by restrictive temperature. While TS20 has been used to elucidate the degradation of many important proteins including p53, p27, HIF-1α and ornithine decarboxylase, the molecular basis of its temperature sensitivity has not been fully determined. We cloned full-length E1 cDNA from TS20. Sequencing analysis revealed two point mutations (nt736G to A and nt2313G to C) that lead to substitution of aa189A to T and aa714W to C, respectively. Transient transfection assays revealed that mutant E1 was less stable than its wild-type counterpart, and restrictive temperature (39°C) accelerated its degradation. Under permissive temperature, reverting aa714C to W significantly improved E1 stability and activity. Under restrictive temperature, reverting of both substitutions was required to fully restore E1 stability. Similar results were observed when the mutants were expressed in non-TS20 cells, indicating the mutations are sufficient for its temperature sensitive degradation observed in TS20 cells. Functionally, reverting aa714C to W was sufficient to facilitate the monoubiquitination of H2A and to support TS20 growth at 39°C. It also significantly improved the ubiquitination-dependent disposal of HIF-1α. Our data conclusively demonstrate that mutations introgenic to UVBE1 cause E1 instability, which leads to deficiency of E1 function. Our data establish the molecular basis for unambiguous interpretation of experimental data based on TS20 cells, and provide new insight into the structural determinants of E1 stability.
Keywords: Ubiquitination, Proteasome, Protein degradation, Protein misfolding, HIF-1α, Protein quality control
Introduction
Covalent attachment of one or more ubiquitin (Ub) monomers to proteins expands proteomic diversity and provides a way to control protein levels and function. It has been well established that ubiquitination plays critical roles in modulating a variety of cellular processes, including cell cycle progression, immune responses, differentiation, signal transduction, protein trafficking and quality control (Schulman and Harper, 2009). The most prominent function of ubiquitination is manifested by ubiquitination-proteasome pathway for protein degradation within cells, where a target protein is sequentially labeled with Ub molecules, yielding a polyubiquitin chain for rapid proteolysis via the 26S proteasome (Schwartz and Ciechanover, 1999). The proteolytic role of polyubiquitination depends on the specific linkages between Ub monomers through amino acid residue 48-Lys. HIF-1α and P53 are among the best known targets of the U.P.S. Defects in this pathway have been associated with various diseases, such as cancers, diabetes and neurodegenerative disorders (Ciechanover, 2005; Goldberg, 2007).
In addition to regulating protein stability, ubiquitination also participates in a variety of non-proteolytic functions with the attachment of a single Ub molecule, also known as monoubiquitination, or the assembly of polyubiquitin chain through alternative lysyl residues within Ub. Monoubiquitination has been implicated in endocytic signaling, introcelluar vesicle dynamics, signal transduction, protein translocation, DNA replication, and transcription regulation (Hicke, 2001; Mukhopadhyay and Riezman, 2007; Terrell et al., 1998). Monoubiquitination of H2A, for example, has been associated with DNA replication and cell proliferation (Wang et al., 2004). Polyubiquitination through Lys63 linkage has been implicated in signaling activation, receptor endocytosis, intracellular protein trafficking and DNA damage repair (Elsasser and Finley, 2005; Friedberg et al., 2005; Galan and Haguenauer-Tsapis, 1997; Pickart and Fushman, 2004).
Ubiquitination processes are mediated sequentially through Ub-activating (E1), Ub-conjugating (E2), and Ub-protein ligase (E3) multienzyme cascades (Hershko and Ciechanover, 1998; Hershko et al., 1983). In mammalian cells, there are dozens of E2s, and several hundred E3s. However, there is a single E1 enzyme for the entire array of downstream reactions. Ub is first activated by E1, forming a high energy thioester bond with the E1. The E1-Ub thioester complex is then recognized by an E2, and the thioester-linked Ub is transferred to E2. Finally, E2-Ub thioester complex uses the energy stored in the E2-Ub thioester bond to ubiquitinate target proteins, either directly or through substrate specific E3s complex (Haas and Rose, 1982). Therefore, by catalyzing the initial step of ubiquitination E1 plays an essential role in all ubiquitination processes.
TS20 is a Balb3T3-derived temperature sensitive cell line which arrests at S phase of the cell cycle with dramatically reduced protein ubiquitination at restrictive temperature (39°C) (Chowdary et al., 1994; Zeng et al., 1984). Previous study showed that shifting TS20 cells from 35°C to 39°C caused rapid degradation of E1 proteins (Chowdary et al., 1994; Salvat et al., 2000). Since exogenous expression of human E1 restored normal ubiquitination and rescued TS20 cell growth at 39°C, TS20 has been assumed to carry a temperature sensitive E1 (Chowdary et al., 1994). Since its establishment, this cell line has contributed to the elucidating of the degradation pathways for many important proteins. Based on studies with TS20 cells, the turnover of p53 and HIF-1α, for example, were demonstrated to depend on the ubiquitination-proteasome pathway (Chowdary et al., 1994; Salceda and Caro, 1997). In recent years, ubiquitination-independent proteasomal degradation pathway and its important roles have been discovered (Asher et al., 2002; Kong et al., 2006; Liang et al., 2006; Rosenberg-Hasson et al., 1989; Shirane et al., 1999). Since complete lack of E1 activity leads to cell death, the temperature sensitive TS20 cell line provides a particularly important tool to study the ubiquitination-independent degradation pathway. However, the molecular nature of the mutation in TS20 cell has not been determined. The complexity of biological system and lack of direct evidence of E1 mutation complicate the interpretation of experimental observations based on this cell line. Among several alternative explanations, one example is that the temperature sensitive phenotype could be caused by a mutation extragenic to E1 gene (UBA1 or UBE1), but regulates stability of proteins including E1.
We here report the identification of two point mutations on E1 cDNA isolated from this cell line, and provide conclusive evidence to support that those two mutations are sufficient to cause temperature sensitive phenotype. We further show that reverting of these mutations is sufficient to correct the temperature sensitive phenotype. Our study provides the molecular basis for unambiguous interpretation of experimental data based on TS20 cells.
Materials and Methods
Cell culture
Balb/c 3T3 mouse embryo fibroblast derived TS20 cells were kindly provided by Dr. H. Ozer (UMDNJ, New Jersey). TS20 and HEK 293T lines were maintained in DMEM medium supplemented with 10% fetal bovine serum, L-glutamine (Invitrogen), and mixture of other amino acids (Mediatech). TS20 cells were maintained at 35°C (permissive temperature).
Cloning, sequencing and site-directed mutagenesis
Total RNA was isolated from TS20 cells by using RNAeasy Mini kit (Qiagen). E1 whole length cDNA was synthesized by reverse transcription and further PCR-amplified by E1 specific primers (5′-GGAATTCCAATGTCCAGCTCGCCGCTGTCC-3′ and 5′-CCGCTCGAGTCAGCGAATGGTATATCGGAC-3′, EcoRI and XhoI sites were underlined respectively). The PCR product which was first cloned into pGEM T-Easy vector (Promega), sequenced and compared with wild-type sequenced in database (GeneBank). The E1 cDNA containing two mutations in the pGEM T-Easy vector was digested with Eco RI and Xho I and inserted into pCDNA3-FLAG to generate pflag-E1189T, 714C in which E1189T, 714C is expressed as a fusion with Flag.
To revert aa189T to A in E1, site-directed mutagenesis PCR was performed by using primers (Forward: 5′-GTATCAAGCTAGTGGTGGCAGATACAAGAGGCCTG-3′; Reverse: 5′-CAGGCCTCTTGTATCTGCCACCACTAGCTTGATAC-3′). Similarly, primers (Forward: 5′-CCTGCCACCACTGGCACACCCAGTACT-3′; Reverse: 5′-AGTACTGGGTGTGCCAGTGGTGGCAGG-3′) were used to revert aa714C to W. Prior to transformation, the resulting PCR product was digested with Dpn I to remove template pflag-E1189T, 714C plasmid DNA. The mutant plasmids expressing flag-E1714C, flag-E1189T or flag-E1wt which was derived from a double mutation of E1189T, 714C, were confirmed by DNA sequencing.
Plasmid preparation and transfection
Plasmid DNA used for transfection was isolated by using a Maxiprep kit (Qiagen). Cells were transfected using Lipofectamine 2000 (Invitrogen) by following the manufacturer’s instructions. Cells were pre-plated in 100-mm plate the day before transfection. When cells reach about 90% confluence the next day, 6 μg (for TS20 cells) or 10 μg (for 293T cells) of plasmid DNA were mixed with 18 μl or 30 μl Lipofectamine 2000, respectively, and added to the cells. Cells were trypsinized 24 h after transfection, divided equally, and cultured in 100-mm plates at indicated temperatures.
Antibodies, cell lysate preparation, and western blotting
Mouse anti-Flag and anti-α-tubulin antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Rabbit anti-E1, anti-HIF-1α, anti-C-terminal Ubiquitin and anti-Ubiquitin antibodies were from Cell Signaling (Beverly, MA), Novus Biologicals (Littleton, CO), Epitomics (Burlingame, CA) and Enzo Life Sciences (Plymouth Meeting, PA), respectively. Horseradish peroxidase-coupled secondary antibodies were from Sigma-Aldrich (St. Louis, MO) and Invitrogen (Carlsbad, CA).
For Western blot analyses, cells were lysed in urea buffer (8 M urea, 10 mM Tris, 10% glycerol, 1% SDS, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor mix, pH 6.8) on ice with an Ultra-Turrax T8 homogenizer (IKA GmbH & Co.) for 60 s. Proteins in the lysates were separated on a 4–20% gradient SDS-PAGE (Bio-Rad), and then electrotransferred onto a polyvinylidene difluoride membrane. The membrane was processed in subsequent steps with blocking with 5% milk in TBST, incubation with specific primary antibody, washing in TBST and incubation with horseradish peroxidase-labeled secondary antibody. The membranes were finally developed with the ECL Plus system (Amersham Biosciences).
Immunoprecipitation assays
TS20 cells (3×106 cells per 10 cm plate, 2 plates) were transfected with a total of 14 μg of each pflag-E1 constructs. After 24 h, cells were consolidated and redistributed into 4 plates. After 20 hr, 2 plates were cultured at 35 and 2 plate were cutlured at 39 for 6 h. Cells were lysed with IP buffer (50 mM Tris-HCl, 300 mM NaCl, 1% triton-×-100, 5 mM EDTA, 50 mM NaF, Na3VO4 and Protease Inhibitor Cocktail, (Roche Applied Science, Indianapolis, IN)). Flag-E1 proteins were immunoprecipitated by using anti-flag antibody and protein G agarose gel (Thermo Fisher Scientific, Rockford, IL), followed by immunoblot assay with an antibody against E1.
In vitro transcription-translation and protein stability analysis
In vitro E1 protein translation was achieved by using TNT T7-Coupled Wheat Germ Extract System (Promega) by following the manufacturer’s instructions. 1 μg of each E1 constructs were added to the reaction mixture to generate mRNA, and the translation was carried out in the mixture with [35S] methionine at 30°C for 1 h. The translation products were incubated with TS20 cell lysate for 1, 2, and 4 hours, respectively. The turnover of E1 proteins was compared by quantification of [35S] methionine-labeled polypeptides following SDS-polyacrylamide gel electrophoresis separation.
Establishment of stable cell lines
TS20 cells were transfected with pflag-E1189T or pflag-E1wt constructs using the method provided above. After culturing at 39°C for 3 weeks, survived clones from each transfection were selected and expanded as stable cell lines, named E1189T and E1wt cells, respectively.
Cell proliferation analysis
Cells were plated in triplicate in 96-well plates at a density of 1×104 cells/well. Cell numbers were examined after 1, 3, 6 and 7 days using the CyQUANT® NF-Cell Proliferation Kit (Invitrogen) with reference to standard curves obtained from each of the tested cell line.
Fluorescent staining and microscope
TS20, E1189T and E1wt cells were plated in 8-well chamber slides (BD Biosciences). Cells were incubated at 35°C or 39°C for 18 and 24 hours, respectively. Viability and induction of cell death were examined by using apoptosis/necrosis detection kit (Enzo) where cells was stained with annexin V-Cyanine-3/7-AAD according to the manufacturer’s recommendations. Cells were visualized by confocal microscopy (Fluoview 1000; Olympus).
Results
Identification of two missense mutations in E1 isolated from TS20 cells
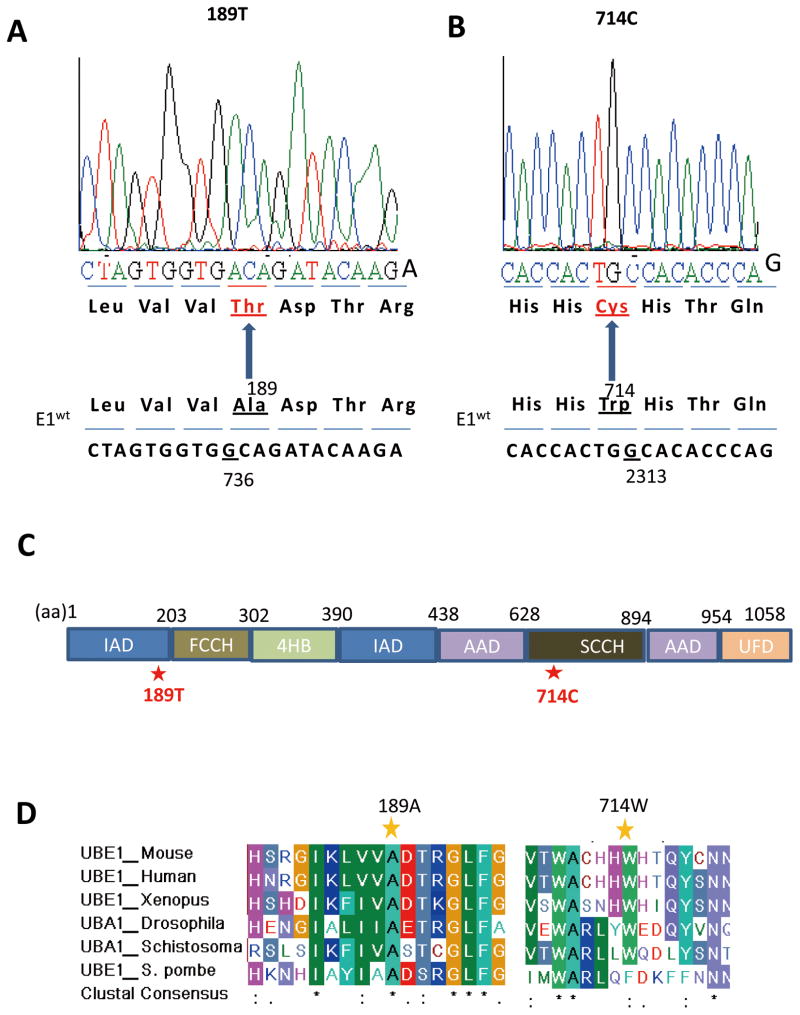
It has been shown that transfection of human E1 gene rescued TS20 cells at 39°C (Chowdary et al., 1994), suggesting that the temperature-sensitive phenotype might be caused by the defect of endogenous E1 function. However, it is not clear whether the temperature sensitive phenotype was caused directly by E1 mutation, or by a mutation extragenic to E1 gene which affected the stability and function of murine E1 protein. To precisely define the genetic defect that impairs E1 function in TS20 cells, we cloned E1 cDNA from TS20 cells by RT-PCR. Sequencing analysis showed two missense mutations at nt736 and nt2313 (NCBI Reference Sequence: NM_001136085.1), causing two amino acid substitutions (from aa189A to T and aa714W to C, respectively) (Fig. 1A and B). These two mutations have been confirmed by repeated RT-PCR and sequencing analysis of independently isolated cDNA clones. In addition, sequencing analysis of genomic DNA isolated from TS20 cells confirmed the mutations in one allele of E1 gene. Thus, these two mutations unlikely represent artifacts caused by the RT-PCR cloning procedures. We noted that all cDNA clones sequenced showed the same mutations, consistent with the established notion that due to X-chromosomal inactivation the other allele of E1 gene is not transcribed.
Fig. 1. Identification of two missense mutations in E1 from TS20 cells.
A and B: Several independently isolated E1 cDNA clones derived from TS20 cells were sequenced. Two point mutations, nt736A to G and nt2313C to G were identified, which change aa189T to A and aa714C to W, respectively. Representative sequencing data were shown. C: The schematic structure of E1 protein showed. E1 protein consists of four functional domains: adenylation domains is composed of two sub-domains inactive and active andenylation domain, IAD and AAD, respectively; catalytic cysteine domains is composed of two half-domains named first and second catalytic cysteine half-domain, FCCH and SCCH, respectively; a four-helix bundle (4HB); and C-terminal ubiquitin-fold domain (UFD). D: Protein sequences of mouse E1 were aligned with that of other species. Results reveal that both aa189A and aa714W residues are highly conserved among evolutionarily distant species. Alignments were generated using BioEdit.
Structurally E1 protein consists of four functional domains (Fig. 1C): adenylation domains, which is composed of two sub-domains named inactive and active andenylation domains (IAD and AAD, respectively); two catalytic cysteine half-domains which are named first and second catalytic cysteine half-domains (FCCH and SCCH, respectively); a four-helix bundle (4HB) domain; and C-terminal ubiquitin-fold domain (UFD)(Lake et al., 2001; Lee and Schindelin, 2008; Lois and Lima, 2005; Szczepanowski et al., 2005; Walden et al., 2003). Specifically, the A to T substitution lies within the IAD, and the W to C substitution lies within the second catalytic cysteine half-domain (Fig. 1C). Both amino acid residues are highly conserved among species range from invertebrates to vertebrates (Fig. 1D), underlying their biological importance.
The 714W to C substitution reduces E1 stability
To determine whether these two substitutions directly cause the temperature sensitive phenotype of TS20 cells, we performed site-directed mutagenesis to revert these two mutations. The reverted E1 constructs are named as pflag-E1714C, pflag-E1189T and pflag-E1wt.
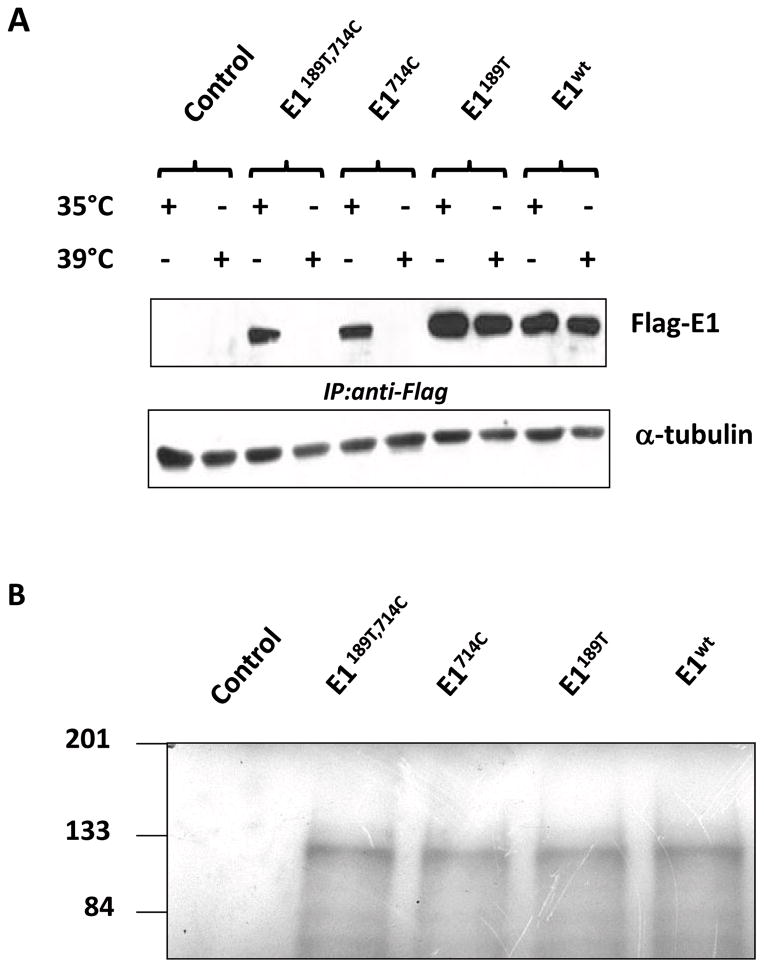
We then examined the expression levels of flag-E1 proteins in TS20 cells at permissive (35°C) or restrictive temperature (39°C for 6 h) in cultured cells. At 35°C, all four flag-tagged E1 proteins (pflag-E1189T, 714C, pflag-E1714C, pflag-E1189T and pflag-E1WT) were readily detected (Fig. 2A). However, shifting cells to 39°C remarkably reduced the protein levels of E1189T, 714C and E1714C. When cells were cultured at 39°C, protein level of E1189T was also reduced while E1wt protein remained at similar level (Fig. 2A). Reverse-transcription-RCR assays indicated all constructs are transcribed in cells, and their mRNA levels were generally comparable (data not shown). We also tested the in vitro translation capability of those constructs by using wheat germ extract transcription/translation system. In this system, all constructs were transcribed and translated at similar efficiency (Fig. 2B). Taken together, these data showed that 714W to C substitution is the major cause of reduced stability of E1 in TS20 cells, whereas 189A to T substitution plays a secondary role.
Fig. 2. 714W to C substitution affects E1 stability in TS20 cells.
A: Four E1 constructs: pflag-E1189T, 714C pflag-E1714C, pflag-E1189T, and pflag-E1wt together with empty control vector were transfected in TS20 cells, and each transfection was split into two equal plates 24 h after transfection. At 44 h post transfection, one plate was shifted to 39°C for 8 h. Cell lysates were collected. Since the transient expression levels of mutant E1 in TS20 cells was extremely low, E1 proteins were enriched by immunoprecipitation using anti-flag antibody and detected using anti-UBE1 antibody. Aliquotes of cell lysates (10%) were used for normalization whereα-tubulin was detected by anti-α-tubulin antibody. B: All four E1 constructs were in vitro transcribed and translated by using TNT T7-coupled Wheat germ extract system. Proteins levels of E1, indicated by incorporated [35S] methionine, were detected by autoradiography after resolving on SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Mutation triggers proteasome–dependent degradation of E1 proteins
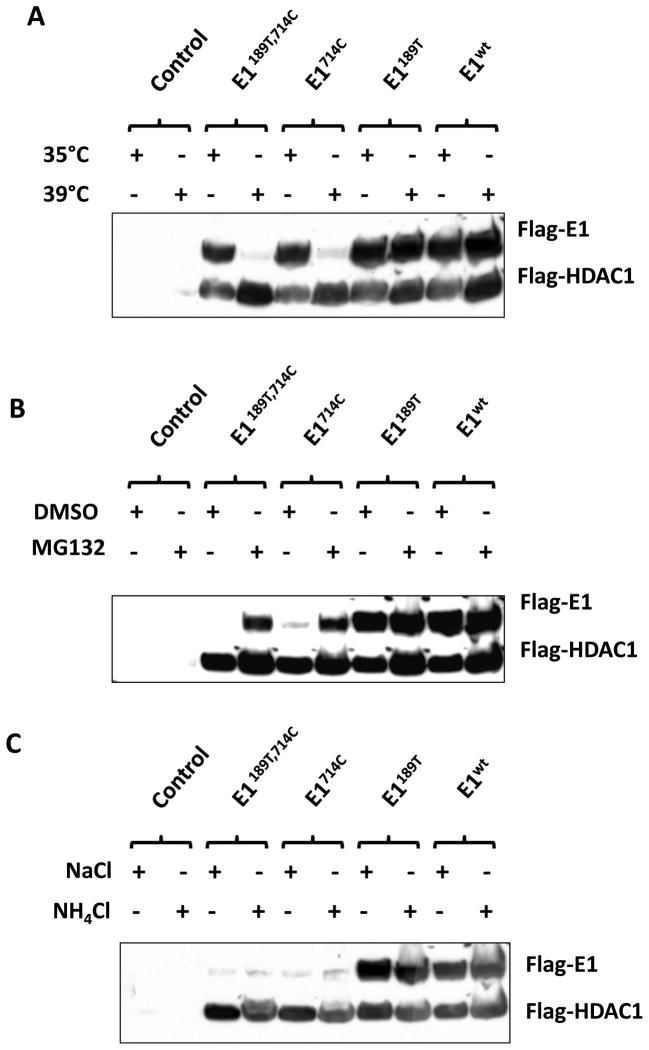
To explore if the identified E1 mutations were sufficient to cause E1 instability and to exclude any involvement of other genetic alteration extragenic to E1, we tested the stability of those constructs in HEK293T cells. Similarly, the expression levels of E1 mutants with W to C substitution were remarkably reduced when cultured at 39°C (Fig. 3A).
Fig. 3. E1 mutations are sufficient to trigger proteasome–dependent degradation.
A: Four E1 constructs: pflag-E1189T, 714C pflag-E1714C, pflag-E1189T, and pflag-E1WT were transfected into 293 cells. Plasmid pflag-HDAC1 was co-transfected as a control for transfection efficiency. Each transfected dish was split into two equal plates 24 h after transfection. At 44 h post transfection, one dish was shifted to 39°C for 8 h. Transiently expressed flag-E1 and flag-HDAC1 proteins were detected by using anti-flag antibody. B: Transiently transfected cells were cultured at 39°C for 8 h with either DMSO (control) or proteasome inhibitor MG132. Protein levels of Flag-E1 were examined by western blotting. C: Transiently transfected cells were cultured at 39°C for 8 h with either NaCl (control) or lysosome inhibitor NH4Cl. Protein levels of flag-E1 were examined by Western blotting.
In living cells, proteins are commonly degraded by either ubiquitin-proteosomal system or the lysosomal system. To examine which cellular mechanism is involved in the temperature-triggered degradation of E1 mutant proteins, we treated cells with proteasome inhibitor MG132 or lysosome inhibitor ammonia chloride at 39°C. Data shown in Fig. 3B indicated that the inhibition of proteasome activity restored the level of mutant E1 proteins. However, inhibition of lysosome function showed no effect on E1 levels (Fig. 3C). These data demonstrated that W to C substitution triggers a proteasome-dependent degradation of the mutant E1.
Reverting mutations rescue TS20 proliferation at restrictive temperature
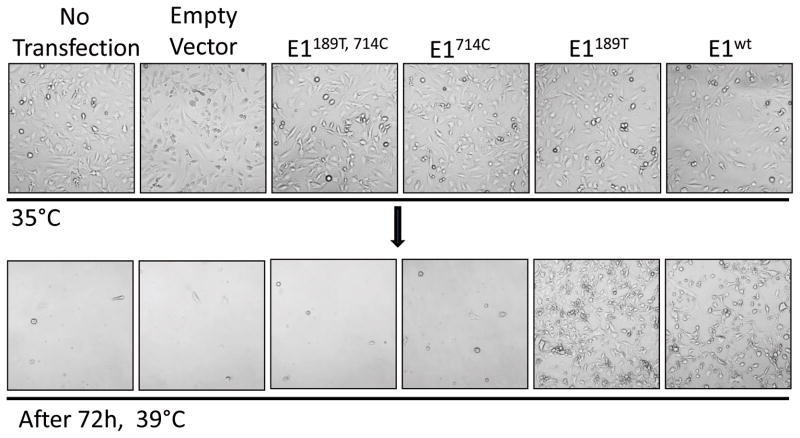
Restrictive temperature inactivates E1 and causes cell death and growth arrest. We next examined whether expression of E1 revertants was able to rescue TS20 growth at 39°C. We transfected TS20 cells to express the wild-type and mutant E1 proteins. Two days after transfection, cells were shifted from 35°C to 39°C and incubated for another 3 days. We observed that cells transfected with wild-type E1 (E1wt) or 714C to W revertant E1 (E1189T) survived and continued proliferating. However, TS20 cells over expressing either E1714C or E1189T, 714C retained their temperature sensitive phenotype and no cells survived at 39°C after three days (Fig. 4A). Together, we conclude that the 714W to C substitution is the major determinant of the temperature sensitive phenotype.
Fig. 4. Reverting 714C to W rescues TS20 survival and proliferation at restrictive temperature.
TS20 cells were transiently transfected with each of the E1 constructs to overexpress the corresponding E1 proteins. Two days after transfection, cells were shifted from permissive temperature to 39°C, and incubate for another 3 days. Cell morphology was photographed immediately before, or after 72 h incubating at 39°C.
Reverting aa714C to W restore E1 function and cell proliferation
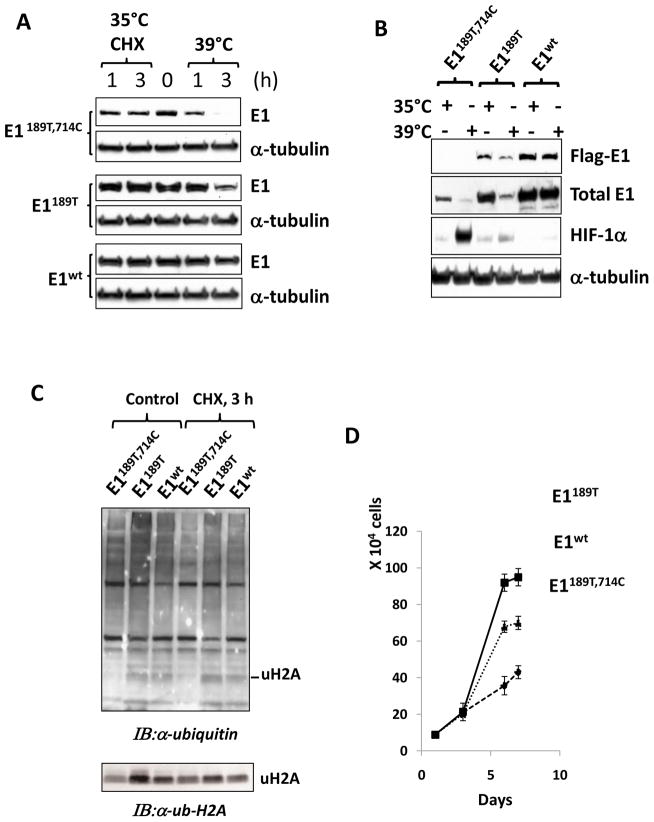
To examine whether reverting aa714C to W restore E1 function in facilitating the ubiquitin-proteasome pathway, we established two cell lines expressing mutant E1 by transfecting TS20 cells first at 35°C followed by selecting survived colonies at 39°C. After incubation at 39°C for 4 weeks, colonies were formed from cells transfected with either E1189T or wild-type E1. Transfection of either E1714C or E1189T, 714C failed to establish any colony, again suggesting 714C mutation is the critical contributor to the temperature sensitive phenotype. Next, established individual colonies for E1189T and wild-type E1 were expanded and analyzed for E1 stability and activity. We observed that the protein levels of E1189T in stable cell lines were slightly affected by incubating at 39°C (Fig. 5A), suggesting that the 189T mutation also causes E1 instability, but it is less severe than 714C.
Fig. 5. Stable expression of 714C to W revertant restores E1 function.
A: Two newly established cell lines E1189T and E1wt together with their TS20 cell line, which contains E1189T,714C mutations, were analyzed. Cells were culture at permissive 35°C or restrictive 39°C for 1 or 3 h. Cycloheximide (Sigma) were used to block de novo translation and test the protein stability. Total E1 protein levels were detected by using an anti-UBE1 antibody. α-tubulin was detected as a loading control by using an anti-α-tubulin antibody. Note that in stable transfected cell lines, the exogenous E1 represent the major part of total E1. B: The three cell lines used in A were cultured at both temperature and restrictive temperature points. Flag-E1 and total E1 protein levels were detected using anti-flag and anti-UBE1 antibodies, respectively. Ubiquitin proteasome pathway targeted protein, HIF-1α level was determined as an indicator of the polyubiquitination-proteasomal function while α-tubulin levels were examined as a loading control. C: Ubiquitination profile in the three cell lines were examined by using anti-ubiquitin antibody, and a band representing monoubiquitinated H2A (uH2A) was indicated. Its identity was further confirmed by anti-Ub-H2A specific antibody. D: The proliferation rates of the three cell lines were assayed at permissive temperature. Cell numbers (determined by DNA contents) were determined at days 0, 1, 3, 6 and 7. Standard errors were indicated.
One of the biological consequences of TS20 cells at restrictive temperature was the accumulation of short-life proteins, such as HIF-1α and p53, whose levels are mainly regulated through the ubiquitination-proteasome pathway (Chowdary et al., 1994; Salceda and Caro, 1997). To test whether the stable cell lines have sufficient E1 activity to support the polyubiquitination and the degradation of short-life proteins at 39°C, we examined HIF-1α levels in those cell lines under different culture conditions. Consistent with previous reports (Salceda and Caro, 1997), TS20 cells accumulated HIF-1α when cultured at 39°C, and cells stably expressing E1wt cells didn’t accumulate HIF-1α at either 35°C or 39°C (Fig. 5B), suggesting expressing of wild-type E1 recovered sufficient E1 activity to fully support the polyubiquitination and degradation of HIF-1α under the normoxic condition, a normal cellular function essential for oxygen sensing and homeostasis (Semenza et al., 1994; Semenza and Wang, 1992). At 39°C, the level of HIF-1α slightly increased in E1189T cells, suggesting that E1189T improved the polyubiquitination of HIF-1α, but it is less effective than WT.
In addition to polyubiquitination, monoubiquitination of proteins also requires E1 activity. H2A has been used previously as a marker in studying E1 function (Wang et al., 2004). To examine the ability of E1 constructs in supporting monubiquitination of H2A, we performed ubiquitination profiling analysis using anti-ubiquitin antibody. Our data revealed that cells expressing either E1wt or E1189T possessed significantly increased levels of monoubiquitined H2A (Wang et al., 2004), which was further confirmed by Western blotting by using anti-ubiquitinated H2A antibody (Fig. 5C).
E1-dependent ubiquitination-proteasome system controls some major events in cell cycle progression, and it has been reported that TS20 cells were arrested at S1 phase shortly after shifting to 39°C (Zeng et al., 1985). Anaphase promoting complex-dependent events, for example, requires polyubiquitination of protein regulators demised to degradation. In addition, H2A monoubiquitination plays a critical role in chromosome replication and cell cycle progression as well. We next asked how cell proliferating rate is related to E1 activity conferred by the mutant constructs. We compared the growth curves of these cell lines with TS20 cells and observed that cells expressing either E1189T or E1wt grew faster than the parental TS20 cell line (Fig. 5D), suggesting that insufficient level of E1 activity may affect the cell proliferation rate.
Defects in E1 cause apoptotic death of TS20 cells
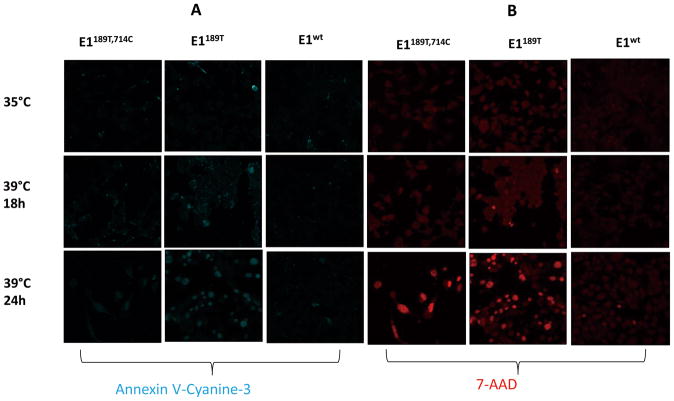
Cell death may be caused by either apoptotic or necrotic pathways. To determine which death pathway is involved in TS20 cells at restrictive temperature, we examined morphologic evidence of apoptosis and necrosis. The binding of annexin V with phosphatidylserine on the extracellular face of the plasma membrane is a hallmark of early apoptosis. DNA intercalating dye 7-AAD labeled nuclei represents late stage apoptosis and necrosis. Using annexin V and 7-AAD staining, we confirmed that restrictive temperature caused both apoptosis and necrosis of parental TS20 cells (Fig. 6A, B). Cells expressing E1wt showed resistance to apoptosis or necrosis caused by incubating at 39°C. Cells expressing E1189T could rapidly proliferate at 35°C and survive at restrictive temperature. However, when incubated at 39°C, cells expressing E1189T showed increased apoptotic rate (Fig. 6A, B).
Fig. 6. Defect in E1 causes apoptotic cell death of TS20 at restrictive temperature.
A: Three stable cell lines were tested for apoptosis under restrictive temperature. Apoptosis detection reagent Annexin V-Cyanine-3 (blue) was used to identify apoptotic cells. B: Similar to A, three stable cell lines were tested for necrosis under restrictive temperature. Reagent 7-AAD (red) was used to detect cells undergoing necrosis or late phase of apoptosis.
Stabilization of mutant E1 is not sufficient to rescue TS20 survival at restrictive temperature
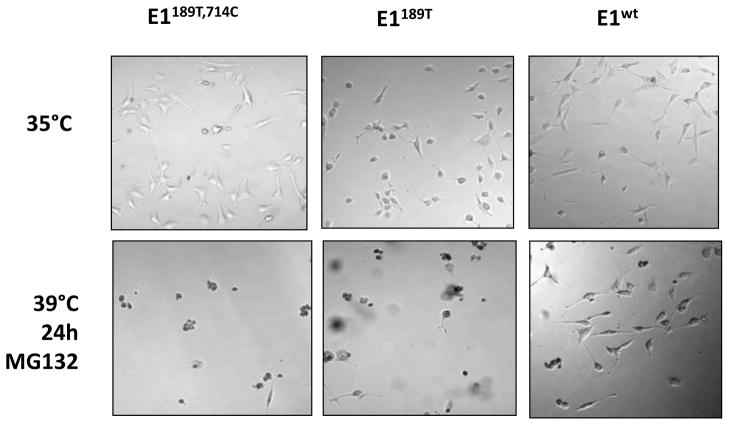
We have shown that the degradation of E1 mutants is proteasomal-dependent, and inhibition of proteasome restored mutant E1 levels at restrictive temperature. It has been well established that restrictive temperature causes cell death of TS20 cells. We asked whether stabilizing mutant E1 by inhibiting its proteasomal degradation would rescue the temperature sensitive phenotype. To address that, we treated E1189T, 714C, E1714C and E1wt cells with proteasomal inhibitor MG132 for 24 h at 39°C. Cell morphology was imaged before or after treatment. Our data indicated that inhibition of proteosomal degradation had a devastating effect on the viability of all three cell lines (Fig. 7). Therefore, restore of mutant E1 levels or E1 activity is not sufficient to rescue cell survival, suggesting that proteosomal degradation of certain types of proteins is required for cell survival and proliferation, and disfunction of ubiquitination-proteosomal degradation system plays a critical role in the temperature sensitive phenotype.
Fig. 7. Stabilizing E1 mutant is not sufficient to rescue the temperature sensitive phenotype.
Three stable cell lines were cultured at restrictive temperature for 24 h with proteasome inhibitor MG132. Cells were imaged using a light microscope before or after incubation at 39°C for 24 h.
Discussion
In this study, we identified two point mutations in E1 transcripts isolated from the temperature sensitive TS20 cells, which lead to aa189A to T and aa714W to C substitutions in deduced E1 protein. Characterization of these mutations reveals that aa714W to C substitution is the major cause of E1 instability. In addition, aa189A to T mutation also affects E1 stability at restrictive temperature. Inhibition of proteasome activity increased the protein levels of E1 mutants while inhibition of lysosomal protease activity showed no effect. Our data demonstrate that E1 mutations are the bona fide cause of the temperature sensitive phenotype of TS20 cells, which is mediated by a proteasome-dependent degradation of mutant E1, and does not require any other genetic alteration extragenic to UBA1 (UBE1) gene.
Previously, we reported that certain chemotherapeutic drugs triggered ubiquitination-independent, but proteasome-dependent degradation of HIF-1α, partly based on data obtained from TS20 cells (Kong et al., 2006). Because of lack of direct evidence of E1 mutation, one major concern raised by other researchers was that the complexity of biological system may provide alternative explanations for E1 inactivation at restrictive temperature. Specifically, it is equally possible a mutation extragenic to the E1 gene may be responsible for the degradation of mutant E1 and HIF-1α. The same concern may be raised for ubiquitination-independent degradation of other proteins demonstrated with TS20 or other cell lines with similar temperature sensitive E1 function. Our data presented here unambiguously validated that E1 mutations are the bona fide cause of the temperature sensitive phenotype of TS20 cell line, thus validating the ubiquitination-independent protein degradation observed in this cell line.
The precise mechanism underlying the temperature-triggered degradation of mutant E1 remains unclear. One possibility is that mutations in E1 increase E1 misfolding, which gives rise to structurally unstable E1 proteins. When cultured at permissive temperature, folding defects may be recognized by protein chaperone complexes, such as heat shock protein 70 and 90 complexes, which facilitate mutant protein stability by refolding or physically interacting with the mutant proteins. Restrictive temperature leads to extensive misfolding of E1 and other cellular proteins, which may eventually overwhelm the folding capacity of cellular chaperone system. Thus, the misfolded E1 is eventually degraded by proteasome. Particularly, the 714W to C mutation turns out to be the major contributor to the instability of mutant E1. As shown in Fig. 1, the 714W to C substitution is located at the catalytic domain with multiple cysteine residues. It is possible that the aberrant cysteine residue may interfere with the folding processes of other sulfhydryl groups at the E1 protein.
An intriguing question is whether the temperature-triggered degradation of mutant E1 itself depends on ubiquitination. While our data support that under restrictive temperature, proteasome activity is required to degrade mutant E1, it is also assumed that under such conditions ubiquitination activity as a whole is generally very limited. One possible explanation is that mutant E1 proteins are preferred substrates for ubiquitination, so even very low residual levels of E1 activity would be sufficient to cause its own ubiquitination. Our data showed that in the presence of MG132, the accumulated mutant E1 proteins did not have an apparent increase in molecular weight, a change caused by ubiquitination. In addition, those E1 mutant proteins also failed to be recognized by anti-ubiquitin antibody in Western blotting. Taken together, those observations may suggest that under restricted temperature, the degradation of mutant E1 proteins are unlikely dependent of ubiquitination. An alternative possibility is that proteasome-mediated degradation of mutant E1 involves the ubiquitination-independent pathway, which has been reported for the degradation of p53, p27, ODC and HIF-1α under certain conditions (Asher et al., 2002; Kong et al., 2006; Rosenberg-Hasson et al., 1989; Shirane et al., 1999). If it is true, the ubiquitination-independent degradation of misfolded E1 may be considered a quality control mechanism of E1 protein.
We also noticed that even under permissive temperature, mutant E1 proteins carrying aa714C are expressed at low levels. Their levels could not be enhanced by exposing cells to proteasome inhibitor MG132 (data not shown), suggesting the existence of other non-proteosomal mechanism that affects mutant E1 expression under this condition. In addition, in vitro translated E1 proteins, both mutant and wild-type forms, are rapidly degraded (data not shown). However, when living cells were analyzed, wild-type E1 obviously has a relatively longer biological half life than in vitro (Fig. 5A), suggesting living cells have a mechanism to actively protect wild-type E1 protein. We are currently investigating if this protective mechanism involves the molecular chaperone systems that facilitate E1 folding.
A comparison of the stable cell lines expressing E1189T or E1wt with TS20 at permissive temperature revealed that both E1189T and E1wt were stable, and the global ubiquitination levels were similar. However, the monoubiquitination level of H2A in TS20 was much lower than that of E1189T and E1wt cells (Fig. 5C), suggesting H2A ubiquitination is more sensitive to E1 insufficiency. E1189T increased the level of monoubiquitinated H2A and promoted cell proliferation rate as wild-type E1 did (Fig. 5C, D). However, repeated experiments indicated that E1189T was less effective than the wild-type E1 in supporting the polyubiquitnation of HIF-1α (Fig. 5B). In addition, cells stably expressing E1189T have a higher apoptotic rate than cells expressing E1wt (Fig. 6). We noticed that E1 functions at the initial step of multiple ubiquitination pathways. Monoubiquitinated H2A (uH2A) has been linked to chromosome replication and cell proliferation (Vassilev et al., 1995). Functionally, it interacts with dozens of E2s and hundreds of E3s. Mutation in E1189T may affect its binding affinity with different E2s or E3s. The H2A E3 ligases Ring1B and RNF8 have been demonstrated to play essential roles in cell proliferation (Plans et al., 2008; Tuttle et al., 2007). These data suggests a possibility that aa189A to T mutation may have a subtle defect altering its substrate specificity in the ubiquitination processes.
It is important to note that the single E1 enzyme is essential for the entire array of downstream ubiquitination reactions. Loss of E1 level in mutant cells at 39°C will result in disfunction of both polyubiquitination-mediated protein degradation processes and monoubiquitination-regulated protein function. Considering the scope of proteins will be affected by E1-insufficiency, it would be difficult to assume one or two protein substrates of ubiquitination are responsible for the temperature sensitive phenotype. In this study, we picked HIF-1α as one example of the UPS substrates, and H2A as an example for the monoubiquitination substrates. Obviously, neither HIF-1α or nor H2A should be considered the ubiquitinated protein factor exclusively responsible for the temperature sensitive phenotype.
In summary, our study provided molecular and genetic evidence that E1 mutation is the determining factor responsible for the associated temperature sensitive phenotype of TS20 cells. The temperature sensitive proteasome-dependent disposal of mutant E1 at restrictive temperature suggests an active quality control system for E1 protein. The TS20 cell line and stable cell lines expressing E1 mutants will become valuable tools in exploration of mechanisms underlying both ubiqutination-dependent and ubiquitination-independent protein degradation pathways. These cell lines will facilitate the study of other cellular processes regulated by protein ubiqitination.
Acknowledgments
We thank Ms. X Sun for her technical help cloning E1 cDNA from TS20 cells. We thank Dr. H. Ozer for providing the TS20 cells. We thank Ms. Anusha Rajan for proofreading this manuscript before submission. This work is supported in part by grant R01-CA129494 (to N.S.) from NCI, National Institutes of Health (NIH) and start-up fund from Drexel University.
Footnotes
The authors declare no conflict of interest.
References
- Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc Natl Acad Sci U S A. 2002;99(20):13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdary DR, Dermody JJ, Jha KK, Ozer HL. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol Cell Biol. 1994;14(3):1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture) Angew Chem Int Ed Engl. 2005;44(37):5944–5967. doi: 10.1002/anie.200501428. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7(8):742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18(5):499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16(19):5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35(Pt 1):12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem. 1982;257(17):10329–10337. [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258(13):8206–8214. [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106(5):527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2006;26(6):2019–2028. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414(6861):325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134(2):268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Liang D, Kong X, Sang N. Effects of histone deacetylase inhibitors on HIF-1. Cell Cycle. 2006;5(21):2430–2435. doi: 10.4161/cc.5.21.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24(3):439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27(10):1355–1365. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C. Degradation of ornithine decarboxylase in mammalian cells is ATP dependent but ubiquitin independent. Eur J Biochem. 1989;185(2):469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- Salceda S, Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272(36):22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- Salvat C, Acquaviva C, Scheffner M, Robbins I, Piechaczyk M, Jariel-Encontre I. Molecular characterization of the thermosensitive E1 ubiquitin-activating enzyme cell mutant A31N-ts20. Requirements upon different levels of E1 for the ubiquitination/degradation of the various protein substrates in vivo. Eur J Biochem. 2000;267(12):3712–3722. doi: 10.1046/j.1432-1327.2000.01404.x. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269(38):23757–23763. [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane M, Harumiya Y, Ishida N, Hirai A, Miyamoto C, Hatakeyama S, Nakayama K, Kitagawa M. Down-regulation of p27(Kip1) by two mechanisms, ubiquitin-mediated degradation and proteolytic processing. J Biol Chem. 1999;274(20):13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- Szczepanowski RH, Filipek R, Bochtler M. Crystal structure of a fragment of mouse ubiquitin-activating enzyme. J Biol Chem. 2005;280(23):22006–22011. doi: 10.1074/jbc.M502583200. [DOI] [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1(2):193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Tuttle RL, Bothos J, Summers MK, Luca FC, Halazonetis TD. Defective in mitotic arrest 1/ring finger 8 is a checkpoint protein that antagonizes the human mitotic exit network. Mol Cancer Res. 2007;5(12):1304–1311. doi: 10.1158/1541-7786.MCR-07-0388. [DOI] [PubMed] [Google Scholar]
- Vassilev AP, Rasmussen HH, Christensen EI, Nielsen S, Celis JE. The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J Cell Sci. 1995;108 (Pt 3):1205–1215. doi: 10.1242/jcs.108.3.1205. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422(6929):330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Zeng GC, Donegan J, Ozer HL, Hand R. Characterization of a ts mutant of BALB/3T3 cells and correction of the defect by in vitro addition of extracts from wild-type cells. Mol Cell Biol. 1984;4(9):1815–1822. doi: 10.1128/mcb.4.9.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng GC, Zannis-Hadjopoulos M, Ozer HL, Hand R. Defective DNA topoisomerase I activity in a DNAts mutant of Balb/3T3 cells. Somat Cell Mol Genet. 1985;11(6):557–569. doi: 10.1007/BF01534721. [DOI] [PubMed] [Google Scholar]