Abstract
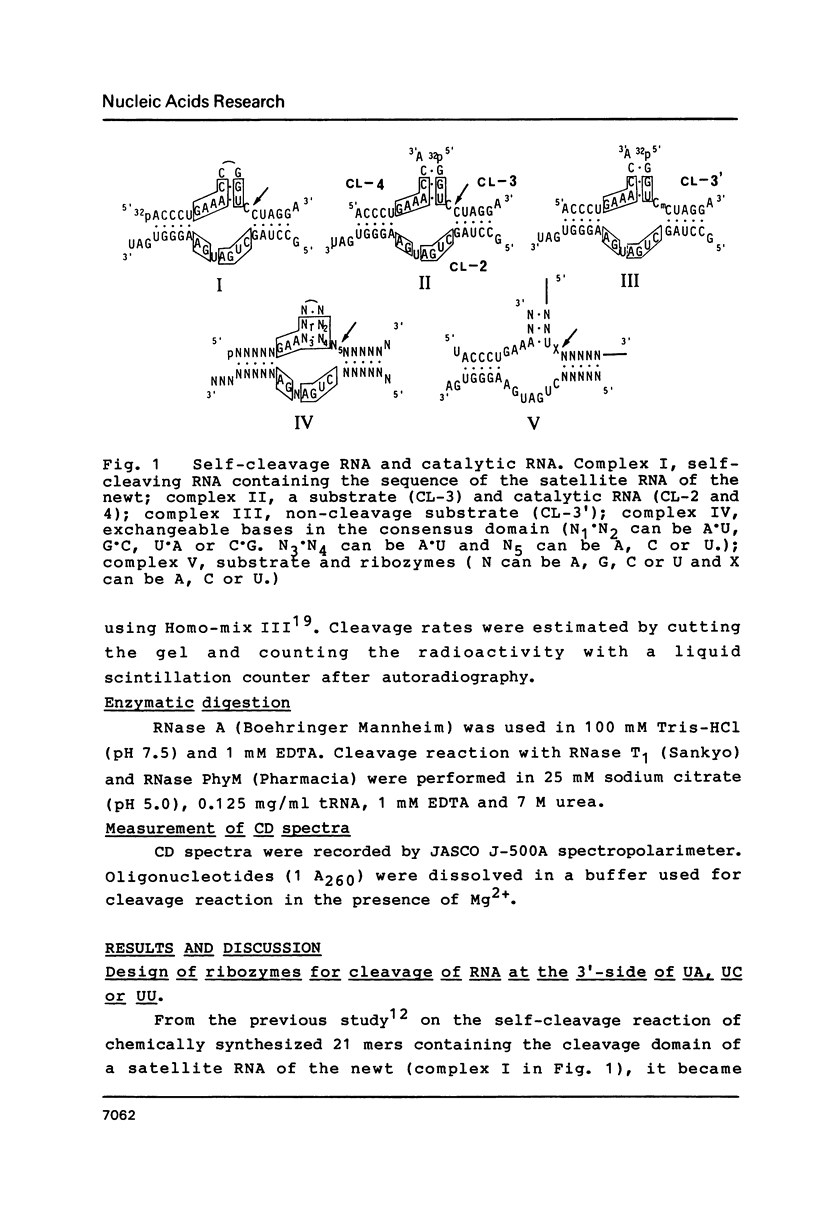
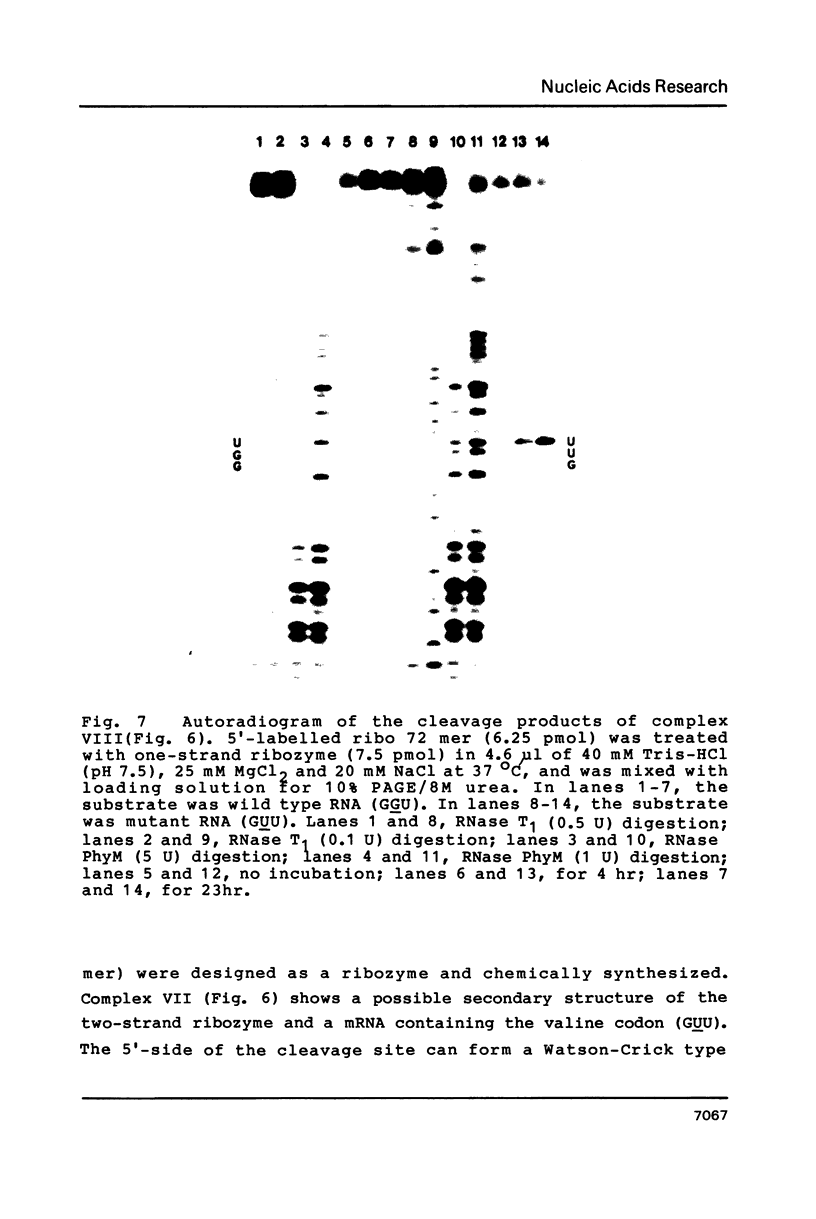
RNA enzymes (ribozymes) which can cleave RNA by recognizing sequences of 9-15 bases are described. Substrates must contain UX (X = U, C or A). A ribozyme consisting of two oligoribonucleotides (19 mer and 15 mer) was shown to cleave a ribo 11 mer catalytically with Km and kcat values of 0.53 microM and 0.03 min-1, respectively. A non-cleavable substrate-ribozyme complex containing 2'-O-methylnucleoside was prepared and CD spectra were compared at different temperature. In order to obtain an efficient ribozyme, a one-strand RNA with a chain length of 37 was prepared. The ribozyme was shown to distinguish a single base mutation in mRNA's which were prepared by transcription of two synthetic DNA duplexes coding for positions 7-26 of c-Ha-ras protein. The mutant (Val-12) mRNA which had GUU was cleaved but the wild type mRNA which contained GGU was not changed, when treated by the ribozymes in the presence of Mg2+.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown R. S., Dewan J. C., Klug A. Crystallographic and biochemical investigation of the lead(II)-catalyzed hydrolysis of yeast phenylalanine tRNA. Biochemistry. 1985 Aug 27;24(18):4785–4801. doi: 10.1021/bi00339a012. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Zaug A. J., Grabowski P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981 Dec;27(3 Pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Imura A., Iwai S., Miura K., Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2'-O-methyl)ribonucleotides. Nucleic Acids Res. 1987 Aug 11;15(15):6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Iwai S., Ohtsuka E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987 May 11;215(2):327–330. doi: 10.1016/0014-5793(87)80171-0. [DOI] [PubMed] [Google Scholar]
- Inouye M. Antisense RNA: its functions and applications in gene regulation--a review. Gene. 1988 Dec 10;72(1-2):25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries A. C., Symons R. H. A catalytic 13-mer ribozyme. Nucleic Acids Res. 1989 Feb 25;17(4):1371–1377. doi: 10.1093/nar/17.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Cleavage of specific sites of RNA by designed ribozymes. FEBS Lett. 1988 Nov 7;239(2):285–288. doi: 10.1016/0014-5793(88)80935-9. [DOI] [PubMed] [Google Scholar]
- Koizumi M., Iwai S., Ohtsuka E. Construction of a series of several self-cleaving RNA duplexes using synthetic 21-mers. FEBS Lett. 1988 Feb 15;228(2):228–230. doi: 10.1016/0014-5793(88)80004-8. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Inoue Y., Nakamori H., Iwai S., Ohtsuka E., Ikehara M., Noguchi S., Nishimura S. Synthesis and expression of a synthetic gene for the activated human c-Ha-ras protein. Jpn J Cancer Res. 1986 Jan;77(1):45–51. [PubMed] [Google Scholar]
- Miura K., Kamiya H., Tominaga M., Inoue Y., Ikehara M., Noguchi S., Nishimura S., Ohtsuka E. Overproduction of cellular and activated Ha-ras proteins by mutating a synthetic gene. Chem Pharm Bull (Tokyo) 1987 Dec;35(12):4878–4882. doi: 10.1248/cpb.35.4878. [DOI] [PubMed] [Google Scholar]
- Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986 Mar 28;231(4745):1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Shibahara S., Mukai S., Nishihara T., Inoue H., Ohtsuka E., Morisawa H. Site-directed cleavage of RNA. Nucleic Acids Res. 1987 Jun 11;15(11):4403–4415. doi: 10.1093/nar/15.11.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin C. J., Bradley S. M., Bargmann C. I., Weinberg R. A., Papageorge A. G., Scolnick E. M., Dhar R., Lowy D. R., Chang E. H. Mechanism of activation of a human oncogene. Nature. 1982 Nov 11;300(5888):143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- Toulmé J. J., Hélène C. Antimessenger oligodeoxyribonucleotides: an alternative to antisense RNA for artificial regulation of gene expression--a review. Gene. 1988 Dec 10;72(1-2):51–58. doi: 10.1016/0378-1119(88)90127-8. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986 Jan 31;44(2):225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]