Abstract
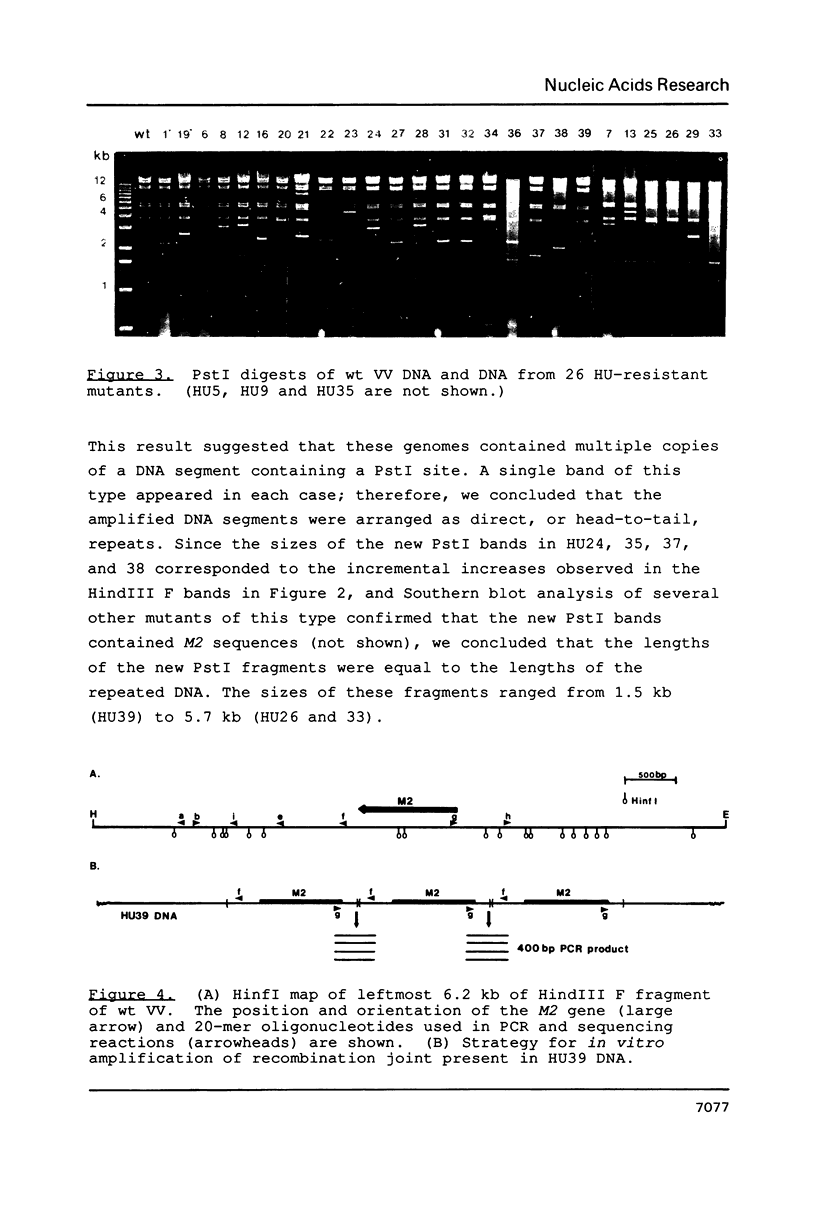
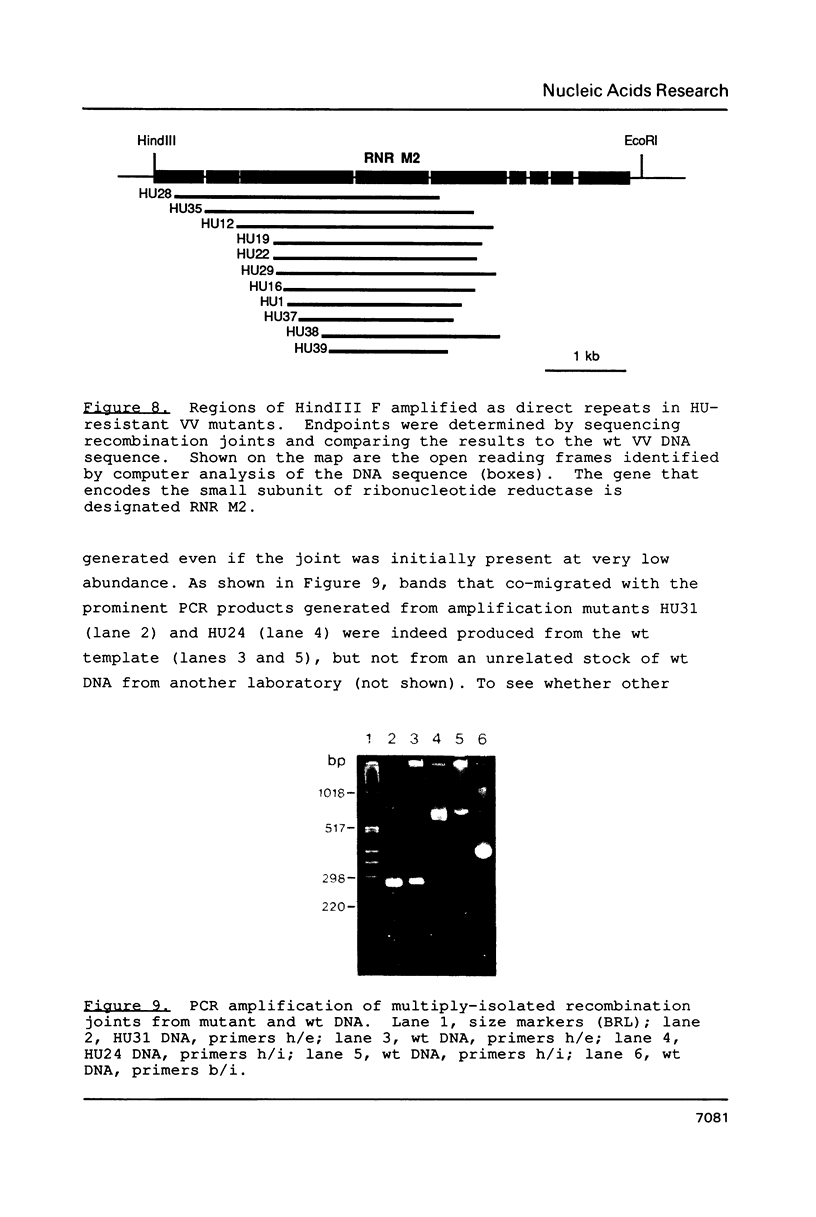
Amplification of the M2 gene encoding the small subunit of ribonucleotide reductase (EC 1.17.4.1) was analyzed in a collection of vaccinia virus (VV) isolates selected for resistance to 5 mM hydroxyurea (HU). Most of the mutants harbored tandem direct repeat arrays of the M2 gene, but several had duplicated M2 as an inverted repeat by genomic rearrangements involving the chromosomal termini. Novel joints formed by direct repeats were mapped, amplified in vitro, and sequenced. The junctions were simple fusions between DNA downstream and upstream of the M2 gene. Lack of sequence homology at the breakpoints indicated that the initial genomic rearrangements leading to gene amplification were due to nonhomologous recombination events.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Archard L. C., Mackett M. Restriction endonuclease analysis of red cowpox virus and its white pock variant. J Gen Virol. 1979 Oct;45(1):51–63. doi: 10.1099/0022-1317-45-1-51. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Esposito J. J., Cabradilla C. D., Nakano J. H., Obijeski J. F. Intragenomic sequence transposition in monkeypox virus. Virology. 1981 Mar;109(2):231–243. doi: 10.1016/0042-6822(81)90495-5. [DOI] [PubMed] [Google Scholar]
- Esposito J., Condit R., Obijeski J. The preparation of orthopoxvirus DNA. J Virol Methods. 1981 Feb;2(3):175–179. doi: 10.1016/0166-0934(81)90036-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986 May 9;45(3):425–430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- Giulotto E., Saito I., Stark G. R. Structure of DNA formed in the first step of CAD gene amplification. EMBO J. 1986 Sep;5(9):2115–2121. doi: 10.1002/j.1460-2075.1986.tb04474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., von Beroldingen C. H., Sensabaugh G. F., Erlich H. A. DNA typing from single hairs. Nature. 1988 Apr 7;332(6164):543–546. doi: 10.1038/332543a0. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. A hotspot for novel amplification joints in a mosaic of Alu-like repeats and palindromic A + T-rich DNA. EMBO J. 1987 Aug;6(8):2401–2408. doi: 10.1002/j.1460-2075.1987.tb02518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Debatisse M., Buttin G., de Saint Vincent B. R. The multicopy appearance of a large inverted duplication and the sequence at the inversion joint suggest a new model for gene amplification. EMBO J. 1988 Feb;7(2):407–417. doi: 10.1002/j.1460-2075.1988.tb02828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. N., Feder J., Hill A. B., Sherwood S. W., Schimke R. T. Transient inhibition of DNA synthesis results in increased dihydrofolate reductase synthesis and subsequent increased DNA content per cell. Mol Cell Biol. 1986 Oct;6(10):3373–3381. doi: 10.1128/mcb.6.10.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal G. J., Moss B. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology. 1988 Dec;167(2):524–537. [PubMed] [Google Scholar]
- Latt S. A., Lalande M., Donlon T., Wyman A., Rose E., Shiloh Y., Korf B., Müller U., Sakai K., Kanda N. DNA-based detection of chromosome deletion and amplification: diagnostic and mechanistic significance. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):299–307. doi: 10.1101/sqb.1986.051.01.035. [DOI] [PubMed] [Google Scholar]
- Looney J. E., Ma C., Leu T. H., Flintoff W. F., Troutman W. B., Hamlin J. L. The dihydrofolate reductase amplicons in different methotrexate-resistant Chinese hamster cell lines share at least a 273-kilobase core sequence, but the amplicons in some cell lines are much larger and are remarkably uniform in structure. Mol Cell Biol. 1988 Dec;8(12):5268–5279. doi: 10.1128/mcb.8.12.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Looney J. E., Leu T. H., Hamlin J. L. Organization and genesis of dihydrofolate reductase amplicons in the genome of a methotrexate-resistant Chinese hamster ovary cell line. Mol Cell Biol. 1988 Jun;8(6):2316–2327. doi: 10.1128/mcb.8.6.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer R. W., Brown G. D., Graves R. L. The white pock mutants of rabbit poxvirus. II. The early white pock (mu) host range (hr) mutants of rabbit poxvirus uncouple transcription and translation in nonpermissive cells. Virology. 1980 Oct 30;106(2):234–249. doi: 10.1016/0042-6822(80)90247-0. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., Yezzi M. J. Association of high rate of recombination with amplification of dominant selectable gene in human cells. Somat Cell Mol Genet. 1988 May;14(3):273–286. doi: 10.1007/BF01534588. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Meuth M. DNA amplification--deletion in a spontaneous mutation of the hamster aprt locus: structure and sequence of the novel joint. Nucleic Acids Res. 1986 Nov 11;14(21):8361–8371. doi: 10.1093/nar/14.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Meuth M. DNA amplification--deletion in a spontaneous mutation of the hamster aprt locus: structure and sequence of the novel joint. Nucleic Acids Res. 1986 Nov 11;14(21):8361–8371. doi: 10.1093/nar/14.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Phear G., Meuth M. DNA sequence analysis of spontaneous mutations at the aprt locus of hamster cells. Mol Cell Biol. 1987 Apr;7(4):1445–1449. doi: 10.1128/mcb.7.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passananti C., Davies B., Ford M., Fried M. Structure of an inverted duplication formed as a first step in a gene amplification event: implications for a model of gene amplification. EMBO J. 1987 Jun;6(6):1697–1703. doi: 10.1002/j.1460-2075.1987.tb02420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup D. J., Ink B. S., Parsons B. L., Hu W., Joklik W. K. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6817–6821. doi: 10.1073/pnas.81.21.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R. A., Anderson P. Structures of spontaneous deletions in Caenorhabditis elegans. Mol Cell Biol. 1988 Sep;8(9):3748–3754. doi: 10.1128/mcb.8.9.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Groves R., Giulotto E., Rolfe M., Stark G. R. Evolution and stability of chromosomal DNA coamplified with the CAD gene. Mol Cell Biol. 1989 Jun;9(6):2445–2452. doi: 10.1128/mcb.9.6.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood S. W., Schumacher R. I., Schimke R. T. Effect of cycloheximide on development of methotrexate resistance of Chinese hamster ovary cells treated with inhibitors of DNA synthesis. Mol Cell Biol. 1988 Jul;8(7):2822–2827. doi: 10.1128/mcb.8.7.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1989 May;86(10):3489–3493. doi: 10.1073/pnas.86.10.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh M. B., Johnson T. L., Mathews C. K. Vaccinia virus induces ribonucleotide reductase in primate cells. J Virol. 1984 Nov;52(2):507–514. doi: 10.1128/jvi.52.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh M. B., Mathews C. K. Hydroxyurea-resistant vaccinia virus: overproduction of ribonucleotide reductase. J Virol. 1986 Nov;60(2):506–514. doi: 10.1128/jvi.60.2.506-514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabaugh M., Roseman N., Davis R., Mathews C. Vaccinia virus-encoded ribonucleotide reductase: sequence conservation of the gene for the small subunit and its amplification in hydroxyurea-resistant mutants. J Virol. 1988 Feb;62(2):519–527. doi: 10.1128/jvi.62.2.519-527.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G. R., Debatisse M., Giulotto E., Wahl G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989 Jun 16;57(6):901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- Whoriskey S. K., Nghiem V. H., Leong P. M., Masson J. M., Miller J. H. Genetic rearrangements and gene amplification in Escherichia coli: DNA sequences at the junctures of amplified gene fusions. Genes Dev. 1987 May;1(3):227–237. doi: 10.1101/gad.1.3.227. [DOI] [PubMed] [Google Scholar]
- Wright J. A., Alam T. G., McClarty G. A., Tagger A. Y., Thelander L. Altered expression of ribonucleotide reductase and role of M2 gene amplification in hydroxyurea-resistant hamster, mouse, rat, and human cell lines. Somat Cell Mol Genet. 1987 Mar;13(2):155–165. doi: 10.1007/BF01534695. [DOI] [PubMed] [Google Scholar]